Abstract
Genetic studies have identified thousands of variants associated with complex traits. However, most association studies are limited to populations of European descent and a single phenotype. The Population Architecture using Genomics and Epidemiology (PAGE) Study was initiated in 2008 by the National Human Genome Research Institute to investigate the epidemiologic architecture of well-replicated genetic variants associated with complex diseases in several large, ethnically diverse population-based studies. Combining DNA samples and hundreds of phenotypes from multiple cohorts, PAGE is well-suited to address generalization of associations and variability of effects in diverse populations; identify genetic and environmental modifiers; evaluate disease subtypes, intermediate phenotypes, and biomarkers; and investigate associations with novel phenotypes. PAGE investigators harmonize phenotypes across studies where possible and perform coordinated cohort-specific analyses and meta-analyses. PAGE researchers are genotyping thousands of genetic variants in up to 121,000 DNA samples from African-American, white, Hispanic/Latino, Asian/Pacific Islander, and American Indian participants. Initial analyses will focus on single nucleotide polymorphisms (SNPs) associated with obesity, lipids, cardiovascular disease, type 2 diabetes, inflammation, various cancers, and related biomarkers. PAGE SNPs are also assessed for pleiotropy using the “phenome-wide association study” approach, testing each SNP for associations with hundreds of phenotypes. PAGE data will be deposited into the National Center for Biotechnology Information's Database of Genotypes and Phenotypes and made available via a custom browser.
Keywords: cardiovascular diseases, cohort studies, genome-wide association study, multifactorial inheritance, neoplasms, obesity, population characteristics, reproducibility of results
Genome-wide association studies (GWAS) have been successful in confirming and identifying numerous loci related to complex human traits, resulting in the compilation of hundreds of unique single nucleotide polymorphism (SNP)-trait associations at the level of genome-wide significance (P ≤ 5 × 10−8) (1). Though these GWAS successes are considerable, most originate from populations of European descent (2, 3), and it is not yet clear to what extent associations confirmed in one population are generalizable to other populations such as African Americans and Hispanics. Differences in genetic background and environment may alter the effect of causal variants. Further, differences in linkage disequilibrium patterns may modify observed associations of nonfunctional SNPs (i.e., index signals). Given these factors, the determination of causal variants, their roles in gene function, their connections to complex traits, their interaction with known risk factors, and their potential for clinical translation requires making substantial progress beyond GWAS (4–6). Laying the initial groundwork includes evaluation of the full breadth of phenotypic associations of highly replicated GWAS-defined variants and their allele frequencies on a population basis, particularly in populations of non-European ancestry.
The Population Architecture using Genomics and Epidemiology (PAGE) Study (http://www.pagestudy.org) is a National Human Genome Research Institute (NHGRI)-created consortium of large, well-characterized population-based studies that provides an unprecedented opportunity to investigate the epidemiologic architecture of well-replicated genetic variants associated with complex diseases. Just as genetic architecture describes the genomic influences underlying a phenotypic trait, epidemiologic architecture describes population-level phenotypes, exposures, and ancestry that modify a specific genetic effect and influence its population impact.
PAGE investigators have expertise in epidemiology, genetics, biostatistics, bioinformatics, and various common complex diseases. The PAGE consortium consists of 4 large, ongoing population-based studies or consortia: Epidemiologic Architecture for Genes Linked to Environment (EAGLE), which is based on data from 3 National Health and Nutrition Examination Surveys (NHANES; https://chgr.mc.vanderbilt.edu/eagle) (7); the Multiethnic Cohort Study (http://www.crch.org/multiethniccohort/) (8); the Women's Health Initiative (http://www.whi.org) (9); and Causal Variants Across the Life Course (CALiCo), a consortium of 5 cohort studies—Atherosclerosis Risk in Communities (ARIC; http://www.cscc.unc.edu/aric/) (10), Coronary Artery Risk Development in Young Adults (CARDIA; http://www.cardia.dopm.uab.edu/) (11), the Cardiovascular Health Study (http://www.chs-nhlbi.org/) (12), the Hispanic Community Health Study/Study of Latinos (http://www.cscc.unc.edu/hchs/) (13), and the Strong Heart Study (http://strongheart.ouhsc.edu/) (14, 15). A coordinating center provides operational and scientific support, while the NHGRI Office of Population Genomics participates in study design, scientific support, and assessment of progress. With over 121,200 African-American, white, Hispanic/Latino, Asian/Pacific Islander, and American Indian participants available for study across the cohorts, PAGE investigators are well poised to address the critical research questions that follow the establishment of GWAS associations through large-scale replication and generalization.
In this paper, we describe the PAGE consortium, including its goals, organization, data sets, methods, and study design, and highlight how PAGE can contribute to understanding the genetic and epidemiologic architecture of confirmed, associated genetic variants.
MATERIALS AND METHODS
PAGE Study goals
The PAGE Study is designed to refine knowledge on the epidemiologic architecture of common genetic variants associated with human diseases and traits. To address this need, PAGE investigators will evaluate the index signals from GWAS or biologically relevant alleles (i.e. “causal” alleles) according to these objectives:
Assessing the generalizability of the phenotype-variant association to other populations.
Comparing the strength of the effects in various subgroups. These subgroups are defined by race/ethnicity and other demographic characteristics; exposures, risk profiles, and disease characteristics; and social contexts.
Estimating the burden of disease, including relative risks of incident disease, associated with genetic variants in population-based settings.
Characterizing effect modification by genetic and environmental factors, including lifestyle, comorbidity, and medication use.
Extending results to disease subtypes, related biomarkers, intermediate phenotypes, and precursors.
Assessing pleiotropic effects by investigating associations with phenotypes unrelated to those reported in the original studies.
Addressing these objectives will help PAGE researchers determine whether a variant is causal and select candidate variants for in-depth functional studies. PAGE may also identify phenotypic characteristics that provide clues to the underlying biologic effects of the variants under study.
To accomplish these goals, the PAGE investigators have developed approaches to streamline prioritization of participants and SNPs to be genotyped, harmonize phenotypes across studies, assess quality control for genotype and phenotype data, and explore new analytic methods. Because these considerations are relevant to ongoing and future studies of population genomics, particularly in the context of multisite meta-analyses, an additional goal of PAGE researchers is to describe PAGE approaches so they may aid the scientific community.
The breadth of phenotype data available in each PAGE data set permits hundreds of phenotypes to be tested against each genotyped SNP in a “phenome-wide association study.” In contrast to a GWAS, which tests many SNPS for association with 1 phenotype or a few phenotypes, a phenome-wide association study explores the relations of many phenotypes with 1 SNP or a few SNPs (16). This type of study has the potential to identify pleiotropic associations that can provide clues to functional implications of GWAS-defined SNPs and may provide other insights into pathobiology. Finally, to help synthesize results across studies and present these data in a visually intuitive manner, PAGE investigators are developing customized graphical browser software for exploring these results.
PAGE Study organization
The PAGE Study is a collaborative effort between 4 projects representing 8 large population-based studies, a coordinating center, and the NHGRI Office of Population Genomics. PAGE began in 2008 in response to NHGRI solicitations for applications for study sites and a coordinating center (17, 18). PAGE genotyping and analyses are conducted in approximately annual cycles.
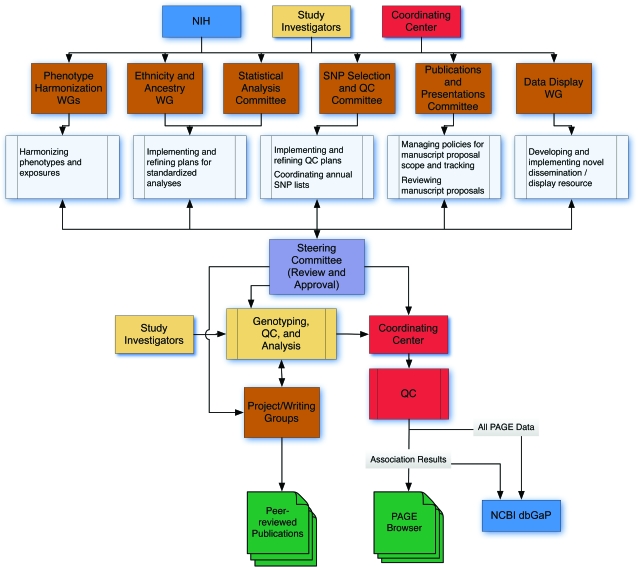
Figure 1 shows the overall organization of the PAGE Study. The Steering Committee meets often (by teleconference or in person) to review priorities and progress, discuss the study design and analyses, and resolve problems. The NHGRI Office of Population Genomics participates in working groups and subcommittees and helps set research priorities, develop program policies, and coordinate activities among PAGE investigators. The Coordinating Center assists with integration, synthesis, quality control, and dissemination of study results and data via the National Center for Biotechnology Information's Database of Genotypes and Phenotypes (dbGaP) (19). The Coordinating Center also develops novel data display tools, facilitates collaborations, and manages program logistics. Working groups and committees are organized as needed to make optimal use of expertise among all PAGE participants. All working groups and committees comprise representatives from all participating studies, with focused efforts to include postdoctoral fellows and new investigators.
Figure 1.
Organizational structure of the Population Architecture Using Genomics and Epidemiology (PAGE) Study, 2008–present. The PAGE Steering Committee (purple box, center) comprises principal investigators from the individual studies and the Coordinating Center and the National Human Genome Research Institute project scientist. Multiple investigators from each of these groups participate in working groups (WGs) and committees (brown boxes) which are tasked with specific responsibilities (light blue boxes) to facilitate analysis, publication, and dissemination of PAGE results. Genotyping, quality control (QC), and statistical analysis are performed by researchers in the individual studies with guidance from the Steering Committee and assistance from the Coordinating Center. Project and writing groups are organized to produce topic-specific manuscripts for the purpose of publication in peer-reviewed journals; the minor distinction between the 2 groups deals with the scope of the analysis (project groups tend to include more studies and produce multiple manuscripts). In parallel, the Coordinating Center performs an additional level of QC on the association-level data from each study, preparing these aggregate data for dissemination via the PAGE data browser, a user-friendly resource for viewing PAGE results. PAGE data will also be available in the National Center for Biotechnology Information's (NCBI) Database of Genotypes and Phenotypes (dbGaP). NIH, National Institutes of Health; SNP, single nucleotide polymorphism.
Study populations
The PAGE network includes 4 population-based projects. Each study includes 15,000–160,000 participants, for whom extensive phenotype, covariate, and exposure data, as well as high-quality DNA, are available for analysis (Table 1). The PAGE studies include participants from a variety of ethnic populations (Table 2). The researchers in all PAGE studies obtained institutional review board approval, and all subjects provided written informed consent. These studies have evaluated thousands of phenotypes, with substantial overlap across the PAGE studies (Table 3) and ancestry groups (Table 4). The individual study Web sites provide additional descriptive details.
Table 1.
Characteristics of Cohorts Included in the Population Architecture Using Genomics and Epidemiology (PAGE) Study, 2008–present
| Study | Study Type | Focus of Study | Years of Data Collection | Length of Follow-up, yearsb | Mean Age, years | Age Range, years | % Women | Genotyping Platform(s) Used |
| EAGLE Study | Cross-sectionala | American health | 1991–1994, 1999–2002 | N/A | 35 | 12–95 | 54 | Applied Biosystems TaqMan (Applied Biosystems, Foster City, California), TaqMan OpenArray (Applied Biosystems), Sequenom iPlex (Sequenom, San Diego, California), Illumina BeadXpress (Illumina, Inc., San Diego, California) |
| Multiethnic Cohort Study | Nested case-control | Cancer | 1993–1996 | 17 | 60 | 45–78 | 55 | Applied Biosystems TaqMan, TaqMan OpenArray |
| Women's Health Initiative | Cohort and clinical trials | Women's health | 1993–1998 | 17 | 63 | 50–79 | 100 | Illumina BeadXpress |
| CALiCo Consortium | ||||||||
| ARIC Study | Longitudinal | Cardiovascular disease | 1987–present | 24 | 54 | 45–64 | 55.1 | Applied Biosystems TaqMan, Sequenom iPlex |
| CARDIA Study | Longitudinal | Cardiovascular disease | 1986–present | 25 | 25 | 18–30 | 54.4 | Applied Biosystems TaqMan, Sequenom iPlex |
| Cardiovascular Health Study | Longitudinal | Cardiovascular disease | 1988–1999 | 18 | 73 | 65–100 | 57.6 | Applied Biosystems TaqMan, Sequenom iPlex |
| Strong Heart Study | Longitudinal | Cardiovascular disease | 1988–present | 23 | 40 | 14–91 | 59.3 | Applied Biosystems TaqMan, Sequenom iPlex |
| Hispanic Community Health Study/Study of Latinos | Longitudinal | Cardiovascular disease | 2008–present | 3 | 55 | 18–72 | 65 | Applied Biosystems TaqMan, Sequenom iPlex |
Abbreviations: ARIC, Atherosclerosis Risk in Communities; CALiCo, Causal Variants Across the Life Course; CARDIA, Coronary Artery Risk Development in Young Adults; EAGLE, Epidemiologic Architecture for Genes Linked to Environment; N/A, not applicable; NHANES, National Health and Nutrition Examination Survey.
The EAGLE study analyzes data from phase 2 of the Third National Health and Nutrition Examination Survey (NHANES III) and NHANES 1999–2002.
Participant follow-up is ongoing; years shown are the maximum number of follow-up years as of the date of publication.
Table 2.
Number of Participants in Each Study Included in the Population Architecture Using Genomics and Epidemiology (PAGE) Study, by Race/Ethnicity, 2008–present
| Ethnic/Racial Group | CALiCo Consortiuma |
EAGLE Study |
Women's Health Initiative |
Multiethnic Cohort Study |
Total |
|||||
| Total No. With DNAb | No. of Persons Analyzedc | Total No. With DNA | No. of Persons Analyzed | Total No. With DNA | No. of Persons Analyzed | Total No. With DNA | No. of Persons Analyzed | Total No. With DNA | No. of Persons Analyzed | |
| White | 18,093 | 18,093 | 6,634 | 6,634 | 118,347 | 26,246 | 14,699 | 6,441 | 157,773 | 57,414 |
| Hispanic/Latinod | 6,500e | 0e | 3,950 | 3,950 | 5,367 | 2,430 | 18,086 | 8,909 | 33,903 | 15,289 |
| African-American | 7,203 | 7,203 | 3,458 | 3,458 | 11,924 | 5,218 | 11,279 | 8,030 | 33,864 | 23,909 |
| Asian/Pacific Islanderf | 47 | 47 | 3,474 | 1,954 | Japanese: 24,676 | 11,774 | 33,760 | 16,975 | ||
| Native Hawaiian: 5,563 | 3,200 | |||||||||
| American Indian | 7,283 | 7,283 | 597 | 388 | 7,880 | 7,671 | ||||
| Other/not specified | N/A | N/A | 956 | 0 | 1,936 | 0 | N/A | N/A | 2,892 | 0 |
| Total | 39,126 | 32,626 | 14,998 | 14,042 | 141,645 | 36,236 | 74,303 | 38,354 | 270,072 | 121,258 |
Abbreviations: CALiCo, Causal Variants Across the Life Course; EAGLE, Epidemiologic Architecture for Genes Linked to Environment; N/A, not applicable.
For studies included in the CALiCo consortium, see Table 1.
Total number of participants with DNA available.
Total number of persons analyzed in year 1 and/or year 2. Similar numbers are expected for years 3 and 4.
All subjects who specified any Hispanic or Latino heritage are tabulated in this row only.
Data on the CALiCo Hispanic/Latino participants became available starting in year 3.
In the Multiethnic Cohort Study, Native Hawaiians and Japanese are considered separate racial/ethnic groups.
Table 3.
Availability of Phenotype Data Across Studies in the Population Architecture Using Genomics and Epidemiology (PAGE) Study, by Study, 2008–present
| Phenotype Domain | EAGLE Study | Multiethnic Cohort Study | Women's Health Initiative | CALiCo Consortium |
||||
| ARIC Study | CARDIA Study | Cardiovascular Health Study | Strong Heart Study | Hispanic Community Health Study/Study of Latinos | ||||
| Alcohol, tobacco, and other substances | ||||||||
| Alcohol use | X | X | X | X | X | X | X | X |
| Smoking history | X | X | X | X | X | X | X | X |
| Anthropometrics | X | X | X | X | X | X | X | X |
| Biomarkers | X | X | X | X | X | X | X | X |
| Cancer | ||||||||
| Breast | X | X | X | X | X | X | ||
| Prostate | X | X | X | X | X | |||
| Colorectal | X | X | X | X | X | X | ||
| Lung | X | X | X | X | X | X | ||
| Cardiovascular | ||||||||
| Hypertension | X | Xa | X | X | X | X | X | X |
| Prevalent coronary heart disease | X | Xa | X | X | X | X | X | X |
| Mean systolic and diastolic blood pressures | X | X | X | X | X | X | X | |
| Diabetes and renal function | X | X | X | X | X | X | X | X |
| General biochemistry tests | X | X | X | X | X | X | X | X |
| Hematology | X | X | X | X | X | X | X | |
| Infectious diseases and immunity | X | Xa | X | X | X | X | X | X |
| Inflammation | X | X | X | X | X | X | X | X |
| Neurology | X | X | X | |||||
| Nutrition and dietary supplements | X | X | X | X | X | X | X | X |
| Ocular | X | Xa | X | X | X | |||
| Physical activity | X | X | X | X | X | X | X | X |
| Psychiatric | X | X | X | X | X | X | ||
| Reproductive health | X | X | X | X | X | X | X | X |
| Respiratory | X | X | X | X | X | X | X | |
| Skin, bone, muscle, and joint | X | Xa | X | X | X | X | X | |
| Speech and hearing | X | X | X | X | X | X | ||
Abbreviations: ARIC, Atherosclerosis Risk in Communities; CALiCo, Causal Variants Across the Life Course; CARDIA, Coronary Artery Risk Development in Young Adults; EAGLE, Epidemiologic Architecture for Genes Linked to Environment.
Phenotypes are self-reported.
Table 4.
Sizes of Population Samples Available in the Population Architecture Using Genomics and Epidemiology (PAGE) Study, by Phenotype and Ancestry, 2008–present
| Phenotype Domain | Ethnic/Racial Group |
Total | ||||
| White | African- American | Hispanic | American Indian | Asian, Pacific Islander, or Native Hawaiian | ||
| Alcohol, tobacco, and other substances | ||||||
| Alcohol use | 53,884 | 19,526 | 18,648 | 7,739 | 11,229 | 111,026 |
| Smoking history | 56,508 | 21,622 | 20,757 | 7,739 | 11,457 | 118,083 |
| Anthropometrics | 56,567 | 21,671 | 20,851 | 7,739 | 11,470 | 118,298 |
| Biomarker | 37,481 | 15,056 | 13,014 | 7,577 | 1,884 | 75,012 |
| Cancer | ||||||
| Breast | 24,830 | 8,563 | 7,918 | 69 | 1,032 | 42,412 |
| Prostate | 18,597 | 7,970 | 7,819 | 0 | 959 | 35,345 |
| Colorectal | 24,306 | 8,498 | 7,961 | 66 | 1,080 | 41,911 |
| Lung | 24,467 | 8,133 | 7,540 | 64 | 334 | 40,538 |
| Cardiovascular | ||||||
| Hypertension | 47,138 | 16,105 | 12,524 | 7,739 | 6,218 | 89,724 |
| Prevalent coronary heart disease | 31,894 | 12,055 | 9,811 | 7,518 | 1,583 | 62,861 |
| Mean systolic and diastolic blood pressures | 38,157 | 14,899 | 13,067 | 7,518 | 942 | 74,583 |
| Diabetes and renal function | 37,168 | 15,270 | 12,750 | 7,518 | 3,313 | 76,019 |
| General biochemistry tests | 52,685 | 17,316 | 14,199 | 7,736 | 2,707 | 94,643 |
| Hematology | 52,371 | 16,766 | 13,879 | 7,736 | 2,024 | 92,776 |
| Infectious diseases and immunity | 36,392 | 9,614 | 13,892 | 446 | 2,039 | 62,383 |
| Inflammation | 28,250 | 12,367 | 12,171 | 7,399 | 1,187 | 61,374 |
| Neurology | 4,861 | 1,059 | 268 | 0 | 0 | 6,188 |
| Nutrition and dietary supplements | 53,884 | 19,526 | 11,531 | 7,739 | 11,229 | 103,909 |
| Ocular | 44,765 | 11,511 | 4,177 | 446 | 2,687 | 63,586 |
| Physical activity | 35,044 | 12,483 | 18,131 | 7,739 | 11,068 | 84,465 |
| Psychiatric | 32,407 | 8,533 | 4,299 | 4,142 | 2,039 | 51,420 |
| Reproductive health | 51,401 | 17,477 | 15,029 | 7,739 | 6,434 | 98,080 |
| Respiratory | 48,560 | 15,482 | 12,015 | 4,142 | 2,039 | 82,238 |
| Skin, bone, muscle, and joint | 31,809 | 7,806 | 4,893 | 4,142 | 2,984 | 51,634 |
| Speech and hearing | 29,186 | 6,694 | 10,662 | 446 | 2,039 | 49,027 |
The large sample sizes allow for a variety of powerful analyses that include both replication and discovery opportunities. For example, a PAGE-wide analysis of low density lipoprotein cholesterol levels (measured in mg/dL) excluding nonfasting participants under 18 years of age includes 21,986 European Americans, 9,328 African Americans, 6,144 American Indians, and 2,532 Mexican Americans/Hispanics. Using Quanto (20) and assuming an additive genetic model with α = 0.005, we have at least 80% power to replicate associations with effect sizes (β) as low as 1.26 (European Americans), 2.08 (African Americans), 2.07 (American Indians), and 3.46 (Mexican Americans/Hispanics), depending on allele frequency. These genetic effect sizes are similar to the effect estimates reported in most GWAS for low density lipoprotein cholesterol (see Web Table 1, available on the Journal’s Web site (http://aje.oxfordjournals.org/)). Power will vary from analysis to analysis given the range of phenotypes, the list of exclusions, the number of strata, the minor allele frequency of the SNP(s), the number of PAGE sites that genotype each SNP, and the effect sizes expected for each association.
Phenotype harmonization and subject selection
Collectively, PAGE investigators identify the phenotypes to focus on for each genotyping and analysis cycle. The focus phenotypes are selected to strike a balance between those that are common to multiple studies and those that are specific to individual studies. Higher priority is considered for phenotypes with a major health impact, diseases that have differing prevalences across ethnic groups, and phenotypes with robustly replicated SNP associations. Studywide, in the first 2 years, PAGE investigators are analyzing phenotypes associated with cardiovascular disease, lipids, stroke, type 2 diabetes, a variety of cancers, and smoking; anthropometric and electrocardiographic traits; ages at menopause and menarche; and a variety of biomarkers.
Prior to analyses, phenotype harmonization must occur, whereby phenotypic variables are identified that have had data collected using similar methods across all or most participating studies. For covariate and exposure phenotypes (such as smoking, alcohol consumption, diet, medical history, family history, medication use, and socioeconomic status), an investigator from each study creates an inventory of variables related to that phenotype, including assay parameters and/or specific questionnaires. Each study investigator also participates in a working group to identify commonly ascertained variables and to recommend specific variables for use in analyses. Trait distributions are compared across studies to define new harmonized variables as appropriate.
Subject selection varies by site, since each study must balance the number of variants and participants genotyped within available budgets. EAGLE and CALiCo are genotyping all participants in the data set who have information on the studied phenotype and DNA available. In the larger cohorts, such as the Women's Health Initiative and the Multiethnic Cohort Study, the investigators select subsets of participants and maximize sample sizes for available phenotypes and ethnic groups. Subsets are identified that will provide a substantial sample size (cases and controls) for each of the PAGE focus phenotypes while preferentially including participants who also have relevant biomarker measurements.
SNP selection and genotyping methods
Once focus phenotypes are determined for a genotyping and analysis cycle, the published literature is searched to identify associated SNPs to use for PAGE analysis. The NHGRI GWAS catalog (1) is especially useful for these searches. Since the studies vary in genotyping capacity and in available phenotypes, researchers in each study then choose a custom SNP set from the jointly determined SNPs of interest.
Genotyping in PAGE is performed separately by the investigators in each study, which requires detailed guidelines for consistent quality control. These guidelines include thresholds for SNP and sample call rates (>90%), concordance of blinded replicates (>98%), and no clear evidence of Hardy-Weinberg disequilibrium (P > 0.001), which is evaluated within and across studies and in each racial/ethnic population. For each cycle, each laboratory genotypes 360 HapMap samples (from populations most relevant to PAGE) to serve as cross-laboratory and cross-platform quality control samples (http://hapmap.ncbi.nlm.nih.gov/). The Coordinating Center performs quality control analyses on the HapMap genotypes, including concordancy with published HapMap data and across PAGE studies, detection of Mendelian errors in trios, and evaluation of Hardy-Weinberg equilibrium. Quality control results are shared across studies and are discussed within the SNP selection/quality control working group in order to identify problematic assays.
Analysis methods
The Statistical Analysis Committee develops recommendations for statistical analyses and ensures that these are comparable among PAGE project groups. A writing group develops the statistical analysis plan for a defined cluster of phenotypes, which is then applied by investigators in each study to their own data. Project groups formed around specific phenotypes develop meta-analysis plans that are then used for a PAGE-wide analysis. PAGE does not pool individual-level data (with the exception of the MetaboChip Pilot Study; see below). Meta-analyses are logistically simpler and capitalize on analysts’ familiarity with their local data set, allowing them to analyze their own study's data using harmonized variables while allowing the participation of studies with restrictions on sharing of individual-level data. Lin and Zeng (21) have shown that there is no efficiency loss in performing meta-analysis instead of pooled analysis. PAGE investigators attempt to address common criticisms of meta-analysis, such as publication bias, varying quality control, and heterogeneity of statistical analyses, by sharing unpublished data and standardizing quality control of genotyping and statistical analyses.
The PAGE studies have each collected data, much of it longitudinal, on thousands of phenotypes that PAGE has classified into tiers. One tier contains targeted high-interest phenotypes that are carefully harmonized and subjected to detailed cross-study analyses that may include multiple models and covariates. Results of these analyses will appear in primary research papers. The larger tier of phenotypes, consisting of virtually all of the variables collected by each study, regardless of a priori scientific interest, will not be harmonized and will be subject only to routine analysis performed separately by investigators in each study. These analyses will adopt additive genetic models and incorporate minimal adjustment for covariates. Quantitative variables will be analyzed both with and without logarithmic transformation, using the amount of skewness to determine the preferred result.
Ancestry
PAGE comprises highly ethnically diverse US populations, including several that are historically of mixed ancestry (Table 2). The prevalences of many traits of interest in the PAGE collaboration, such as hyperlipidemia, type 2 diabetes, and most cancers, display important population differences, as do the allele frequencies for many SNPs. Careful attention must be paid to ancestry, and the associated concern for population stratification, to avoid false-positive association results. The individual studies in PAGE all collect some information about ancestry but in varying degrees of detail. In 3 of the PAGE studies, researchers are genotyping the Kosoy et al. (22) panels of 128 ancestry informative markers, while in the fourth they are deriving ancestry from existing GWAS to adjust association analyses for varied ancestry. Genetic data will be used separately by each study within strata of self-reported race/ethnicity, using a principal component approach, to adjust for admixture between continental ancestral groups (African, European, and Asian) (23). The leading principal components will be included as continuous adjustment variables in linear or logistic regression analysis relating SNPs of interest to the phenotypes.
MetaboChip Pilot Study
In 2009, PAGE investigators received supplemental funding from NHGRI under the American Recovery and Reinvestment Act to undertake a pilot study in African Americans using the MetaboChip (http://www.sph.umich.edu/csg/kang/MetaboChip/), a high-density custom Illumina iSelect array of 196,725 SNPs (Illumina, Inc., San Diego, California) that focuses on variants associated with atherosclerotic-cardiovascular and metabolic traits (M. Boehnke, University of Michigan, personal communication, 2010). The chip has the added value of including thousands of 1,000 Genome SNPs (http://www.1000genomes.org/) that will facilitate transethnic fine mapping around key loci, complementing PAGE's goal of assessing whether findings for GWAS-identified variants are generalizable to populations of non-European descent. The MetaboChip Pilot Study was initiated when we realized that there was a lack of generalizability of SNPs chosen in European and European-American GWAS to the ethnically diverse PAGE sample. For the PAGE MetaboChip Pilot Study, approximately 6,000 African-American participants (3,500 from CALiCo/ARIC, 2,200 from the Women's Health Initiative, and 580 from the Multiethnic Cohort Study) evaluated for metabolic and cardiovascular phenotypes will be genotyped. The fine mapping of risk loci for multiple adiposity-, lipid-, and diabetes-related traits will allow us to evaluate whether fine mapping will aid in generalization and replication of genetic effects across populations and may allow us to more fully and accurately evaluate population differences (via fine mapping) for a limited number of loci.
Data-sharing
In accordance with National Institutes of Health (NIH) policy on large-scale genomic data, PAGE data will be shared with the scientific community, primarily via dbGaP. Recognizing that not all studies are suitable for widespread sharing of the genetic data of individual participants, NHGRI established PAGE with the expectation of distributing summary association data, which should present few, if any, obstacles (17, 18). These summary data include study protocols and questionnaires, phenotype variable dictionaries, summary phenotype and genotype data, and the results of PAGE analyses. These data are submitted to the Coordinating Center, examined for consistency and for confirmation that values are within expected bounds, reformatted according to dbGaP specifications, and submitted to dbGaP. Although initial plans for PAGE specified dissemination of aggregate data via an open-access Web site, subsequent changes to NIH policy (24) (see the NIH's GWAS Web site (http://grants.nih.gov/grants/gwas/)) require nearly all PAGE data to be accessible only via controlled access to approved users, such as that provided through dbGaP. Individual-level data for the ARIC, Cardiovascular Health Study, Multiethnic Cohort Study, and Women's Health Initiative components of PAGE will also be available through dbGaP. PAGE investigators retain exclusive rights to submit publications developed with the data and samples for a period of 1 year after data are released through dbGaP.
Novel data display tools
The comprehensive range of genotype-phenotype associations that PAGE investigators will analyze does not lend itself to graphical browsing using most existing tools or resources. Therefore, a novel Web-based display browser is being developed to facilitate exploration of the analysis results and will be available to approved users. Results in PAGE can be classified along several axes: SNP, phenotype, study, gender, and self-described race and ethnicity. The browser includes a flexible query interface with many display options and filters. The current browser provides 3 types of displays: a heat map for the simultaneous presentation of large numbers of results (P values or effect sizes) (Web Figure 1); box plots and bar plots to display summary statistics for quantitative and categorical variables, respectively (Web Figure 2); and a forest plot to represent a moderate number of effect sizes, along with their confidence intervals (Web Figure 3). These graphical views will be available for all tiers of PAGE analyses and will help identify previously unknown associations between PAGE phenotypes and PAGE variants.
DISCUSSION
The PAGE Study is designed to advance our understanding of how well-replicated risk variants affect phenotypes. PAGE has great potential for innovative contributions that bridge genomics and population-based research. The breadth of phenotypic data across the PAGE network and the inclusion of large samples from several ethnic/racial groups allow us to move beyond discovery toward characterizing each variant, by defining its epidemiologic architecture.
Defining the epidemiologic architecture begins with characterizing the genetic association across populations. Prior candidate gene research supports the hypothesis that genetic effects of mostly coding variation are usually consistent across human populations (25). To date, however, GWAS are performed mainly in populations of European descent (2); rely on linkage disequilibrium to a greater extent than most candidate gene studies; and often identify associations in noncoding regions and nearly always with weak effect (1). Thus, the issue of “transferability” of GWAS findings to diverse populations remains understudied and is a major topic of post-GWAS research (2, 3, 26, 27).
Another area of GWAS follow-up research is estimating the true population impact mature variants have on traits (28). The design of discovery efforts generally precludes the accurate estimation of genetic effect size or magnitude of risk. This inaccuracy is due to the “winner's curse” phenomenon, whereby initial assessments of risk may be overestimated, rendering subsequent studies of similar sizes underpowered (29). PAGE counters this by using very large sample sizes to estimate the effects of GWAS-determined associations in European Americans (Table 2) and to extend these findings to diverse populations. Ascertainment bias, potentially skewing effect estimates determined in case-control studies, is mitigated in PAGE's population-based cohorts because of the use of population-based sampling and adjudicated phenotypes. In 3 of the 4 PAGE projects (CALiCo, the Multiethnic Cohort Study, and the Women's Health Initiative), investigators employ rigorous definitions of incident disease and conduct extensive longitudinal follow-up, facilitating studies based on repeated measures and, perhaps, prognosis. The fourth project (EAGLE/NHANES) is a nationally representative cross-sectional survey of the US population and includes meticulous measures of risk factors and traits related to common chronic conditions. The PAGE team includes expert epidemiologic investigators who are intimately familiar with these data sets, including participant recruitment, exposure assessment, follow-up, and study design. Their involvement in every aspect of the PAGE analyses ensures that the complexities of these data sets and any potential biases will be weighed in data-analysis plans and interpretation of results.
Investigators in the PAGE cohorts have a wealth of data, including phenotypic subtypes and intermediate phenotypes, covariates, biomarkers, and lifestyle variables. These data are appropriate for detailed exploration of their impact on disease pathways, potentially elucidating mechanisms and functions. These data also present unprecedented opportunities to assess the roles of well-characterized modifiers in complex traits, particularly gene-environment interaction models. PAGE offers a unique opportunity to explore these lines of research in several large racially and ethnically diverse populations, an opportunity that is unavailable in existing single-site projects. We expect that the future of genomics will include more collaborative efforts of this magnitude across different large studies to allow for making discoveries, interpreting findings, and drawing generalizable conclusions relevant to public health.
Finally, PAGE Study findings will also help inform our knowledge of complex trait biology. PAGE can examine pleiotropic effects by moving beyond disease-SNP associations and exploring the role of these SNPs in intermediate disease phenotypes (30). In doing so, we may be able to gain insight not only into disease pathogenesis but also into gene function. For example, it would help to know whether a variant associated with myocardial infarction is also associated with C-reactive protein or another inflammatory marker, or whether a breast cancer susceptibility locus is also associated with circulating estrogen concentrations. Such exploration of disease loci may help us understand their role in disease development.
Research beyond the scope of PAGE, such as intensive fine mapping of index SNPs, discovery of variants responsible for novel GWAS peaks, and (finally) functional studies aimed at elucidating mechanisms, will be important contributions to the field. Nonetheless, PAGE will help fill in the gaps between focused candidate gene association studies and GWAS. The PAGE Study advances efforts to determine the population-based impact of SNPs identified in GWAS discovery efforts. With its enormous breadth of phenotypes and sample sizes, PAGE promises to address unanswered questions regarding the epidemiologic architecture of GWAS-identified variants, gene-gene and gene-environment interactions, pleiotropic variants, and intermediate phenotypes to help elucidate their function. Not only will PAGE investigators publish the study's results, they will also disseminate aggregate and individual-level data via dbGaP and share aggregate results via a novel display tool. PAGE is designed to complement and extend initial GWAS findings, most notably in the areas of diversity, phenotypic breadth, and epidemiologic context. Coupled with its data and sharing of results, PAGE is poised to be a key contributor to population-level annotation of the human genome.
Supplementary Material
Acknowledgments
Author affiliations: Department of Genetics, School of Arts and Sciences, Rutgers University, Piscataway, New Jersey (Tara C. Matise, Steven Buyske); Information Sciences Institute, University of Southern California, Marina del Rey, California (Jose Luis Ambite); Department of Statistics, School of Arts and Sciences, Rutgers University, Piscataway, New Jersey (Steven Buyske); Department of Genetics, Southwest Foundation for Biomedical Research, San Antonio, Texas (Shelley A. Cole); Department of Molecular Physiology and Biophysics and Center for Human Genetics Research, Vanderbilt University, Nashville, Tennessee (Dana C. Crawford, Marylyn D. Ritchie, Jonathan L. Haines); Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California (Christopher A. Haiman); Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Gerardo Heiss, Kari E. North); Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, Washington (Christopher S. Carlson, Charles Kooperberg, Ulrike Peters); Epidemiology Program, Cancer Research Center of Hawaii, University of Hawaii, Honolulu, Hawaii (Loic Le Marchand); and Office of Population Genomics, National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland (Teri A. Manolio, Lucia A. Hindorff).
The Population Architecture using Genomics and Epidemiology (PAGE) Study is a collaboration between the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) Study, the Multiethnic Cohort Study, the Women's Health Initiative (WHI), and the Genetic Epidemiology of Causal Variants Across the Life Course (CALiCo) consortium, which comprises the Atherosclerosis Risk in Communities (ARIC) Study, the Coronary Artery Risk Development in Young Adults (CARDIA) Study, the Cardiovascular Health Study, the Hispanic Community Health Study/Study of Latinos (HCHS/SOL), and the Strong Heart Study. A complete list of members of the PAGE consortium can be found at http://www.pagestudy.org.
PAGE is funded by the National Human Genome Research Institute (NHGRI) and is supported by National Institutes of Health grants U01HG004803 (CALiCo), U01HG004798 (EAGLE), U01HG004802 (Multiethnic Cohort Study), U01HG004790 (WHI), and U01HG004801 (Coordinating Center).
The EAGLE Study is funded through the NHGRI PAGE program (grant U01HG004798). The study participants were derived from the National Health and Nutrition Examination Surveys, which are supported by the Centers for Disease Control and Prevention. (The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention).
Characterization of epidemiological architecture in the Multiethnic Cohort Study is funded through the NHGRI PAGE program (grant U01HG004802). The Multiethnic Cohort Study is funded by the National Cancer Institute (grants R37CA54281, R01 CA63, P01CA33619, U01CA136792, and U01CA98758).
Funding for the study of the epidemiology of putative genetic variants in the WHI is provided through the NHGRI PAGE program (grant U01HG004790). The WHI is funded by the National Heart, Lung, and Blood Institute (NHLBI) and the US Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. (The authors thank the WHI investigators and staff for their dedication). A full listing of WHI investigators can be found at http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf.
Funding for the CALiCo consortium was provided through the NHGRI PAGE program (grant U01HG004803). The ARIC Study is carried out as a collaborative study supported by NHLBI contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, and N01-HC-55022. The CARDIA Study is supported by the following NHLBI contracts: N01-HC-95095, N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050, N01-HC-45134, N01-HC-05187, and N01-HC-45205. The Cardiovascular Health Study is supported by contracts N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01-HC-15103, N01 HC-55222, N01-HC-75150, and N01-HC-45133 and grants U01HL080295 and R01 HL087652 from the NHLBI, with an additional contribution from the National Institute of Neurological Disorders and Stroke. Genome-wide association study DNA handling and genotyping in the Cardiovascular Health Study was supported in part by National Center for Research Resources grant M01-RR00425 to the Cedars-Sinai General Clinical Research Center genotyping core and by National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The HCHS/SOL is supported by NHLBI contracts N01-HC65233, N01-HC65234, N01-HC65235, N01-HC65236, and N01-HC65237. The following institutes/centers/offices contribute to the HCHS/SOL through a transfer of funds to the NHLBI: the National Center on Minority Health and Health Disparities, the National Institute on Deafness and Other Communication Disorders, the National Institute of Dental and Craniofacial Research, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Neurological Disorders and Stroke, and the Office of Dietary Supplements. The Strong Heart Study is supported by NHLBI grants U01 HL65520, U01 HL41642, U01 HL41652, U01 HL41654, and U01 HL65521. (The opinions expressed in this paper are those of the author(s) and do not necessarily reflect the views of the Indian Health Service).
Assistance with phenotype harmonization; single nucleotide polymorphism selection and annotation; data cleaning; data management, integration, and dissemination; and general study coordination is provided by the PAGE Coordinating Center (grant U01HG004801). The National Institute of Mental Health also provides financial support for the Coordinating Center.
The PAGE investigators thank the staff of all PAGE studies for their important contributions. The authors thank Julia Higashio for editing assistance.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Conflict of interest: none declared.
Glossary
Abbreviations
- ARIC
Atherosclerosis Risk in Communities
- CALiCo
Causal Variants Across the Life Course
- CARDIA
Coronary Artery Risk Development in Young Adults
- dbGaP
Database of Genotypes and Phenotypes
- EAGLE
Epidemiologic Architecture for Genes Linked to Environment
- GWAS
genome-wide association study(ies)
- NHANES
National Health and Nutrition Examination Surveys
- NHGRI
National Human Genome Research Institute
- NIH
National Institutes of Health
- PAGE
Population Architecture using Genetics and Epidemiology
- SNP
single nucleotide polymorphism
References
- 1.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Need AC, Goldstein DB. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25(11):489–494. doi: 10.1016/j.tig.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg NA, Huang L, Jewett EM, et al. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11(5):356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322(5903):881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frazer KA, Murray SS, Schork NJ, et al. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 6.McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17(R2):R156–R165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Center for Health Statistics, Centers for Disease Control and Prevention. Plan and Operation of the Third National Health and Nutrition Examination Survey, 1988–94. (Vital and health statistics, series 1, no. 32). Hyattsville, MD: National Center for Health Statistics; 1994.
- 8.Kolonel LN, Henderson BE, Hankin JH, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151(4):346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Women's Health Initiative Study Group. Design of the Women's Health Initiative clinical trial and observational study. Control Clin Trials. 1998;19(1):61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 10.The ARIC investigators. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol. 1989;129(4):687–702. [PubMed] [Google Scholar]
- 11.Hughes GH, Cutter G, Donahue R, et al. Recruitment in the Coronary Artery Disease Risk Development in Young Adults (Cardia) Study. Control Clin Trials. 1987;8(4 suppl):S68–S73. doi: 10.1016/0197-2456(87)90008-0. [DOI] [PubMed] [Google Scholar]
- 12.Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1(3):263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 13.Sorlie PD, Avilés-Santa LM, Wassertheil-Smoller S, et al. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol. 2010;20(8):629–41. doi: 10.1016/j.annepidem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee ET, Welty TK, Fabsitz R, et al. The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132(6):1141–1155. doi: 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- 15.North KE, Howard BV, Welty TK, et al. Genetic and environmental contributions to cardiovascular disease risk in American Indians: the Strong Heart Family Study. Am J Epidemiol. 2003;157(4):303–314. doi: 10.1093/aje/kwf208. [DOI] [PubMed] [Google Scholar]
- 16.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Human Genome Research Institute. Epidemiologic Investigation of Putative Causal Genetic Variants—Study Investigators (U01) (Request for Applications (RFA) no. RFA-HG-07-014). Bethesda, MD: National Human Genome Research Institute; 2007. ( http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-07-014.html). (Accessed September 28, 2007)
- 18.National Human Genome Research Institute. Epidemiologic Investigation of Putative Causal Genetic Variants—Coordinating Center (U01) (Request for Applications (RFA) no. RFA-HG-07-015). Bethesda, MD: National Human Genome Research Institute; 2007. ( http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-07-015.html). (Accessed September 28, 2007)
- 19.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155(5):478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 21.Lin DY, Zeng D. Meta-analysis of genome-wide association studies: no efficiency gain in using individual participant data. Genet Epidemiol. 2010;34(1):60–66. doi: 10.1002/gepi.20435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosoy R, Nassir R, Tian C, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30(1):69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 24.Homer N, Szelinger S, Redman M, et al. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4(8):e1000167. doi: 10.1371/journal.pgen.1000167. (doi: 10.1371/journal.pgen.1000167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36(12):1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 26.Keebler ME, Sanders CL, Surti A, et al. Association of blood lipids with common DNA sequence variants at 19 genetic loci in the multiethnic United States National Health and Nutrition Examination Survey III. Circ Cardiovasc Genet. 2009;2(3):238–243. doi: 10.1161/CIRCGENETICS.108.829473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shriner D, Adeyemo A, Gerry NP, et al. Transferability and fine-mapping of genome-wide associated loci for adult height across human populations. PLoS One. 2009;4(12):e8398. doi: 10.1371/journal.pone.0008398. (doi: 10.1371/journal.pone.0008398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraft P, Wacholder S, Cornelis MC, et al. Beyond odds ratios—communicating disease risk based on genetic profiles. Nat Rev Genet. 2009;10(4):264–269. doi: 10.1038/nrg2516. [DOI] [PubMed] [Google Scholar]
- 29.Zollner S, Pritchard JK. Overcoming the winner's curse: estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80(4):605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Baldassano R, Zhang H, et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum Mol Genet. 2010;19(10):2059–2067. doi: 10.1093/hmg/ddq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.