Abstract
Cell patterning methods enable researchers to control specific homotypic and heterotypic contact-mediated cell-cell and cell-ECM interactions and to impose defined cell and tissue geometries. To micropattern individual cells to specific points on a substrate with high spatial resolution, we have developed a cell deposition microscope based on the laser guidance technique. We discuss the theory of optical forces for generating laser guidance and the optimization of the optical configuration (NA ≈ 0.1) to manipulate cells with high speed in three dimensions. Our cell deposition microscope is capable of patterning different cell types onto and within standard cell research devices and providing on-stage incubation for long-term cell culturing. Using this cell deposition microscope, rat mesenchymal stem cells from bone marrow were micropatterned with cardiomyocytes into a substrate microfabricated with polydimethylsiloxane on a 22mm × 22 mm coverglass to form a single-cell coculturing microenvironment, and their electrophysiological property changes were investigated during the coculturing days.
1. Introduction
Mimicking the in vivo microenvironment in a cell culture is important for restoration of the functions of isolated cells and in vivo relevant cell-cell interactions. This microenvironment includes the spatial arrangement of multiple cell types. Correct cell arrangements will allow in vivo-like cellular communication among multiple cell types, which is essential for retaining native cell function. Micropatterning techniques provide feasible approaches to achieve defined heterotypic cell arrangements. Various surface patterning techniques have been employed to control spatial cell arrangements through chemically or physically modifying the substrate’s surface based on cell adhesion properties[1, 2]. These techniques are applicable for creating layers of cells and engineer tissue in vitro but cannot place individual cells with high accuracy for systematic studies of in vivo-relevant cell-cell interactions. In addition, the heterotypic cell patterning achieved by surface patterning is typically limited to placing two types of cells onto the same coculture substrate. Furthermore, the dependence of these techniques on promoting or inhibiting cell adhesion prohibits controlling or monitoring the temporal features of cell-cell interactions prior to complete cell attachment. Besides, cell shape changes and cell migrations after patterning are constrained by the chemical and physical treatments of the surface, which may prohibit certain cell-cell interaction effects, such as cell expelling after contact. To achieve accurate cell arrangement at the single-cell level, we have developed an optical force-based cell deposition microscope with high spatial resolution and precision for selecting individual cells and patterning them to specific points on a substrate. With this system, individual cells can be precisely positioned to create reproducible patterns with minimal variation for systematic and statistical study of in vivo relevant cell-cell interactions.
Optical force exerted on a particle is the result of momentum change between photons and the particle during the process of light scattering. Optical force has been introduced into biological applications for precise manipulation of living cells[3, 4]. Researchers have reported the use of laser tweezers (formed by a strongly focused laser beam) to study a variety of cellular and subcellular activities, such as vesicle transport[5], motion of single kinesin molecules along a microtubule track[6], and measurement of tether formation associated with cell membranes[7]. A weakly focused laser beam has been combined with microfluidics in optical chromatography[8], optical lattice[9], and optical stretcher[10] for noninvasive cell-type analysis and sorting. The laser guidance technique associated with weakly focused laser beams has been used in optics-based tissue engineering. Laser direct writing technique[11] based on optical guidance has made it possible to study developmental processes, cell signaling, pathogenesis, and tissue repair mechanisms using in vitro cell culture scenarios that mimic or modify in vivo conditions. In our research, we applied the laser guidance technique to design a cell deposition microscope for studying heterotypic cell-cell interactions at the single-cell level. Here we report 1) optimization of the microscope’s optical configuration, according to optical force simulation, to achieve high-speed cell manipulations in three dimensions; 2) design of the cell deposition microscope with microinjection and on-stage incubation mechanisms for heterotypic cell micropatterning and long-term coculture and 3) application of this microscope to micropattern individual rat mesenchymal stem cells (rMSCs) into a microstructure with cardiomyocytes. We demonstrate the formation of a single-cell coculture microenvironment for studying the electrical coupling of stem cells with cardiomyocytes.
2. Optical force theory
To describe the theory behind the optical force phenomenon, Ashkin[12] first introduced the geometric optics method, in which a beam of parallel rays focused to one point is assumed. When an individual ray impinges on the particle, the momentum exchange is induced by reflection and refraction of the ray. The reflection-refraction can be analyzed using Snell’s law to estimate the optical force. However, the diffraction effects, which also contribute to the momentum exchanges, especially when the particle size is of the same order as the laser wavelength, were neglected in Ashkin’s theory. Gauesbet[13] has introduced a Generated Lorenz-Mie Theory (GLMT) to calculate optical forces with a full electromagnetic description, which incorporates all the effects of light-particle interactions, including reflections, refractions, and diffractions. In this report, the GLMT method is used to calculate the optical force exerted by a beam on a spherical particle (e.g., a biological cell) by estimating the momentum removed from the incident beam by particle absorption and scattering.
According to classical optical theory, optical force can be calculated in Cartesian coordinates using the following equation:
| (1) |
where i = x, y, z, ω0 is the laser beam’s waist; P is the laser power; c is the light’s speed, and nmedium is the refractive index of the medium; Cpr, i is the reduced radiation pressure cross-section, which equals to the light’s energy loss per unit time due to the interaction with particles. The Poynting vector Sr represents the energy flux (W/m2) of a laser beam. Integrating the Sr on the surface of the particle ∫SrdS can be used to calculate Cpr, i, which provides a measurement of the amount of energy lost per unit time during light-particle interaction. In the GLMT method, this integration is evaluated with 1) the classical Mie coefficients that are dependent on the particle size and refractive index and 2) beam-shape coefficients that are dependent on the wavelength and beam waist. The beam-shape coefficients can be obtained by expressing the incident wave as a series of spherical wave expansions and can be numerically evaluated with reasonable speed by use of improved localized approximation, which has been justified rigorously in the case of a Gaussian beam. As a result, the optical force Fi can be calculated in three dimensions (i = x, y, z) from the integration of each component of the Poynting vector Sr.
3. Materials and methods
3.1 Optimized optical configuration
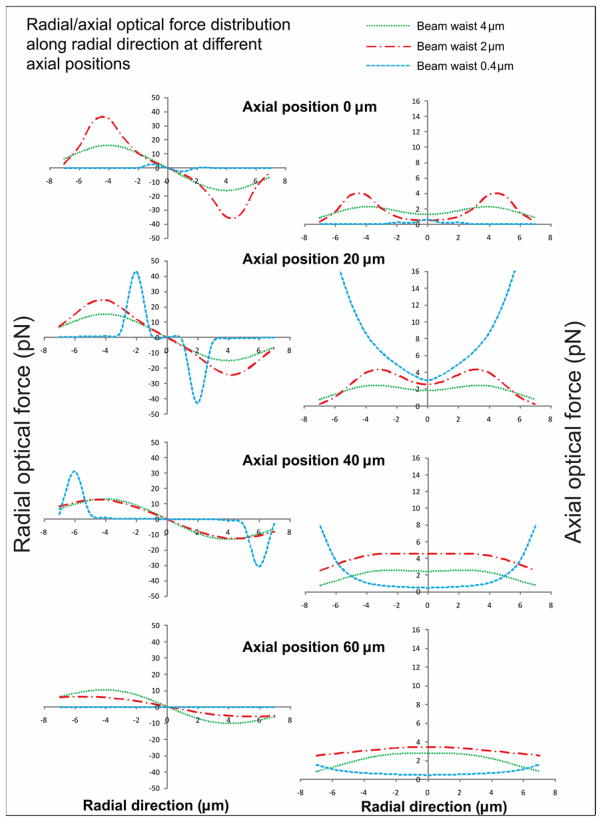
The entire cell deposition microscope was built around a stationary downward-propagating laser beam, which was expanded, collimated, and focused into the cell deposition chamber for cell guidance. At the beginning of the cell deposition process, a cell was trapped at the center of the laser beam because of the radial component of the optical force (radial force). Due to the axial component of the optical force (axial force), the cell was pushed downward along the laser beam propagation direction (Z direction) to the substrate. The trapped cell should be moved back to the object plan of the imaging system during guidance for continuous imaging tracing. Therefore, the chamber was simultaneously lifted up with a motion speed equal to the cell-guidance speed determined by the axial force. To guide cells with a high transverse speed, the radial optical force should have a sufficient peak-value to trap the cell in the beam center. Simultaneously, to reduce individual cell patterning time, the axial optical force should be strong enough to provide a relatively high vertical guidance speed. In addition to these two force peak-value requirements, the forces should also attenuate gradually in space to provide an effective range that would allow cells to follow the movement of the trap during guidance. However, the maximal peak value and the broadest effective range of the force distribution would not occur in the same spatial region for a given optical beam configuration. Therefore, the cell should be kept in a guidance region that compromises among maximum axial force, maximum radial force and broadest effective force range so that it would be guided onto the substrate with a maximum speed. To reveal an optical configuration that would generate an optimal guidance region, the optical force distribution associated with different beam waists (0.4, 2, and 4 μm) was numerically calculated using the GLMT method. The other parameters used in our calculation were wavelength = 830 nm, power = 200 mW, refractive index of the medium = 1.33, refractive index of the cell = 1.36, and cell diameter = 10 μm.
The optical forces (radial and axial) were simulated along the radial direction at different axial positions, as shown in figure 1. The beam with a 4 μm waist would not provide a radial force as high as that provided by a 2 μm beam for trapping a cell in the beam center. On the other hand, although the radial force generated from the beam with a 0.4 μm waist was stronger than that generated by the 2 μm beam in a relatively broad range, the axial force in the same range generated by this beam was not sufficient for effective guidance. Therefore, the 2 μm beam could provide enough peak force and effective force range, which would be ideal for laser cell deposition. In addition, according to the simulation results, the best guidance region centered at the focal plane of laser beam. Therefore, the image plane should be adjusted to the laser beam’s focal plane.
Figure 1.
Radial and axial optical forces were simulated along the radial direction at different axial positions to locate the guidance region for the cell deposition microscope.
To validate our simulation experimentally, we constructed the laser beams with different NA by changing the combination of three lenses: The first two lenses were used for beam expansion and alignment, and the third one was used as the guidance lens to focus the laser beam into the chamber for cell guidance. According to the equation, , the lenses were chosen to match the beam waists used in the simulation. To evaluate different beam configurations for our cell deposition microscope, we moved a cell in transversal directions at a speed of 50 μm/s and recorded the effective regions where the control of the cell was not lost. Shown in table 1, the laser beam with NA = 0.1 (beam waist close to 2 μm) could provide the optimized manipulation on biological cells for our cell deposition microscope, which agreed with the simulation results.
Table 1.
The effective regions were evaluated for different laser beam’s configurations
| NA | Beam waist | Lens combination | Effective regions |
|---|---|---|---|
| ≈ 0.7 | ≈ 0.4 | 8mm-200mm-30mm | Loss of control |
| ≈ 0.1 | ≈ 2 | 30mm-200mm-30mm | ≈ 80 μm |
| ≈ 0.06 | ≈ 4 | 30mm-60mm-30mm | Loss of control |
3.2 System design
The system design for cell deposition is shown in figure 2. The laser source was a single-transverse-mode diode laser (200 mW, 830 nm, CW, Intense Inc.) attached to a diode laser mount with an aspheric collimating lens. Anamorphic prism pairs were used immediately outside the laser mount to transform the elliptical beam into a circular one. Then the beam was expanded and focused into the cell-deposition chamber by a group of NIR achromatic lenses (f = 15 mm, 100 mm and 30 mm) to provide an optical configuration optimized for cell deposition (NA ≈ 0.1). The illumination source was a collimated green LED (530 nm, 200 mW), which was focused onto the substrate inside the chamber. Before the image carried by the illumination beam was captured by the CCD camera with a long-working-distance objective (20x, Mitutoyo Plan Apo Infinity-corrected), it was passed through several IR filters to remove artifacts from the guidance beam. The CCD camera was mounted on a 3D translational stage to align the CCD’s center to the laser beam’s focus point so that the guidance region of the beam coincided with the object plane of the imaging system. A commercial on-stage incubator (Okolab Electric CO2 microscope stage incubator, Warner Instrument Inc.) was modified to serve as the cell-deposition chamber, which can regulate temperature, CO2, and humidity for long-term cell culturing during and after the cell deposition procedure. The modified on-stage incubator was also compatible with a 35 mm petri dish and a standard multi-electrode array with a ring. The laser/imaging window was made from a 10 mm circle coverslip to seal the chamber and reduce the convection force disturbing the cell deposition procedure.
Figure 2.
Schematic of the cell deposition microscope. The red line indicates the laser beam focused into the cell deposition chamber.
The control software was written in LabVIEW8.5, which allowed for manual assignment of specific processes to individual processor cores. The system was initialized before cell deposition procedure by issuing the parameters to laser, imaging, stage, and microinjection and enabling them “ready for operation”. The real-time image and video of cell deposition could be captured, recorded, and stored in a preselected folder. The real-time executive (RTX) Aerotech stage communicated with the computer through an IEEE1394 port. The motion of the motorized stage was regulated by restricting the maximum horizontal (100 μm/s) and vertical (25 μm/s) guidance speeds to ensure that the cell would be within the guidance region. Therefore, the stage could avoid moving the chamber so quickly that the cell would be left behind. The opening/closing of the laser shutter, the laser intensity adjustment, and the microinjection command were controlled via serial port/RS232 access through VISA in LabVIEW. Cell suspension could be injected into the media-filled chamber through a hollow fiber coupled to a 50μL microsyringe. The cell feeding ratio was optimized for single-cell flow by adjusting the volume (50 nL) and speed (25 nL/s) of the microinjection. Cell manipulation and navigation were primarily controlled by an Xbox360 controller because of Windows compatibility and long-term availability. The analog thumbsticks were used to maneuver in X, Y, and Z directions. The horizontal plane projection of the vertical laser beam was marked on the computer screen as a cross, and the deposition destination on the substrate was marked as a circle. The injection of cells into the chamber triggered the motorized stage to bring those cells into the field-of-view. Then the user maneuvered a cell into the center of the screen (the cross) and opened the laser shutter. Once the cell was captured in the center of the field-of-view, an on-screen direction indicator pointed towards the previously marked circle. Following the indicator, the user navigated to the circle using the thumbsticks, carrying the cell along in the laser guidance region with transversal manipulation speed at 100 μm/s. This process was repeated until the intended cell pattern was formed.
3.3 Cell preparation
3.3.1. Neonatal rat cardiomyocytes
Cardiomyocytes were isolated and collected from three-day neonatal rats using a two-day protocol. Ten neonatal rats were dissected, and the hearts were collected and minced in Moscona’s Saline. The heart tissue was transferred into 50 mL Dulbecco’s Phosphate Buffered Saline (DPBS, HyClone, Thermo Scientific Inc.) with 4 mg Trypsin (Worthington Inc.) and 50 mg Neutral Protease (Worthington Inc.) and stored in the −4 °C refrigerator overnight. The next day, the heart tissue was transferred into 50 mL Kreb’s Ringers Bicarbonate Buffer (KRB) with 10 mg Collagenase type I (Worthington Inc.) and 30 mg Collagenase type II (Worthington Inc.), and then shaken in a water bath at 50 RPM for 1 hour. The cardiomyocytes were isolated and cultured in the DMEM high glucose medium (HyClone, Thermo Scientific Inc.) supplement with 20% fetal bovine serum (FetalClone III, HyClone, Thermo Scientific Inc.) and 1% penicillin streptomycin (Cellgro, Mediatech Inc.).
3.3.2. Rat mesenchymal stem cells from bone marrow (rMSCs-bm)
Commercial rMSCs were purchased from ScienCell™ research laboratories. The rMSCs were cryopreserved at passage-one culture and delivered frozen. They were characterized by the immunofluorescent method with antibodies to CD73, CD90, CD105 and Oil Red staining after adipodifferentiation. The rMSCs were cultured using mesenchymal stem cell medium provided by the same company and used for coculture before the fifth passage.
3.4 Microstructure fabrication
The cell-culture substrate with designed microstructures was fabricated using polydimethylsiloxane (PDMS) with the standard procedure of photolithography and soft lithography. SU-8 2050 photoresist was spun on a silicon wafer and polymerized under UV light through an AutoCAD-designed mask to form the mold. PDMS was spun onto the mold to create the PDMS membrane. For example, the substrate used to obtain the results demonstrated here had through holes for forming rectangular cell deposition microwells. Each microwell was designed at 50 μm in length and 25μm in width, and the distance between two neighboring microwells was 200 μm. The PDMS membrane was spun at a particular speed to achieve the height of 40 μm, which restricted cell growth to inside the microwells and allowed micropipettes to reach the cells during patch clamp experiments. The PDMS membrane was attached to a 22 mm × 22 mm coverglass, treated with oxygen plasma for 10 minutes, UV sterilized for at least 15 minutes and coated with fibronectin to form a cell culture substratum. This substratum was placed at the bottom of a cell culture dish mounted into the on-stage incubator built on the cell deposition microscope.
3.5 Patch clamp experiments
After cells were deposited into the substratum with the microstructure, patch-clamp experiments were conducted on the cocultured rMSCs from Day 1 to Day 4. Borosilicate glass electrodes were pulled with Brown-Flaming puller (P-97, Sutter Instrument Co.). Tip resistance was 3–4MΩ when it was filled with pipette solution. The pipette solution contained 140 mM K-gluconate, 1 mM EGTA, 2 mM MgCl2, 2 mM Na2ATP, 10 mM HEPES (pH7.2, 330mOsm). The bath solution contained 139 mM NaCl, 3 mM KCl, 17 mM NaHCO3, 12 mM Glucose, 3 mM CaCl2, 1 mM MgCl2, 10 mM HEPES (pH 7.2, 330mOsm). The patch clamp experiments were conducted using a computer-controlled current/voltage clamp (Multichannel 700A, Axon Instruments Co.) and a 16-bit data acquisition system (Digidata 1322A, Axon Instruments Co.) at room temperature. The tip potentials were compensated for before the pipette touched the cell. After a giga-seal was obtained by negative suction with the leakage current tested by the amplifier at less than 30pA, the pipette capacitances were compensated for. Then the cell membrane was ruptured by gentle suction to establish the whole-cell configuration. The membrane potential was held at −70 mV with a 90 mV voltage pulse added on it, and current signals were recorded.
4. Results and discussions
Normal cardiomyocytes are highly dependent on the functional expression of the ion channels to form action potentials and electrical coupling with other cells. To fully determine the scientific and therapeutic potential of stem cells for cardiovascular disease treatment, it is necessary to assess comprehensively a stem cell’s electrical phenotype upon differentiation. Potential stem and progenitor cells include mesenchymal stem cell (MSCs)[14], hematopoietic stem cells (HSCs)[15], embryonic stem cells (ESCs)[16], and resident cardiac stem cells[17]. These cells can be obtained allogenically from human donors, or xenogenically from other species, like porcine, rodent, or canine animals. Although all of these cell types are being considered for future clinical trials without any standardized, well-controlled in vitro assays available to compare their electrical coupling efficiency with host cardiomyocytes[18]. Therefore, development of a reproducible coculture model for stem cells and cardiomyocytes is essential to systematically and statistically study their electrical coupling under a highly-controlled microenvironment. Using the cell deposition microscope, individual rMSCs and cardiomyocytes were micropatterned into a fabricated microstructure to form a single-cell-level coculture model, which was used for the study of electrical coupling under a contact-mediated microenvironment. Since inward currents were crucial to the excitation and contraction of cardiomyocytes, they were considered as a specific marker for rMSCs differentiation and functional electrical coupling with cardiomyocytes. Through such a single-cell-level coculture model, phenotypic variations at the single-cell level that would be masked in population assays can be uncovered in the study of electrical coupling between stem cells and cardiomyocytes.
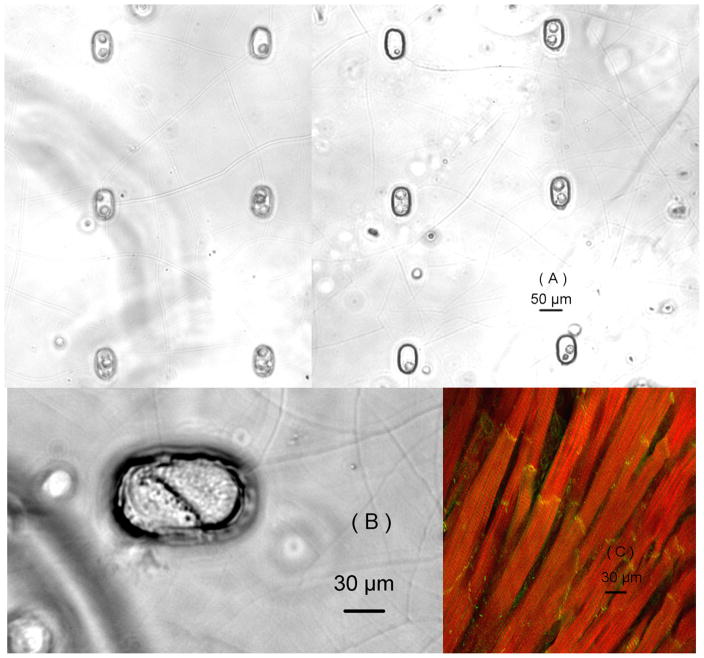
To create a heterotypic cell coculture model with single-cell resolution, cardiomyocytes were first deposited into the microwells on the substrate, and then the rMSCs were deposited into the microwells containing the cardiomyocytes, shown in figure 3(a). Similar culturing models were also created with pure cardiomyocytes or rMSCs to serve as the control groups for the coculturing model. The individual stem cell and cardiomyocyte filled the entire microwell and formed a broad contact between their cell membranes one day after heterotypic cell patterning, as shown in figure 3(b). This contact-mediated microenvironment forced the rMSCs to form a broad contact directly with the cardiomyocytes, which mimics the in vivo structure of cardiomyocytes with end-to-end cellular connections, shown in figure 3(c). The inward currents were recorded on different cells using patch clamp technique under whole-cell configuration by changing the patched cell’s membrane potential from −70 mV to 20mV. The percentage of stem cells with inward currents and their magnitudes were compared on different coculturing days (table 2). No inward current was recorded from noncocultured rMSCs even after four days of culturing. However, cocultured rMSCs showed electrophysiological changes at different temporal scales. On Day 1, cocultured rMSCs did not exhibit inward currents, but the percentage of cocultured rMSCs with currents increased from 13.3% on Day 2 and to 29.6% on Day 4. The magnitudes of inward currents from cocultured rMSCs were much smaller than those from cardiomyocytes (−1117 ± 185 pA), but they significantly increased from Day 2 (−214 ± 62 pA) to Day 4 (−435 ± 77 pA).
Figure 3.
(a)The single-cell-level coculturing model created using the cell deposition microscope and captured using a 10X objective immediately after cell depositions; (b) Two cells formed a broad contact between their cell membrane captured using a 40X objective after one day of coculture; (c) Actual structure of cardiomyocytes with end-to-end contact. Actins are stained red, and gap junctions are stained yellow.
Table 2.
Electrophysiological changes on cocultured rMSCs
| Coculturing days | Day 1 | Day 2 | Day 3 | Day 4 |
|---|---|---|---|---|
| Percentage of rMSCs with inward currents | 0/22 0% |
6/35 13.3% |
10/47 21.3% |
8/27 29.6% |
| Magnitude of inward currents | 0pA | −214 ± 62 pA | −350 ± 59 pA | −435 ± 77 pA |
Electrical coupling and conduction normally begin with the generation of an action potential by cardiac pacemaker cells; this causes depolarization of surrounding cells and consequently, the initiation of a propagation wavefront through the tissue. Stem cell transplantation as a cell-based treatment aims at regenerating functional tissue within the damaged heart, where nonfunctional tissue can lead to disruption of electrical coupling and conduction. It has been reported that stem cells were able to enhance function of ischemic myocardium through improvement of myocardial perfusion and contractile performance[19]. However, such approaches are not immune to electrical coupling problems, because the stem cells must support a uniform depolarization wavefront by acquiring electrophysiological properties and integrating with cardiomyocytes to provide an electrical conduction. The stem cells cocultured with cardiomyocytes have been reported to generate a cardiac-like action potential[20], and to exhibit several inward and outward ion currents[21]. However, traditional in vitro studies on electrical coupling between stem cells and cardiomyocytes did not provide a geometrically identified microenvironment, which makes it difficult to interpret and quantify the obtained results and perform statistical studies on their electrical coupling. Through our microfabrication technique, we created a highly controlled microenvionment with geometrical restrictions and homogeneous fibronectin coating. Individual rMSCs and cardiomyocytes were deposited into this microenvironment to form a heterotypic cell coculture model with single-cell resolution. Direct cellular contact forced by the geometrical restriction will stimulate the stem cells to form in vivo-like cellular contact with cardiomyocytes. In this coculture model, the capability of rMSC differentiation towards functional cardiomyocytes was systematically and statistically examined under the identical microenvironment. Comparing the patch-clamp results on different coculture days, we discovered that rMSCs regulated themselves to gradually acquire electrophysiological properties that resembled the cardiac phenotype. This contact-mediated microenvironment stimulated stem cells to electrically couple with the cardiomyocytes and promoted their differentiation.
5. Conclusions
Our cell deposition microscope based on the laser guidance technique was developed to pattern cells with a resolution that exceeds the error caused by inherent cell variation and irregularity. To manipulate the cells with high speed in transverse directions, the optical configuration was optimized through optical force simulations and laser guidance experiments. We found that the optical system with NA ≈ 0.1 generated adequate force magnitude and effective force range. The cell deposition microscope was designed to be compatible with a variety of substrates, including commercially available multi-electrode arrays (MEAs) with glass culture rings, 35 mm cell culture petri dishes, and microstructures microfabricated with PDMS. The microscope also provided on-stage incubation to maintain temperature, CO2, and humidity for long-term cell culturing during and after the cell deposition procedure. Unlike conventional cell patterning techniques, which constrain cells within the protein patterned area, the system allows natural cell migration after initial deposition. Therefore cell-cell and cell-ECM interactions can be studied without specific cell or ECM confinement. We observed neuronal axon guidance by glial cells after the glial cells spread following the neuron-glia deposition. The rat mesenchymal stem cells from bone marrow and rat neonatal cardiomyocytes were deposited into a microstructure to study their electrical coupling using patch clamp technique at single-cell resolution. This coculturing assay provided a highly controlled microenvironment for cardiomyocytes and rMSCs to ensure reproducibility in the spatial distribution of cocultured cells and statistical study on their interactions. We found that during the contact-mediated interactions with cardiomyocytes, rMSCs acquired the characteristics of the cardiomyocyte phenotype and exhibited gradual change in their electrophysiological properties. The developed portable cell deposition microscope allows cell biologists to systematically study this phenomenon in an advanced biology laboratory
Acknowledgments
This work has been partially supported by NIH (SC COBRE P20RR021949 and Career Award 1k25hl088262-04); NSF (MRI, CBET-0923311 and SC EPSCoR RII EPS-0903795 through SC GEAR program); and the Program for Key International S&T Cooperation Projects of the Ministry of Science of China (2008DFA30590). JXY and BZG would also like to acknowledge the support from the grant established by the State Key Laboratory of Precision Measuring Technology and Instruments (Tianjin University).
References
- 1.Andersson H, van den Berg A. Microfabrication and microfluidics for tissue engineering: state of the art and future opportunities. Lab Chip. 2004;4:98. doi: 10.1039/b314469k. [DOI] [PubMed] [Google Scholar]
- 2.Park TH, Shuler ML. Integration of cell culture and microfabrication technology. Biotechnol Prog. 2003;19:243. doi: 10.1021/bp020143k. [DOI] [PubMed] [Google Scholar]
- 3.Dholakia K, MacDonald MP, Zemanek P, Cizmar T. Cellular and colloidal separation using optical forces. Methods Cell Biol. 2007;82:467. doi: 10.1016/S0091-679X(06)82017-0. [DOI] [PubMed] [Google Scholar]
- 4.Hsu HY, Ohta AT, Chiou PY, Jamshidi A, Neale SL, Wu MC. Phototransistor-based optoelectronic tweezers for dynamic cell manipulation in cell culture media. Lab Chip. 2010;10:165. doi: 10.1039/b906593h. [DOI] [PubMed] [Google Scholar]
- 5.Singer W, Frick M, Haller T, Bernet S, Ritsch-Marte M, Dietl P. Mechanical forces impeding exocytotic surfactant release revealed by optical tweezers. Biophys J. 2003;84:1344. doi: 10.1016/S0006-3495(03)74950-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta AD, Rief M, Spudich JA, Smith DA, Simmons RM. Single-molecule biomechanics with optical methods. Science. 1999;283:1689. doi: 10.1126/science.283.5408.1689. [DOI] [PubMed] [Google Scholar]
- 7.Titushkin I, Cho M. Distinct membrane mechanical properties of human mesenchymal stem cells determined using laser optical tweezers. Biophys J. 2006;90:2582. doi: 10.1529/biophysj.105.073775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terray A, Arnold J, Hart SJ. Enhanced optical chromatography in a PDMS microfluidic system. Opt Express. 2005;13:10406. doi: 10.1364/opex.13.010406. [DOI] [PubMed] [Google Scholar]
- 9.MacDonald MP, Spalding GC, Dholakia K. Microfluidic sorting in an optical lattice. Nature. 2003;426:421. doi: 10.1038/nature02144. [DOI] [PubMed] [Google Scholar]
- 10.Remmerbach TW, Wottawah F, Dietrich J, Lincoln B, Wittekind C, Guck J. Oral cancer diagnosis by mechanical phenotyping. Cancer Res. 2009;69:1728. doi: 10.1158/0008-5472.CAN-08-4073. [DOI] [PubMed] [Google Scholar]
- 11.Nahmias Y, Odde DJ. Micropatterning of living cells by laser-guided direct writing: application to fabrication of hepatic-endothelial sinusoid-like structures. Nat Protoc. 2006;1:2288. doi: 10.1038/nprot.2006.386. [DOI] [PubMed] [Google Scholar]
- 12.Ashkin A. Forces of a single-beam gradient laser trap on a dielectric sphere in the ray optics regime. Methods Cell Biol. 1998;55:1. doi: 10.1016/s0091-679x(08)60399-4. [DOI] [PubMed] [Google Scholar]
- 13.Ren KF, Grehan G, Gouesbet G. Prediction of reverse radiation pressure by generalized Lorenz-Mie theory. Applied Optics. 1996;35:2702. doi: 10.1364/AO.35.002702. [DOI] [PubMed] [Google Scholar]
- 14.Pittenger MF, Martin BJ. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ Res. 2004;95:9. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 15.Balsam LB, Wagers AJ, Christensen JL, Kofidis T, Weissman IL, Robbins RC. Haematopoietic stem cells adopt mature haematopoietic fates in ischaemic myocardium. Nature. 2004;428:668. doi: 10.1038/nature02460. [DOI] [PubMed] [Google Scholar]
- 16.Kolossov E, et al. Identification and characterization of embryonic stem cell-derived pacemaker and atrial cardiomyocytes. Faseb J. 2005;19:577. doi: 10.1096/fj.03-1451fje. [DOI] [PubMed] [Google Scholar]
- 17.Barile L, Chimenti I, Gaetani R, Forte E, Miraldi F, Frati G, Messina E, Giacomello A. Cardiac stem cells: isolation, expansion and experimental use for myocardial regeneration. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl1):S9. doi: 10.1038/ncpcardio0738. [DOI] [PubMed] [Google Scholar]
- 18.Pedrotty DM, Klinger RY, Badie N, Hinds S, Kardashian A, Bursac N. Structural coupling of cardiomyocytes and noncardiomyocytes: quantitative comparisons using a novel micropatterned cell pair assay. Am J Physiol Heart Circ Physiol. 2008;295:H390. doi: 10.1152/ajpheart.91531.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joggerst SJ, Hatzopoulos AK. Stem cell therapy for cardiac repair: benefits and barriers. Expert Rev Mol Med. 2009;11:e20. doi: 10.1017/S1462399409001124. [DOI] [PubMed] [Google Scholar]
- 20.Pijnappels DA, Schalij MJ, van Tuyn J, Ypey DL, de Vries AA, van der Wall EE, van der Laarse A, Atsma DE. Progressive increase in conduction velocity across human mesenchymal stem cells is mediated by enhanced electrical coupling. Cardiovasc Res. 2006;72:282. doi: 10.1016/j.cardiores.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 21.de Boer TP, et al. Human cardiomyocyte progenitor cell-derived cardiomyocytes display a maturated electrical phenotype. J Mol Cell Cardiol. 2010;48:254. doi: 10.1016/j.yjmcc.2009.05.004. [DOI] [PubMed] [Google Scholar]