Abstract
In most organisms, the segregation of chromosomes during the first meiotic division is dependent upon at least one crossover (CO) between each pair of homologous chromosomes. COs can result from chromosome double-strand breaks (DSBs) that are induced and preferentially repaired using the homologous chromosome as a template. The PCH2 gene of budding yeast is required to establish proper meiotic chromosome axis structure and to regulate meiotic interhomolog DSB repair outcomes. These roles appear conserved in the mouse ortholog of PCH2, Trip13, which is also involved in meiotic chromosome axis organization and the regulation of DSB repair. Using a combination of genetic and physical assays to monitor meiotic DSB repair, we present data consistent with pch2Δ mutants showing defects in suppressing intersister DSB repair. These defects appear most pronounced in dmc1Δ mutants, which are defective for interhomolog repair, and explain the previously reported observation that pch2Δdmc1Δ cells can complete meiosis. Results from genetic epistasis analyses involving spo13Δ, rad54Δ, and mek1/MEK1 alleles and an intersister recombination reporter assay are also consistent with Pch2 acting to limit intersister repair. We propose a model in which Pch2 is required to promote full Mek1 activity and thereby promotes interhomolog repair.
DNA double-strand breaks (DSBs) that occur during vegetative growth are preferentially repaired via homologous recombination in which the Rad51 recombinase and its partner Rad54 mediate strand exchange with the sister chromatid. This intersister repair occurs even in diploid cells where a homologous chromosome template is available and is thought to help prevent chromosome rearrangements (Kadyk and Hartwell 1992; M. Shinohara et al. 1997; Arbel et al. 1999; Krogh and Symington 2004). In meiosis, formation of programmed DSBs and their repair using the homologous chromosome as a template is essential for the production of viable gametes (Roeder 1997). Although Rad51 and Rad54 are still present, meiotic interhomolog strand exchange is accomplished by their respective orthologs, Dmc1 and Rdh54 (Dresser et al. 1997; Klein 1997; A. Shinohara et al. 1997; M. Shinohara 1997; Arbel et al. 1999; Hollingsworth 2010). Interhomolog DSB repair creates linkages provided by genetic exchanges, or crossovers (COs), between homologous chromosomes. In many organisms, these COs are required for reductional chromosome segregation at the meiosis I (MI) division, which lowers cell ploidy by one-half, allowing for the generation of haploid gametes (Roeder 1997). If any pair of homologous chromosomes fails to receive a CO, MI nondisjunction can occur and produce aneuploid gametes, which cause conditions such as Down syndrome or infertility in humans (Hassold et al. 2007).
During meiotic prophase I in budding yeast, ∼140–170 DNA DSBs are introduced into the genome by a group of 10 proteins, of which Spo11 is the catalytic component (Blitzblau et al. 2007; Buhler et al. 2007; Mancera et al. 2008). Although COs are the only repair products known to promote MI disjunction, only ∼50% of DSBs in yeast meiosis are repaired as interhomolog COs. Some DSBs are repaired using the homologous chromosome without producing a CO; this is known as a noncrossover (NCO). Obligate CO formation, CO interference, and CO homeostasis are manifestations of interhomolog DSB repair regulation acting to ensure each pair of homologous chromosomes disjoins at MI (Bishop and Zickler 2004; Borner et al. 2004; Martini et al. 2006; Blitzblau et al. 2007; Buhler et al. 2007; Chen et al. 2008; Mancera et al. 2008; Berchowitz and Copenhaver 2010). The obligate CO refers to the observation that all homologous chromosome pairs receive at least one CO. CO interference describes the nonrandom, evenly spaced distribution of CO events, and CO homeostasis describes the finding that CO levels are maintained as DSB frequencies decrease (reviewed in Jones and Franklin 2006; Berchowitz and Copenhaver 2010). Little is known about the mechanisms or relatedness of the different aspects of CO control, although one mutant, pch2Δ, has decreased CO interference and may also be defective for CO homeostasis (Joshi et al. 2009; Zanders and Alani 2009).
The ∼10–33% of meiotic DSBs estimated to not be repaired using a homologous chromosome are repaired by homologous recombination using the sister chromatid as a template (e.g., Goldfarb and Lichten 2010). The shift in DSB repair template preference from the sister chromatid in the mitotic cell cycle to the homologous chromosome in meiosis is referred to as “interhomolog bias” (Jackson and Fink 1985; Schwacha and Kleckner 1994, 1997; Wan et al. 2004; Webber et al. 2004; Niu et al. 2005, 2007, 2009; Goldfarb and Lichten 2010). Interhomolog bias is established shortly after DSB formation and requires components of the axial elements, which are linear structures that form along each pair of sister chromatids early in meiotic prophase (Hollingsworth 2010). An early step in instituting interhomolog bias is phosphorylation of Hop1 of the Hop1/Red1 axial element duo by the Mec1 and Tel1 kinases (Hollingsworth and Byers 1989; Hollingsworth et al. 1990; Rockmill and Roeder 1990; Carballo et al. 2008). Red1 and phosphorylated Hop1 are required for the activation of the effector kinase Mek1 (Niu et al. 2005, 2007; Carballo et al. 2008). Mek1 appears to directly promote interhomolog repair (Terentyev et al. 2010). In addition, Mek1 phosphorylates Rad54, which inhibits the interaction between Rad51 and Rad54 (Niu et al. 2009). Phosphorylation of Rad54 contributes to, but is not sufficient for, complete interhomolog bias (Niu et al. 2009). The meiosis-specific protein Hed1 also acts to prevent Rad51–Rad54 complex formation by competing with Rad54 for Rad51 binding, although the role of Hed1 in interhomolog bias is yet to be determined (Tsubouchi and Roeder 2006; Busygina et al. 2008). Interhomolog bias is maintained in haploid meiosis and inhibits DSB repair, suggesting that interhomolog interactions are not required (Demassy et al. 1994; Callender and Hollingsworth 2010). At hemizygous DSB sites in diploid meiosis, intersister repair is constrained by a Mek1-dependent delay, although efficient intersister DSB repair does occur (Goldfarb and Lichten 2010).
The mechanisms promoting interhomolog bias are often studied in dmc1 null mutant backgrounds in which unrepaired DSBs trigger the meiotic recombination checkpoint to arrest cells at pachytene, the last stage of meiotic prophase before cells are committed to undergo the MI division (Hollingsworth 2010). There are two ways in which recombination checkpoint arrest can be overcome in a dmc1 mutant background. The first is to eliminate any of the essential recombination checkpoint genes such as MEC1, RAD17, or RAD24. In such cases, meiosis proceeds with unrepaired breaks to form inviable gametes (Lydall et al. 1996). The second is to eliminate (or reduce; see below) the checkpoint-eliciting DNA lesions either by preventing DSB formation or by allowing inappropriate Dmc1-independent DSB repair (Bishop et al. 1992, 1999; Schwacha and Kleckner 1994, 1997; Xu et al. 1997; Thompson and Stahl 1999). The latter can be accomplished by several mechanisms. Overexpressing RAD51 or RAD54 and/or mutating HED1 in a dmc1 background allows for meiotic progression and the production of moderate to wild-type levels of interhomolog COs and thus of viable spores (Bishop et al. 1999; Tsubouchi and Roeder 2003, 2006; Busygina et al. 2008). Alternatively, when RED1, HOP1, or MEK1 are mutated, interhomolog bias is lost and DSBs are rapidly repaired via Rad51–Rad54-dependent strand exchange using the sister chromatid as a template, and meiosis progresses to produce inviable spores (Bishop et al. 1999; Wan et al. 2004; Niu et al. 2005, 2007).
Pch2 (pachytene checkpoint) is a putative AAA ATPase that promotes the checkpoint arrest/delays observed in zip1, rad17, mms4, and sae2 recombination mutants. The pch2Δ mutation also suppressed the dmc1Δ checkpoint arrest in some but not all studies (San-Segundo and Roeder 1999; Zierhut et al. 2004; Hochwagen et al. 2005; Wu and Burgess 2006; Mitra and Roeder 2007; Zanders and Alani 2009). In budding yeast, Pch2 is also required for wild-type kinetics of meiotic progression, CO interference, and establishing proper organization of Hop1 and Zip1 on meiotic chromosomes (Sym and Roeder 1995; Borner et al. 2008; Joshi et al. 2009; Zanders and Alani 2009). Several of these roles appear conserved in the mouse PCH2 ortholog Trip13, which is required for wild-type levels of DSB repair, wild-type CO distribution, and proper organization of HORMADs (which share homology with Hop1) and the synaptonemal complex central element protein SYCP1 on meiotic axes (Li and Schimenti 2007; Wojtasz et al. 2009; Roig et al. 2010).
Here we investigated the mechanisms by which the pch2Δ mutation suppresses the meiotic arrest/delay phenotypes of dmc1Δ mutations. First, we found that lowering DSB levels in dmc1Δ mutants reduced the fraction of cells that arrest, indicating that the recombination checkpoint is sensitive to DSB levels. Second, we found that Pch2 inhibited some DSB repair in dmc1Δ cells that likely includes, but may not be limited to, intersister recombination. Third, we identified genetic interactions between PCH2 and RAD54 and PCH2 and MEK1 that support a role for Pch2 in limiting intersister repair. Finally, we present a genetic assay that demonstrates an increase in intersister repair at one locus in pch2Δ mutants. We synthesize our data with published results to propose a model in which Pch2 is required for full Mek1 activity.
Materials and Methods
Media and yeast strains
All yeast strains (Table 1) were grown at 30° on yeast peptone dextrose (YPD) supplemented with complete amino acid mix, synthetic complete, or synthetic complete −histidine (Argueso et al. 2004). All strains were sporulated at 30°. The sporulation media and sporulation conditions used to generate the data in Tables 2–4 were described previously (Zanders and Alani 2009). Differences in spore formation and viability were analyzed by a χ2 test in which P-values <0.05 were considered statistically significant. Geneticin (Invitrogen), nourseothricin (Hans-Knoll Institute fur Naturstoff-Forschung), and hygromycin B (Calbiochem) were added in standard concentrations to YPD media when required (Wach et al. 1994; Goldstein and McCusker 1999).
Table 1.
Yeast strains used in this study
| Strain | Genotype |
| NH943/EAY2580 | MATa/α, homozygous for: ho::hisG ade2Δ, ura3(ΔSma-Pst), leu2::hisG, CEN3::ADE2, lys5-P, cyh2r, his4-B |
| EAY2581/EAY2210 | as NH943/EAY2580 except pch2Δ::NATMX4 |
| EAY2582/SKY635 | as NH943/EAY2580 except spo11-HA3His6::KANMX4 |
| EAY2787/EAY2263 | as NH943/EAY2580 except pch2Δ::NATMX4, spo11-HA3His6::KANMX4 |
| EAY2637/EAY2638 | as NH943/EAY2580 except dmc1Δ::KANMX4 |
| EAY2639/EAY2640 | as NH943/EAY2580 except pch2Δ::NATMX4, dmc1Δ::KANMX4 |
| EAY2619/EAY2630 | as NH943/EAY2580 except spo11-HA3His6::KANMX4, dmc1Δ::KANMX4 |
| EAY2631/EAY2632 | as NH943/EAY2580 except pch2Δ::NATMX4, spo11-HA3His6::KANMX4, dmc1Δ::KANMX4 |
| EAY2582/SKY665 | as NH943/EAY2580 except spo11-HA3His6::KANMX4/spo11(Y135F)-HA3His6::KANMX4 |
| EAY2787/EAY2265 | as NH943/EAY2580 except pch2Δ::NATMX4, spo11-HA3His6::KANMX4/spo11(Y135F)-HA3His6::KANMX4 |
| EAY2620/EAY2800 | as NH943/EAY2580 except spo11-HA3His6::KANMX4/spo11(Y135F)-HA3His6::KANMX4, dmc1Δ::KANMX4 |
| EAY2622/EAY2802 | as NH943/EAY2580 except pch2Δ::NATMX4, spo11-HA3His6::KANMX4/spo11(Y135F)-HA3His6::KANMX5, dmc1Δ::KANMX4 |
| EAY2589/EAY2590 | as NH943/EAY2580 except spo13::URA3 |
| EAY2591/EAY2592 | as NH943/EAY2580 except pch2Δ::NATMX4, spo13::URA3 |
| EAY2595/EAY2596 | as NH943/EAY2580 except spo11-HA3His6::KANMX4, spo13::URA3 |
| EAY2593/EAY2594 | as NH943/EAY2580 except pch2Δ::NATMX4, spo11-HA3His6::KANMX4, spo13::URA3 |
| EAY2643/EAY2644 | as NH943/EAY2580 except dmc1Δ::KANMX4, spo13::URA3 |
| EAY2641/EAY2642 | as NH943/EAY2580 except pch2Δ::NATMX4, dmc1Δ::KANMX4, spo13::URA3 |
| EAY2633/EAY2634 | as NH943/EAY2580 except spo11-HA3His6::KANMX4, dmc1Δ::KANMX4, spo13::URA3 |
| EAY2635/EAY2636 | as NH943/EAY2580 except pch2Δ::NATMX4, spo11-HA3His6::KANMX4, dmc1Δ::KANMX4, spo13::URA3 |
| EAY2578/EAY2579 | as NH943/EAY2580 except rad50S::URA3 |
| EAY2585/EAY2586 | as NH943/EAY2580 except pch2Δ::NATMX4, rad50S::URA3 |
| EAY2587/EAY2588 | as NH943/EAY2580 except spo11-HA3His6::KANMX4, rad50S::URA3 |
| EAY2583/EAY2584 | as NH943/EAY2580 except pch2Δ::NATMX4, spo11-HA3His6::KANMX4, rad50S::URA3 |
| EAY2722/EAY2723 | as NH943/EAY2580 except rad54Δ::HPHMX4 |
| EAY2681/EAY2746 | as NH943/EAY2580 except pch2Δ::NATMX4, rad54Δ::HPHMX4 |
| EAY2740/EAY2741 | as NH943/EAY2580 except spo11-HA3His6::KANMX4, rad54Δ::HPHMX4 |
| EAY2726/EAY2727 | as NH943/EAY2580 except pch2Δ::NATMX4, spo11-HA3His6::KANMX4, rad54Δ::HPHMX4 |
| EAY2742/EAY2743 | as NH943/EAY2580 except rad54Δ::HPHMX4, dmc1Δ::KANMX4 |
| EAY2728/EAY2729 | as NH943/EAY2580 except pch2Δ::NATMX4, rad54Δ::HPHMX4, dmc1Δ::KANMX4 |
| EAY2738/EAY2739 | as NH943/EAY2580 except spo11-HA3His6::KANMX4, rad54Δ::HPHMX4, dmc1Δ::KANMX4 |
| EAY2724/EAY2725 | as NH943/EAY2580 except pch2Δ::NATMX4, spo11-HA3His6::KANMX4, rad54Δ::HPHMX4, dmc1Δ::KANMX4 |
| NH942 | MATα, ho::hisG, ade2Δ, can1, ura3(ΔSma-Pst), met13-B, trp5-S, CEN8::URA3, thr1-A, cup1S |
| EAY2209/EAY2210 | as NH942/NH943 except pch2Δ::NATMX4 |
| EAY2681/EAY2685 | as NH942/NH943 except pch2Δ::NATMX4, rad54Δ::HPHMX4 |
| EAY2951/EAY2952 | MATa/α homozygous for ho::hisG, ura3, and his3Δ::KANMX4 hemizygous for his3-Δ5′ his3-Δ3′::URA3 |
| EAY2955/EAY2956 | as EAY2951/2952 except pch2Δ::NATMX4 |
| EAY3077 | MATa, ho::hisG, leu2 |
| EAY3078 | MATα, ho::hisG, leu2 |
| EAY3079 | MATa, ho::hisG, HIS4, lys5-P, mek1Δ::LEU2::mek1Q241G::URA3 |
| EAY3080 | MATα, ho::hisG, HIS4, mek1Δ::LEU2::mek1Q241G::URA3 |
| EAY3081 | MATa, ho::hisG, HIS4, lys5-P, mek1Δ::LEU2::mek1Q241G::URA3, pch2Δ::NATMX4 |
| EAY3082 | MATα, ho::hisG, HIS4, mek1Δ::LEU2::mek1Q241G::URA3, pch2Δ::NATMX4 |
| EAY3083 | MATa, ho::hisG, HIS4, mek1Δ::LEU2::mek1Q241G::URA3, spo11-HA::KANMX4 |
| EAY3084 | MATα, ho::hisG, HIS4, mek1Δ::LEU2::mek1Q241G::URA3, spo11-HA::KANMX4 |
| EAY3085 | MATa, ho::hisG, HIS4, mek1Δ::LEU2::mek1Q241G::URA3, pch2Δ::NATMX4, spo11-HA::KANMX4 |
| EAY3086 | MATα, ho::hisG, his4, mek1Δ::LEU2::mek1Q241G::URA3, pch2Δ::NATMX4, spo11-HA::KANMX4 |
| EAY3087 | MATa, ho::hisG, his4 |
| EAY3088 | MATα, ho::hisG, his4, ade2Δ |
| EAY3089 | MATa, ho::hisG, HIS4, ura3, MEK1-GST-KANMX4 |
| EAY3090 | MATα, ho::hisG, his4, MEK1-GST-KANMX4 |
| EAY3091 | MATa, ho::hisG, his4, MEK1-GST-KANMX4, pch2Δ::NATMX4 |
| EAY3092 | MATα, ho::hisG, his4, MEK1-GST-KANMX4, pch2Δ::NATMX4 |
| EAY3093 | MATa, ho::hisG, his4, ura3, MEK1-GST-KANMX4, spo11-HA::KANMX4 |
| EAY3094 | MATα, ho::hisG, his4, ura3, MEK1-GST-KANMX4, spo11-HA::KANMX4 |
| EAY3095 | MATa, ho::hisG, his4, ura3, ade2Δ, leu2::hisG, MEK1-GST-KANMX4, pch2Δ::NATMX4, spo11-HA::KANMX4 |
| EAY3096 | MATα, ho::hisG, HIS4, ura3, MEK1-GST-KANMX4, pch2Δ::NATMX4, spo11-HA::KANMX4 |
The diploid strain names are composites of the haploid strains used to create them.
Table 2.
Spore formation efficiency and viability in pch2Δ mutants
| Genotype | % sporulation | No. analyzed | % spore viability | Spores analyzed |
| Wild type | 79.1 | 436 | 93.5 | 400 |
| pch2∆ | 80.9 | 429 | 95.3 | 400 |
| spo11-HA | 81.1 | 434 | 92.5 | 400 |
| pch2∆ spo11-HA | 74.9 | 453 | 56.8 | 400 |
| dmc1∆ | 0.0 | 406 | NA | NA |
| pch2∆ dmc1∆ | 4.6 | 431 | 2.9 | 148 |
| spo11-HA dmc1∆ | 0.4 | 239 | NA | NA |
| pch2∆ spo11-HA dmc1∆ | 39.0 | 439 | 1.3 | 160 |
| spo11-HA/spo11yf-HA | 79.6 | 421 | ND | ND |
| pch2∆/pch2∆; spo11-HA/spo11yf-HA | 67.7 | 440 | ND | ND |
| spo11-HA/spo11yf-HA; dmc1∆/dmc1∆ | 4.6 | 415 | ND | ND |
| pch2∆/pch2∆; spo11-HA/spo11yf-HA; dmc1∆/dmc1∆ | 42.8 | 523 | ND | ND |
| rad54∆ | 58.3 | 439 | 59.5 | 400 |
| pch2∆ rad54∆ | 46.0 | 443 | 47.0 | 400 |
| spo11-HA rad54∆ | 63.5 | 425 | 62.8 | 400 |
| pch2∆ spo11-HA rad54∆ | 30.7 | 440 | 38.0 | 400 |
| dmc1∆ rad54∆ | 0.2 | 422 | NA | NA |
| pch2∆ dmc1∆ rad54∆ | 0.0 | 444 | NA | NA |
| spo11-HA dmc1∆ rad54∆ | 0.0 | 409 | NA | NA |
| pch2∆ spo11-HA dmc1∆ rad54∆ | 0.0 | 403 | NA | NA |
| spo13 | 63.3 | 441 | 47.3 | 400 |
| pch2∆ spo13 | 49.2 | 417 | 44.8 | 400 |
| spo11-HA spo13 | 51.8 | 454 | 44.8 | 400 |
| pch2∆ spo11-HA spo13 | 56.9 | 457 | 44.3 | 400 |
| dmc1∆ spo13 | 9.6 | 428 | 7.3 | 400 |
| pch2∆ dmc1∆ spo13 | 43.4 | 422 | 15.5 | 400 |
| spo11-HA dmc1∆ spo13 | 16.6 | 441 | 16.0 | 400 |
| pch2∆ spo11-HA dmc1∆ spo13 | 57.9 | 480 | 25.5 | 396 |
| pch2∆ spo11-HA dmc1∆ rad54∆ spo13 | 14.0 | 222 | 0.0 | 120 |
Sporulation efficiencies for the above strains were counted after 5 days on sporulation media at 30°. Tetrads (for SPO13 strains) or dyads (from spo13 strains) were dissected on YPD and scored for spore viability after 3 days. NA indicates that the percentage of spore viability is not applicable for strains that do not sporulate. ND indicates that spore viability was not assayed.
Table 4.
Spore formation efficiency and viability in mek1-as and MEK1-GST mutants
| Genotype | % sporulation | No. analyzed | % spore viability | Spores analyzed |
| Wild type | 79.1 | 436 | 93.5 | 400 |
| pch2∆ | 80.9 | 429 | 95.3 | 400 |
| spo11-HA | 81.1 | 434 | 92.5 | 400 |
| mek1-as | 84.7 | 163 | 98.8 | 80 |
| MEK1-GST | 66.9 | 178 | 87.5 | 160 |
| pch2∆ spo11-HA | 74.9 | 453 | 56.8 | 400 |
| mek1-as spo11-HA | 87.8 | 164 | 92.3 | 400 |
| mek1-as pch2Δ | 88.5 | 191 | 92.0 | 400 |
| MEK1-GST spo11-HA | 69.8 | 162 | 85.4 | 546 |
| MEK1-GST pch2Δ | 67.5 | 155 | 86.5 | 408 |
| pch2Δ spo11-HA mek1-as | 85.8 | 183 | 40.8 | 400 |
| pch2Δ spo11-HA MEK1-GST | 63.5 | 143 | 89.0 | 552 |
Sporulation efficiencies for the above strains were counted after 5 days on sporulation media at 30°. Tetrads were dissected on YPD and scored for spore viability after 3 days.
Strains described in Tables 2 and 3 are isogenic to the NHY943 or NHY942 SK1 strains described in de los Santos et al. (2003). The spo11 hypomorphic mutants and the NHY943 strains containing these alleles are described in Martini et al. (2006). As in Martini et al. (2006), we refer to spo11-HA3His6 as spo11-HA. The dmc1Δ and rad54Δ alleles used in this work were all complete open reading frame deletions. The pch2Δ allele contains a deletion of amino acids 17–587 (Zanders and Alani 2009). All deletion cassettes were made via PCR, and the deleted regions were replaced with HPHMX4, KANMX4, or NATMX4 as shown in Table 1. A BamHI fragment of pNKY58 was integrated into the genome to create the spo13::hisG-URA3-hisG mutation, and a BglII to EcoRI fragment of pNKY349 was used to replace RAD50 with rad50S::URA3 (Alani et al. 1990). All mutations were initially integrated into the genome using standard transformation techniques (Gietz et al. 1995). Standard genetic crosses were used to generate the various mutant combinations. Details on strain construction and primer sequences are available upon request.
Table 3.
pch2Δ and pch2Δ rad54Δ display similar meiotic crossover levels
| Total spores | Recombinant | Parental | % recombinant | |
| Chromosome III | ||||
| HIS3-LEU2 | ||||
| Wild type | 2711 | 351 | 2360 | 12.9 |
| pch2∆ | 2691 | 389 | 2302 | 14.5 |
| pch2∆ rad54∆ | 458 | 62 | 396 | 13.5 |
| LEU2-CEN3 | ||||
| Wild type | 2711 | 184 | 2527 | 6.8 |
| pch2∆ | 2691 | 241 | 2450 | 9.0 |
| pch2∆ rad54∆ | 458 | 41 | 417 | 9.0 |
| CEN3-MAT | ||||
| Wild type | 2711 | 402 | 2309 | 14.8 |
| pch2∆ | 2691 | 374 | 2317 | 13.9 |
| pch2∆ rad54∆ | 458 | 65 | 393 | 14.2 |
| Chromosome VII | ||||
| TRP5-CYH2 | ||||
| Wild type | 2711 | 908 | 1803 | 33.5 |
| pch2∆ | 2691 | 1149 | 1542 | 42.7 |
| pch2∆ rad54∆ | 458 | 199 | 259 | 43.4 |
| CYH2-MET13 | ||||
| Wild type | 2711 | 260 | 2451 | 9.6 |
| pch2∆ | 2691 | 469 | 2222 | 17.4 |
| pch2∆ rad54∆ | 458 | 81 | 377 | 17.7 |
| MET13-LYS5 | ||||
| Wild type | 2711 | 559 | 2152 | 20.6 |
| pch2∆ | 2691 | 747 | 1944 | 27.8 |
| pch2∆ rad54∆ | 458 | 139 | 319 | 30.3 |
| Chromosome VIII | ||||
| CEN8-THR1 | ||||
| Wild type | 2711 | 606 | 2105 | 22.4 |
| pch2∆ | 2691 | 649 | 2042 | 24.1 |
| pch2∆ rad54∆ | 458 | 116 | 342 | 25.3 |
| THR1-CUP1 | ||||
| Wild type | 2711 | 668 | 2043 | 24.6 |
| pch2∆ | 2691 | 948 | 1743 | 35.2 |
| pch2∆ rad54∆ | 458 | 123 | 335 | 26.9 |
Wild-type (NH942/NH943), pch2Δ (EAY2209/EAY2210), and pch2Δ rad54Δ (EAY2681/EAY2685) strains were sporulated and analyzed for segregation of genetic markers in the NH942/NH943 strain background. Crossover frequencies in this strain were calculated from recombination frequencies in spores as described previously (Zanders and Alani 2009). Data for wild type and pch2Δ are from Zanders and Alani (2009). Spore viability was 91% for wild type (n =743 tetrads dissected), 97% for pch2Δ (n = 707 tetrads), and 45% for pch2Δ rad54Δ (n = 256 tetrads).
The pch2Δspo11-HA mek1-as and pch2Δspo11-HA MEK1-GST strains presented in Table 4 were constructed as follows. The mek1-as strain was constructed by digesting the plasmid pJR2 with RsrII and then by transforming the mek1-Q241G::URA3 segment into the ura3mek1Δ SK1 diploid YTS1 (plasmid and strain provided by Nancy Hollingworth). The diploid was then tetrad-dissected, selecting for Ura+ haploid segregants. The homozygous MEK1-GST diploid SK1 strain SBY2901 (provided by Sean Burgess) was also tetrad-dissected to obtain haploid segregants. The mek1-as and MEK1-GST segregants described above were mated to EAY2581 (pch2Δ), EAY2263 (pch2Δspo11-HA), and SKY635 (spo11-HA). The resulting diploids were sporulated and tetrad-dissected to obtain the haploids in Table 1 that were then mated to create diploids that were tetrad-dissected.
To create the strains used in the sister-chromatid exchange assays, the HIS3 gene was deleted from a haploid segregant of the SK1 diploid EAY28 to create EAY2908. A cross of EAY2908 by EAY2209 (pch2Δ in NHY943) (Zanders and Alani 2009) generated EAY2910 and EAY2913. The HIS3 sister-chromatid recombination reporter assay contained on plasmid pNN287 (provided by Mike Fasullo) was integrated into the genome near TRP1 in EAY2913, as described by Fasullo and Davis (1987), to create EAY2918. Correct integration of the sister-chromatid recombination assay in EAY2918 was confirmed using Southern blot analysis. EAY2918 was then crossed to EAY2910 to generate strains EAY2951 and EAY2952 (wild type) and EAY2955 and EAY2956 (pch2Δ) used in the sister-chromatid recombination experiments.
To measure sister-chromatid recombination, saturated YPD overnight cultures were diluted into 22 ml YPA and grown for 17 hr. A sample of each YPA culture (0.4–2 ml) was plated on synthetic complete −HIS plates to detect early mitotic sister-chromatid recombination events that would skew meiotic analyses. No such His+ jackpots were observed in cells plated from YPA cultures (generally, fewer than one His+ cell/ml plated was observed for all strains). After 17 hr, the YPA cultures were spun down, washed once in 1% potassium acetate, resuspended in 10 ml 1% potassium acetate, and then allowed to sporulate 24 hr. Undiluted sporulated cells were then plated on synthetic complete −HIS, and cell dilutions were plated on synthetic complete media. The frequency of His+ colony-forming units (cfu) (His+ prototrophy in sporulated cells in which spores in asci were not separated) was found by dividing the number of His+ cfu/ml by the total number of colony-forming units per millimeter. Experimental replicates in which <90% of cells sporulated were not included in the data presented.
Meiotic time courses and DSB Southern blotting
For the time courses to analyze meiotic DSB levels, 0.3 ml (for RAD54 strains) or 0.6 ml (for rad54Δ strains) of a saturated YPD overnight culture from each strain to be analyzed was diluted into 200 ml YPA (2% potassium acetate) plus complete amino acid mix and grown for 17 hr. The YPA culture was then spun down, washed once in 1% potassium acetate, and resuspended in 100 ml 1% potassium acetate (Zanders and Alani 2009). All strains were grown in the same batches of media and treated identically. DNA was isolated from meiotic cultures as in Buhler et al. (2007) for dmc1Δ strains and as in Goyon and Lichten (1993) for rad50S strains. The percentage of DSBs was calculated using Image Quant software. In this analysis, a lane profile was generated and used to calculate the total lane signal. Lane background was determined from the blot regions below DSB signals. Only the peaks above lane background were quantified as DSB-specific signals.
Results
pch2Δ suppresses dmc1Δ arrest by allowing DSB repair
We initiated this study in SK1 budding yeast to examine a role for Pch2 in ensuring a meiotic arrest/delay in the absence of Dmc1 (San-Segundo and Roeder 1999; Zierhut et al. 2004; Hochwagen et al. 2005; Wu and Burgess 2006; Mitra and Roeder 2007; Zanders and Alani 2009). On the basis of recent work showing a role for Pch2 in regulating crossing over in meiosis, we and others hypothesized that Pch2 acts directly in DSB repair (Hochwagen et al. 2005; Wu and Burgess 2006; Borner et al. 2008; Joshi et al. 2009; Zanders and Alani 2009). In such a model, pch2Δ relieves the dmc1Δ arrest by allowing Dmc1-independent DSB repair. Below are physical and genetic studies that are consistent with Pch2 inhibiting DSB repair in dmc1Δ cells.
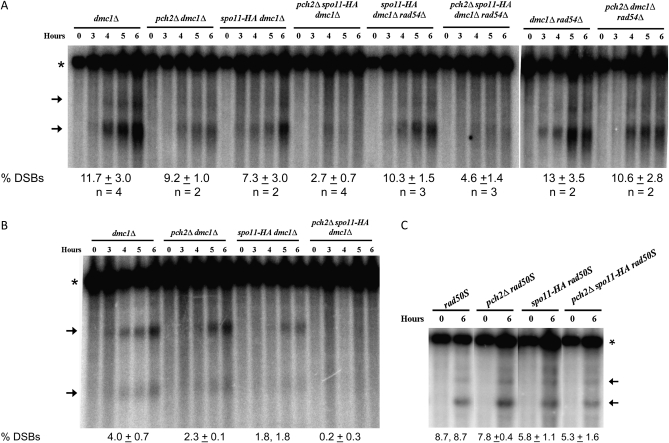
We analyzed visible meiotic DSBs at the YCR048W (chromosome III) and HIS2 (chromosome VI) hotspots in pch2Δ, dmc1Δ, spo11-HA, rad50S, and rad54Δ strain backgrounds. Six hours after meiotic induction, dmc1Δ mutants averaged 11.7 ± 3.0% (±SD; n = 4 independent cultures) DSBs at YCRO48W. pch2Δdmc1Δ (9.2 ± 1.0%, n = 2) and spo11-HA dmc1Δ (7.3 ± 3.0%, n = 2) mutants displayed slightly fewer DSBs at this hotspot. Consistent with previous results, the pch2Δspo11-HA dmc1Δ triple mutant showed lower DSB levels (2.7 ± 0.7%, n = 4; Figure 1A; Zanders and Alani 2009). A similar pattern of DSBs was observed at the HIS2 DSB hotspot on chromosome VI (Figure 1B) (Bullard et al. 1996).
Figure 1.
DSB levels observed at the YCRO48W and HIS2 hotspots. (A) Southern blots were performed on genomic DNA obtained from 0-, 3-, 4-, 5-, and 6-hr postmeiotic induction of the indicated strains to measure DSBs at the YCR048W hotspot on chromosome III (Zanders and Alani 2009). DNA was digested with BglII and probed with a chromosome III fragment (SGD coordinates 215,422–216,703). Representative blots of independent replicates (n = 2 to 4) are shown. Asterisk denotes the 11-kb parental band, and the arrows designate the DSB bands quantified. The percentage of DSBs (percentage of total lane signal) at 6 hr ± standard deviation (SD) is shown for each strain. (B) Methods used in A were performed to measure DSBs at the HIS2 hotspot on chromosome VI. The DNA was digested with BglII and probed as in Bullard et al. (1996). The asterisk denotes the 5-kb parental band and the arrows designate the DSB bands quantified. The percentage of DSBs (percentage of total lane signal) at 6 hr ± SD (n = 2) is shown for each strain. SD is not shown for spo11-HA pch2Δ because the same percentage of DSB value was obtained in two independent experiments. (C) Southern blots were performed on genomic DNA as shown in A at the YCR048W hotspot in rad50S strains. A representative blot (n = 2) is shown with the percentage of DSBs (percentage of total lane signal) at 6 hr (±SD) also shown. An SD is not shown for rad50S because the same percentage of DSB value was obtained in two independent experiments. The asterisk denotes the 11-kb parental band, and the arrows designate the DSB bands quantified. For A–C, similar results were obtained in independent time courses extended to T = 7 hr.
One explanation for the reduced level of breaks observed in pch2Δspo11-HA dmc1Δ mutants is that fewer DSBs are formed in pch2Δ mutants. Previous genetic analyses, however, suggest that pch2Δ mutants do not form fewer DSBs; pch2Δ mutants have increased COs on large chromosomes and increased gene conversion frequencies on chromosomes of all sizes. The opposite effect would be expected from a mutant with reduced DSB frequencies (Zanders and Alani 2009). To formally test if pch2Δspo11-HA dmc1Δ mutants affect DSB formation, we measured DSBs in the rad50S mutant background in which they persist (Alani et al. 1990). At the YCR048w hotspot, rad50S and pch2Δ rad50S mutants showed similar average DSB levels: 8.7% (same value in two independent experiments) and 7.8 ± 0.4%, respectively (n = 2; Figure 1C). The spo11-HA rad50S (5.8 ± 1.1%) and pch2Δspo11-HA rad50S mutants (5.3 ± 1.6%) also showed similar levels of DSBs, although the levels were lower than rad50S alone, as expected because of the presence of spo11-HA (n = 2; Figure 1C). These results suggest that the decrease in DSBs observed in pch2Δspo11-HA dmc1Δ was not due to a decrease in DSB formation.
Low levels of DSBs were observed in pch2Δspo11-HA dmc1Δ. Interestingly, these strains showed a dramatic increase in meiotic completion—39% spore formation, compared to 0% in dmc1Δ strains, which showed high levels of DSBs (Bishop et al. 1999) (Table 2; P < 0.005; Figure 1). The pch2Δ and spo11-HA mutations contribute synergistically to the triple-mutant phenotype because only a small percentage of pch2Δdmc1Δ (4.6%) and spo11-HA dmc1Δ (0.4%) cells formed spores. One explanation for the phenotype is that the recombination checkpoint is sensitive to the level of unrepaired breaks (see Discussion), such that fewer breaks elicit a less robust checkpoint arrest. Consistent with this interpretation, dmc1Δ/dmc1Δ spo11-HA/spo11yf-HA (30% of wild-type DSB levels) have increased meiotic progression [dmc1Δ, 0% spore formation; spo11-HA/spo11yf-HA dmc1Δ/dmc1Δ, 5% spore formation (Table 2; P < 0.005)].
Spores produced by pch2Δdmc1Δ and spo11-HA pch2Δdmc1Δ were mostly inviable (<3% spore viability for each), suggesting that interhomolog recombination was not restored in these mutants (Table 2). We tested whether other types of repair occurred in the spo13 mutant background in which a mixed chromosome division occurs: some chromosomes undergo an equational division whereas others segregate reductionally (Klapholz and Esposito 1980; Hugerat and Simchen 1993). spo13 mutants can produce viable meiotic progeny in the absence of meiotic DSBs or if DSB repair does not yield COs (e.g., Malone and Esposito 1981). Because of this, spore viability analyses in the spo13 mutant background can detect DSB repair that does not facilitate proper MI chromosome segregation (Bishop et al. 1999).
Similar to previous work in SK1 strains, spo13dmc1Δ showed low levels of sporulation (10%) and spore viability (7%) compared to spo13 (63% sporulation, 47% viability; Table 2; P < 0.005 for both sporulation and spore viability) (Bishop et al. 1999). Introducing the spo11-HA allele to dmc1Δspo13 increased spore formation in the resulting triple mutant to 17% and spore viability to 16% (Table 2; P < 0.005 for both sporulation and spore viability). This result is expected because fewer DSBs are produced in spo11-HA strains (Martini et al. 2006; Johnson et al. 2007). Deleting PCH2 in dmc1Δspo13 had a strong effect; sporulation in this triple mutant increased to 43% and spore viability increased to 16% (Table 2; P < 0.005 for both sporulation and spore viability). The spo11-HA pch2Δspo13dmc1Δ quadruple mutant showed even greater sporulation (58%) and spore viability (26%; Table 2; P < 0.005 for both sporulation and spore viability). These results are consistent with some Dmc1-independent repair occurring in pch2Δdmc1Δ mutants.
pch2Δ mutants have an increased dependence on Rad54-mediated repair
Sporulation of pch2Δspo11-HA dmc1Δ mutants and spore viability of the pch2Δspo11-HA dmc1Δspo13 mutants were Rad54-dependent, suggesting that Rad54 is repairing breaks in these mutants (Table 2). During vegetative growth, DSBs are preferentially repaired by homologous recombination involving sister chromatids in steps that are mediated by the Rad51 recombinase and its partner Rad54 (Kadyk and Hartwell 1992; M. Shinohara et al. 1997; Arbel et al. 1999; Krogh and Symington 2004). Consistent with Rad54-dependent recombination in pch2Δ mutants, we observed a reduction in sporulation (46%) and spore viability (47%) in the pch2Δrad54Δ double mutant compared to pch2Δ (81% sporulation and 95% spore viability) and rad54Δ single mutants (58% sporulation and 60% spore viability; Table 2; P < 0.005 for all comparisons). Interestingly, CO levels were not reduced in pch2Δrad54Δ compared to pch2Δ, suggesting the Rad54-dependent recombination in pch2Δ mutants is intersister (Table 3).
An important prediction of the above genetic analyses is that the rad54Δ mutation should result in a restoration of observed DSBs in the pch2Δspo11-HA dmc1Δ mutant. This prediction was not met; 4.6 ± 1.4% DSBs (n = 3) were observed at YCRO48W in pch2Δspo11-HA dmc1Δrad54Δ, compared to 2.7 ± 0.7% (n = 4) in pch2Δspo11-HA dmc1Δ and 10.3 ± 1.5% (n = 3) in spo11-HA dmc1Δrad54Δ 6 hr after meiotic induction (Figure 1). It is possible that both Dmc1- and Rad54-independent repair can occur in the absence of these factors in a pch2Δ background. Another possibility is that hyper-resected DSBs form in pch2Δspo11-HA dmc1Δrad54Δ that cannot be detected by Southern blot. More experimentation is needed to understand this phenotype.
pch2Δ spo11-HA phenotype is modulated by Mek1 activity
Previously, we showed (Zanders and Alani 2009) that pch2Δspo11-HA strains have a spore viability defect and hypothesized that the decreased spore viability was due to defects in CO interference and partner choice. If a compromised interhomolog bias contributes to the pch2Δspo11-HA phenotype, then further undermining interhomolog bias should enhance the phenotype. Alternatively, reinforcing interhomolog bias should suppress the pch2Δspo11-HA phenotype. To test this hypothesis, we utilized two MEK1 alleles: mek1-as (mek1-Q241G hypomorph) (Callender and Hollingsworth 2010) and MEK1-GST (hypermorph) (Wu et al. 2010). mek1-as strains complete meiosis efficiently in the absence of a 1-Na-PP1 inhibitor and display nearly wild-type spore viability; however, Mek1-as has reduced affinity for ATP in vitro, and in one dmc1Δ strain background, the mek1-as mutation conferred phenotypes consistent with defects in interhomolog bias (Wan et al. 2004; Niu et al. 2009; Callender and Hollingsworth 2010). Wu et al. (2010) recently showed that MEK1-GST is a semidominant allele that shows increased interhomolog recombination events, primarily noncrossovers, and fewer intersister events, with no change in DSB levels.
As shown in Table 4, we constructed mek1-as pch2Δspo11-HA and MEK1-GST pch2Δspo11-HA strains and examined their spore formation efficiency and viability (mek1-as strains were analyzed in the absence of a 1-Na-PP1 inhibitor). We found that the mek1-as mutation reduced the spore viability of pch2Δspo11-HA strains from 57 to 41% (P < 0.005). The mek1-as mutation did not significantly affect the spore viability of either single mutant alone (Table 4). The MEK1-GST allele increased spore viability in pch2Δspo11-HA strains from 57 to 89% (P < 0.005); however, as described previously (Wu et al. 2010), there was a general defect in spore formation due to MEK1-GST. MEK1-GST did not increase the spore viability in either single mutant (Table 4). These data are consistent with our hypothesis that excess intersister repair contributes to spore inviablity of pch2Δspo11-HA.
Evidence that Pch2 promotes interhomolog DSB repair
To more directly test if there is an increase in intersister repair in pch2Δ, we adapted an assay developed by Fasullo and Davis (1987) to measure sister-chromatid exchange in meiosis. This assay utilizes a HIS3 reporter gene in which cells become His+ if a sister-chromatid recombination event (either a CO or gene conversion) occurs between two his3 truncations to produce full-length HIS3. The mean frequency of His+ colonies was 1.6 × 10−6 (n = 22 independent cultures) in wild type and 5.3 × 10−6 in cells lacking Pch2 (n = 24). pch2Δ values were significantly higher than wild type (P = 0.008, Mann–Whitney U-test).
Discussion
We provide several independent lines of evidence consistent with a role for Pch2 in inhibiting Dmc1-independent intersister DSB repair in meiosis. First, pch2Δ contributes to a reduction of unrepaired DSBs visible on Southern blots in spo11-HA dmc1Δ mutants (Figure 1). This reduction in detectable DSBs appears to be due to DSB repair because the levels of DSBs formed, as measured in a rad50S background, are not affected by pch2Δ and the spore inviability seen in pch2Δdmc1Δ and pch2Δspo11-HA dmc1Δ mutants is suppressed by spo13Δ (Table 2). We hypothesize that the DSB repair occurring in pch2Δspo11-HA dmc1Δ is intersister because it does not facilitate proper MI chromosome segregation. Our epistasis analysis of pch2Δrad54Δ suggests that pch2Δ mutants are more dependent on Rad54-dependent repair, but that Rad54 does not appear to contribute to interhomolog CO repair in pch2Δ. These data further support the idea that pch2Δ mutants have increased intersister repair. In addition, we demonstrated that a mek1 hypomorph enhanced the spore death phenotype of pch2Δspo11-HA whereas a MEK1 gain-of-function allele suppressed the phenotype. These experiments are consistent with excess intersister DSB repair contributing to spore inviability of pch2Δspo11-HA. Finally, our genetic reporter assay demonstrated an increase in intersister DSB repair at one locus.
Our data and those of other groups are consistent with recombination checkpoint signaling being sensitive to unrepaired DSB levels, such that more DSBs trigger checkpoint arrest in a greater proportion of the cell population (Malkova et al. 1996; Bhalla and Dernburg 2005; MacQueen et al. 2005; Johnson et al. 2007; Callender and Hollingsworth 2010; Goldfarb and Lichten 2010). In wild-type cells, DSBs are quickly repaired and the Mek1-mediated checkpoint delay is transient. In dmc1Δ, DSBs are not repaired and Mek1 elicits checkpoint arrest. When DSBs are reduced in dmc1Δ strains containing spo11 hypomorph alleles, the checkpoint response is less robust and fewer cells arrest. We hypothesize that the spo11-HA and pch2Δ mutations independently contribute to reducing the level of unrepaired DSBs available to trigger the recombination checkpoint in dmc1Δ mutants: spo11-HA forms fewer DSBs and pch2Δ acts by allowing Dmc1-independent repair. In this model, the combination of spo11-HA and pch2Δ in a dmc1Δ background synergistically contribute to DSB repair due to a positive feedback loop wherein fewer DSBs elicit less checkpoint activation, which allows even more DSB repair and meiotic progression. At present we do not have a good sense of the number of DSBs that would be needed to elicit checkpoint activation; however, work from Malkova et al. (1996) showed that a single unrepaired DSB does not arrest meiosis.
Pch2 acts in meiotic CO control to limit CO formation on large chromosomes and promote CO interference (Joshi et al. 2009; Zanders and Alani 2009). This work suggests a broader role for Pch2 in DSB repair that includes inhibiting intersister DSB repair. Although the mechanism of Pch2 function in DSB repair is unknown, there are several avenues worthy of investigation. RTEL-1, the Caenorhabditis elegans homolog of the yeast Srs2 helicase has been shown to be defective in CO interference and CO homeostasis (Youds et al. 2010). Thus one possibility is that Pch2 facilitates access of a helicase to remove inappropriate strand invasion events. Borner et al. (2008) first posited another attractive (and not mutually exclusive) hypothesis that Pch2 somehow promotes Mec1 regulatory action. Indeed, at least one Mek1 effector (a Mec1 target) that promotes interhomolog bias is still unknown (Niu et al. 2009; Hollingsworth 2010). This effector could be Pch2 acting to augment or promote Mek1 activity. Under this model, Mek1 signaling is attenuated in pch2Δ mutants, allowing excess intersister repair. Such an idea is consistent with work from Wu et al. (2010), who showed that an activated MEK1 allele (MEK1-GST) promoted an increase in interhomolog events that were primarily repaired as NCOs and fewer intersister events, with no change in DSB levels (Wu et al. 2010). This idea is also consistent with previous data showing that pch2Δ mutants display increased CO events at the expense of NCOs (Zanders and Alani 2009). Experiments to test these hypotheses are underway.
Acknowledgments
We are grateful to Valentin Boerner, Andreas Hochwagen, Michael Lichten, and Scott Keeney for providing unpublished data and to Chip Aquadro, Joshua Filter, Heather Flores, Tamara Goldfarb, Nancy Hollingsworth, and Michael Lichten for critical comments and technical advice. We thank Mike Fasullo for providing the HIS3 sister-chromatid exchange plasmid and Nancy Hollingsworth and Sean Burgess for providing the mek1-as and MEK1-GST alleles, respectively. Finally, we are grateful to Nancy Hollingsworth for helpful discussions. This work was supported by National Institutes of Health (NIH) grant GM53085 supplemented with an American Recovery and Reinvestment Act award (to E.A.). S.Z. and M.S.B. were supported by NIH training grants awarded to Cornell University.
Literature Cited
- Alani E., Padmore R., Kleckner N., 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61: 419–436 [DOI] [PubMed] [Google Scholar]
- Arbel A., Zenvirth D., Simchen G., 1999. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 18: 2648–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso J. L., Wanat J., Gemici Z., Alani E., 2004. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168: 1805–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L. E., Copenhaver G. P., 2010. Genetic interference: don’t stand so close to me. Curr. Genomics 11: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Dernburg A. F., 2005. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310: 1683–1686 [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Zickler D., 2004. Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117: 9–15 [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Park D., Xu L., Kleckner N., 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456 [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Nikolski Y., Oshiro J., Chon J., Shinohara M., et al. , 1999. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells 4: 425–444 [DOI] [PubMed] [Google Scholar]
- Blitzblau H. G., Bell G. W., Rodriguez J., Bell S. P., Hochwagen A., 2007. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr. Biol. 17: 2003–2012 [DOI] [PubMed] [Google Scholar]
- Borner G. V., Kleckner N., Hunter N., 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Borner G. V., Barot A., Kleckner N., 2008. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc. Natl. Acad. Sci. USA 105: 3327–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler C., Borde V., Lichten M., 2007. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 5: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard S. A., Kim S., Galbraith A. M., Malone R. E., 1996. Double strand breaks at the HIS2 recombination hot spot in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93: 13054–13059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busygina V., Sehorn M. G., Shi I. Y., Tsubouchi H., Roeder G. S., et al. , 2008. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 22: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callender T., Hollingsworth N. M., 2010. Mek1 suppression of meiotic double-strand break repair is specific to sister chromatids, chromosome autonomous and independent of Rec8 cohesin complexes. Genetics 185: 771–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo J. A., Johnson A. L., Sedgwick S. G., Cha R. S., 2008. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132: 758–770 [DOI] [PubMed] [Google Scholar]
- Chen S. Y., Tsubouchi T., Rockmill B., Sandler J. S., Richards D. R., et al. , 2008. Global analysis of the meiotic crossover landscape. Dev. Cell 15: 401–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De los Santos T., Hunter N., Lee D., Larkin B., Loidl J., et al. , 2003. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Massy B., Baudat F., Nicolas A., 1994. Initiation of recombination in Saccharomyces cerevisiae haploid meiosis. Proc. Natl. Acad. Sci. USA 91: 11929–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser M. E., Ewing D. J., Conrad M. N., Dominguez A. M., Barstead R., et al. , 1997. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics 147: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasullo M. T., Davis R. W., 1987. Recombinational substrates designed to study recombination between unique and repetitive sequences in vivo. Proc. Natl. Acad. Sci. USA 84: 6215–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A., 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360 [DOI] [PubMed] [Google Scholar]
- Goldfarb T., Lichten M., 2010. The sister chromatid is frequently and efficiently used for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 8: e1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553 [DOI] [PubMed] [Google Scholar]
- Goyon C., Lichten M., 1993. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol. Cell. Biol. 13: 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T., Hall H., Hunt P., 2007. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 16: R203–R208 [DOI] [PubMed] [Google Scholar]
- Hochwagen A., Tham W. H., Brar G. A., Amon A., 2005. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell 122: 861–873 [DOI] [PubMed] [Google Scholar]
- Hollingsworth N. M., 2010. Phosphorylation and the creation of interhomolog bias during meiosis in yeast. Cell Cycle 9: 436–437 [DOI] [PubMed] [Google Scholar]
- Hollingsworth N. M., Byers B., 1989. HOP1: a yeast meiotic pairing gene. Genetics 121: 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N. M., Goetsch L., Byers B., 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61: 73–84 [DOI] [PubMed] [Google Scholar]
- Hugerat Y., Simchen G., 1993. Mixed segregation and recombination of chromosomes and YACs during single-division meiosis in spo13 strains of Saccharomyces cerevisiae. Genetics 135: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. A., Fink G. R., 1985. Meiotic recombination between duplicated genetic elements in Saccharomyces cerevisiae. Genetics 109: 303–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R., Borde V., Neale M. J., Bishop-Bailey A., North M., et al. , 2007. Excess single-stranded DNA inhibits meiotic double-strand break repair. PLoS Genet. 3: e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. H., Franklin F. C., 2006. Meiotic crossing-over: obligation and interference. Cell 126: 246–248 [DOI] [PubMed] [Google Scholar]
- Joshi N., Barot A., Jamison C., Borner G. V., 2009. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 5: e1000557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Hartwell L. H., 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E., 1980. Isolation of SPO12-1 and SPO13-1 from a natural variant of yeast that undergoes a single meiotic division. Genetics 96: 567–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H. L., 1997. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh B. O., Symington L. S., 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38: 233–271 [DOI] [PubMed] [Google Scholar]
- Li X. C., Schimenti J., 2007. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 3: e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D., Nikolsky Y., Bishop D. K., Weinert T. A., 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383: 840–843 [DOI] [PubMed] [Google Scholar]
- MacQueen A. J., Phillips C. M., Bhalla A., Weiser P., Villeneuve A. M., et al. , 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., Ross L., Dawson D., Hoekstra M. F., Haber J. E., 1996. Meiotic recombination initiated by a double-strand break in rad50 delta yeast cells otherwise unable to initiate meiotic recombination. Genetics 143: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. E., Esposito R. E., 1981. Recombinationless meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1: 891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi R., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E., Diaz R. L., Hunter N., Keeney S., 2006. Crossover homeostasis in yeast meiosis. Cell 126: 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra N., Roeder G. S., 2007. A novel nonnull ZIP1 allele triggers meiotic arrest with synapsed chromosomes in Saccharomyces cerevisiae. Genetics 176: 773–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wan L., Baumgartner B., Schaefer D., Loidl J., et al. , 2005. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell 16: 5804–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Li X., Job E., Park C., Moazed D., et al. , 2007. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol. Cell. Biol. 27: 5456–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wan L., Busygina V., Kwon Y., Allen J. A., et al. , 2009. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell 36: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Roeder G. S., 1990. Meiosis in asynaptic yeast. Genetics 126: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11: 2600–2621 [DOI] [PubMed] [Google Scholar]
- Roig I., Dowdle J. A., Toth A., De Rooj D. G., Jasin M., et al. , 2010. Mouse TRIP13/PCH2 is required for recombination and normal higher order chromosome structure during meiosis. PLoS Genet. 6: e1001062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo P. A., Roeder G. S., 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97: 313–324 [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., 1994. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76: 51–63 [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N., 1997. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135 [DOI] [PubMed] [Google Scholar]
- Shinohara A., Gasior S., Ogawa T., Kleckner N., Bishop D. K., 1997. Saccharomyces cerevisiae recA homologs RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2: 615–629 [DOI] [PubMed] [Google Scholar]
- Shinohara M., Shita-Yamaguchi E., Buerstedde J. M., Shinagawa H., Ogawa H., et al. , 1997. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147: 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., Roeder G. S., 1995. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J. Cell Biol. 128: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terentyev Y., Johnson R., Neale M. J., Khisroon M., Bishop-Bailey A., et al. , 2010. Evidence that MEK1 positively promotes interhomologue double-strand break repair. Nucleic Acids Res. 38: 4349–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. A., Stahl F. W., 1999. Genetic control of recombination partner preference in yeast meiosis: isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics 153: 621–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H., Roeder G. S., 2003. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell 5: 915–925 [DOI] [PubMed] [Google Scholar]
- Tsubouchi H., Roeder G. S., 2006. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 20: 1766–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P., 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808 [DOI] [PubMed] [Google Scholar]
- Wan L., De los Santos T., Zhang C., Shokat K., Hollingsworth N. M., 2004. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol. Biol. Cell. 15: 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber H. A., Howard L., Bickel S. E., 2004. The cohesion protein ORD is required for homologue bias during meiotic recombination. J. Cell Biol. 164: 819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., et al. , 2009. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5: e1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Burgess S. M., 2006. Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr. Biol. 16: 2473–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Y., Ho H. C., Burgess S. M., 2010. Mek1 kinase governs outcomes of meiotic recombination and the checkpoint response. Curr. Biol. 20: 1707–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Weiner B. M., Kleckner N., 1997. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 11: 106–118 [DOI] [PubMed] [Google Scholar]
- Youds J. L., Mets D. G., Mcilwraith M. J., Martin J. S., Ward J. D., et al. , 2010. RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327: 1254–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S., Alani E., 2009. The pch2Delta mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet. 5: e1000571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C., Berlinger M., Rupp C., Shinohara A., Klein F., 2004. Mnd1 is required for meiotic interhomolog repair. Curr. Biol. 14: 752–762 [DOI] [PubMed] [Google Scholar]