Abstract
In order to develop an infection, diarrhogenic Escherichia coli has to pass through the stomach, where the pH can be as low as 1. Mechanisms that enable E. coli to survive in low pH are thus potentially relevant for pathogenicity. Four acid response systems involved in reducing the concentration of intracellular protons have been identified so far. However, it is still unclear to what extent the regulation of other important cellular functions may be required for survival in acid conditions. Here, we have combined molecular and phenotypic analysis of wild-type and mutant strains with computational network inference to identify molecular pathways underlying E. coli response to mild and strong acid conditions. The interpretative model we have developed led to the hypothesis that a complex transcriptional programme, dependent on the two-component system regulator OmpR and involving a switch between aerobic and anaerobic metabolism, may be key for survival. Experimental validation has shown that the OmpR is responsible for controlling a sizeable component of the transcriptional programme to acid exposure. Moreover, we found that a ΔompR strain was unable to mount any transcriptional response to acid exposure and had one of the strongest acid sensitive phenotype observed.
INTRODUCTION
The acidic barrier of the stomach represents a strong challenge for many pathogenic enterobacteria (1). Therefore, although the correlation between acid resistance and pathogenicity is not absolute (2), the ability of some Escherichia coli strains to survive exposure to strong acid conditions is potentially relevant for pathogenicity (3–5). For this reason, the molecular and physiological response to acid stress has been the subject of intense investigation (5). Four acid stress response systems (ARs) that can protect E. coli from low pH are known to date (5–7). Three of these depend on the external supply of amino acids (glutamate, arginine and lysine) and have been proposed to share the same basic mechanism: reductive decarboxylation of the amino acid (consuming a proton) followed by extrusion of the product from the cytoplasm by a dedicated antiporter that also imports the original amino acid (5). One of the acid response systems, AR1, is active in the absence of amino acids (8) and is based on the FoF1 ATPase (6,9). At pH 2.5–3 the pump extrudes protons from the cytoplasm, with the consumption of ATP (7). The details of this system are poorly understood, but it is known to be regulated by both the sigma factor RpoS and the catabolite repressor protein, CRP (10). Also the other acid response systems are regulated by RpoS (10,11), which makes the acid stress response a growth phase dependent process (5,12,13). Other genes found in a 15-kb region around gadA termed the Acid Fitness Island (AFI) have also been shown to be required for acid resistance in E. coli (11,14–16). Among the regulators acting on the AR2 genes are the regulator proteins YdeO, GadE, GadX, GadW and the two-component system EvgA/EvgS (5,17–20). In addition, the system is influenced by cross-talk with other signalling systems including the PhoP/PhoQ system and the RcsB system (21,22). The action of the AR regulators is contingent on the growth medium, the way in which acid shock is induced, and the growth phase, and possibly is affected by strain-specific differences. Escherichia coli also shows acid adaptation, characterized by enhanced resistance to low pH following exposure to mild acidic conditions (23,24). This mechanism is mediated by the up-regulation of acid shock proteins, including components of the acid response systems described above. Thus its study is used as a way of defining important components of the acid resistance mechanisms and their regulation under non-lethal conditions. The regulation of the AR2 system has been intensively studied in E. coli K-12 and is surprisingly complex, with inputs from several different regulatory proteins, including global regulators and small RNAs (25).
Exposure to acid induces a sudden drop at intracellular pH (5), which despite the beneficial effect of the ARs, has profound effects on the physiology of the cell. For example, high intracellular proton concentration may induce uncoupling of oxidative phosphorylation resulting in alteration of energy metabolism (7). More generally, genes involved in energy metabolism, transport and amino-acid biosynthesis are known to be modulated at both mRNA (26) and protein level (27) suggesting that a much broader spectrum of adaptation pathways may be modulated in response to acid exposure. Therefore, the understanding of acid stress response goes beyond the study of the ARs.
A number of genome-wide expression profiling studies representing different acid stress conditions have been recently published (28–30). Although they have contributed to the identification of novel genes transcriptionally regulated during acid exposure, they do not yet provide us with a comprehensive model of E. coli acid resistance.
In order to address this challenge, we have taken a Systems Biology approach to investigating the gene networks involved in acid resistance. We have integrated phenotypic and genome-wide transcriptional data derived from wild-type and mutant strains and by using computational techniques that allow the inference of regulatory networks from observational data (31,32), we have attempted to identify molecular pathways and regulators required for acid adaptation. We show here that failure to modulate the expression of metabolic genes, particularly these encoding enzymes involved in aerobic and anaerobic metabolism, is a dominant feature of strains with a strong acid sensitive phenotype. In addition, the application of network inference techniques led us to hypothesize that OmpR may be a key regulator of the complex transcriptional programme involved in acid adaptation. Experimental validation of our model, based on the analysis of a ΔompR strain, supported this hypothesis.
MATERIALS AND METHODS
Study overview
The overall aim of this study was to identify pathways required for survival in strong acid conditions and to discover key regulators of the transcriptional response to acid exposure. We first used expression profiling with regular sampling over time to characterize the transcriptional response of a wild-type strain of E. coli BW25113 to mild acid conditions under constant growth conditions at an OD600 = 2. This allowed us to characterize the dynamics of the E. coli response to acid and to identify genes differentially regulated over time. By examining the consequences of deleting 38 of these genes we identified genes and pathways that were differentially expressed in mutants with different phenotypic strengths. The subsequent application of a network inference technique (ARACNE) (31,32) allowed the identification of the most likely regulators of these functions. The hypothesis generated from the model was verified by molecular and phenotypic analysis of the strain mutated in the putative regulator.
Bacterial strains
All the experiments described here were based on E. coli K-12 BW25113, which is directly derived from BD792, itself a two-step descendent of the E. coli K-12 ancestral strain (33,34). All mutant strains analysed in this study originated from the Keio collection (35) and were checked by PCR to verify the presence of the deletion before being used. For this purpose, a combination of locus and kanamycin-specific primers were used as described in the original publication (36).
Culture conditions
Bacterial strains were cultured in Luria-Bertani (LB) medium (Sigma Aldrich, USA) supplemented with kanamycin (50 µg/ml). In all our experiments, pH was adjusted to neutrality using sterilized 5 M NaOH. In the acid adaptation and acid shock experiments, pH was adjusted using sterilized 1 M HCl. Media were buffered with 10% MES.
In order to maintain optical density within a narrow range (1.85–2.15) and to keep growth rate constant during the experiment, we used a medium replenishment strategy in which medium and cells were removed and replaced by an equal volume of pre-warmed medium (37°C) at regular intervals (every 5 min). Cultures were grown for 16 h from a single colony at 37°C in a shaking incubator at 200 rpm in 10 ml of LB adjusted to at pH 7. An amount of 200 ml of pre-warmed LB medium at pH 7 in a 1 l conical flask was inoculated with overnight culture to a starting OD600 = 0.1. The culture was grown at 37°C and 200 rpm until it reached OD600 = 2. At this point, we initiated the medium replenishment procedure by removing 10 ml of culture and adding 10 ml of pre-warmed medium at pH 7 every 5 min. Using this procedure we kept the culture at OD600 = 2 and pH 7 for 1 h. This ensures that cells reached a steady state before addition of acid (Supplementary Figure S1). The pH of the culture was then shifted to pH 5.5 by addition of 14 ml of 1 M HCl while maintaining a constant OD for the duration of the experiment (1 h). After addition of acid, replenishment was performed as before but with buffered medium at pH 5.5.
Phenotypic analysis of mutant strains using flow cytometry
Wild-type and mutant strains were phenotypically characterized using flow cytometry analysis (37,38). In this application, we used propidium iodide (PI) and bis-(1,3-diethylthiobarbituric acid) trimethine oxonol (BOX) to, respectively, monitor membrane permeability and polarization. Briefly, bacterial cells were added to 2 ml of freshly made, filtered-sterilized (0.2 µM filter) PBS supplemented with EDTA, PI and BOX (final working concentrations of 4 mM, 5 and 10 µg/ml, respectively). Percentages of healthy and stressed cells were derived from the fluorescence emitted by the cell populations after staining, in relation to reference samples of alive or dead (ethanol-treated) cells (Supplementary Figure S2). The likelihood that the frequency of cells in each quadrant of the Flow Cytometry plots was significantly different between the mutants respect and the wild-type strains has been computed by using Fisher's exact test (39). Contingency tables were constructed using the number of healthy cells in the mutant and wild-type strains.
Plasmid construction
To generate the complementation plasmid pZCompR the complete ompR gene and the native promoter were amplified by PCR from the BW25113 chromosome using primers ompR-348F (GGTTGCTCGAGCGCCCAGACTTGCGGCCCAGG) and ompR + 720R (GGTTGGGATCCTCATGCTTTAGAGCCGTCCGG) that introduce unique XhoI–BamHI restriction sites. The fragment was introduced into the multiple cloning site of the low copy number plasmid, pZC320 (40).
Expression profiling by microarray
The 10 ml samples of cultures were stabilized by adding them to a phenol–ethanol solution (final concentration of 19% phenol and 1% ethanol). Samples were left on ice for 20 min and then centrifuged (4°C and 2600g) for 10 min to recover cell pellets. Stabilized cells were recovered and stored at −80°C. RNA was isolated using the Quiagen RNeasy® kit (Quiagen, USA) according to the manufacturer's instructions. Ten micrograms of input RNA was labelled using Cy5 labelled dCTP using the CyScribe Post-Labelling Kit and purified using CyScribe purification Kit according to the manufacturer's instructions. All kits and dyes were from Amersham Biosciences, USA. Operon E. coli Ultra GAPS microarray slides (Corning, USA) were hybridized overnight with 80 pmol of labelled cDNA. The slides were washed in AdvaWash automated washing station (Adavlytix, USA) and scanned with the ScanArray® GX (PerkinElmer®, USA), using the ScanArray® software.
Quantitative PCR
To validate gene expression profiles results, E. coli cells were grown and stabilized as for the microarray samples. RNA extraction was performed as described in the expression profiling section. An amount of 40 ng of cDNA were analysed with SYBR-Green method, after reverse transcription of the RNA with the 2000 U SuperScript II-Invitrogen kit, according to the manufacturer's instructions. The primers were designed using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and verified for specificity with Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The sequences for the primers were as follows: csrA (left primer CGAGTTGGTGAGACCCTCA, right primer AGCCTGGATACGCTGGTAGA); ompR (left primer CGTCGCTAATGCAGAACAGA, right primer GGTCCACTTCTTCCCCTTTC). Invitrogen primers (25 nmol) were used at a final concentration of 10 µM. SYBR-Green mix from ABGene, with ROX as passive reference, was used in a final volume of 10 µl. The analysis was performed with a 7900HT Fast Real-Time PCR System. Three technical and biological replicates were analysed and the gene expression of ompR was considered relative to the gene expression of csrA, the expression of which does not change under acid conditions (41,42). The three biological replicates were then analysed with the SDS software (Applied Biosystems) and a t-test was performed for significance.
Data analysis
The single channel array data were normalized using quantile normalization (43) in order to correct for systematic errors. Multivariate exploratory analysis of the gene expression data sets was performed using a combination of clustering and principal components analysis (PCA). In order to identify clusters of genes with similar expression profiles, we used SOTA, a clustering methodology based on an appropriate distance function (44) with Pearson correlation coefficient as a similarity measure. The relationships between the transcriptional states of the different cell populations were represented using the first two principal components defined by PCA (45). The number of components was chosen to represent at least 80% of the total sample variance. Bacterial survival and death kinetics, defined by flow cytometry analysis, were visualized using a standard average linkage hierarchical cluster analysis (46) with Pearson correlation as a similarity measure. Both clustering and PCA were performed using the TMEV software application (47,48).
Differentially expressed genes in the time course experiment (Figures 1 and 2) were selected using a fold-based rule (the absolute value of the log2 of the ratio between gene expression intensities between pH 5.5 and pH 7 larger than 1.5 at any time point, which corresponds to a 3-fold increase or decrease in a linear scale). This identified 2137 differentially expressed genes. Whenever experimental replicates were available, differentially expressed genes were identified using Significance Analysis for Microarrays (SAM) (49) as implemented in the TMEV software application (47,48) by employing a 10% FDR threshold, unless otherwise specified in the text.
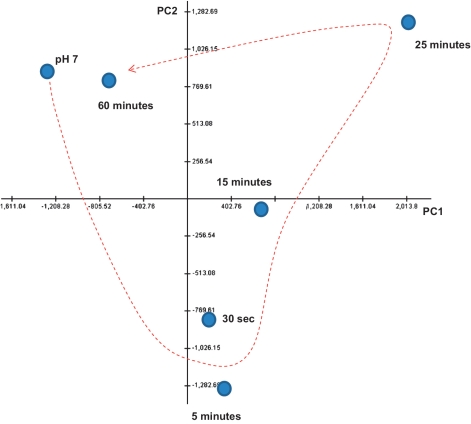
Figure 1.
Principle component analysis of the transcriptional response of BW25113 to acid adaptation. This analysis shows the change in the transcriptional response of BW25113 in the first hour of acid adaptation. The x and y axes represent, respectively, the first and second components in the PCA.
Figure 2.
Cluster analysis of gene expression profiles in response to acid adaptation. This Figure shows the result of cluster analysis performed using SOTA (41) on the most differentially expressed genes. SOTA identified four main clusters of co-ordinately regulated genes. A heatmap representing the expression profile of all genes in each cluster is represented on the left side of the figure (green and red correspond respectively to expression levels below and above the mean of gene expression across the time points). Clusters 1 and 3 show genes that are transiently down-regulated whereas Clusters 2 and 4 show genes that are transiently up-regulated. The table to the right of the heat maps shows GO and KEGG functional terms significantly enriched in each cluster (Count, number of genes for each pathway; Benjamini, P-value correction; ASRs, Acid Stress Response systems genes; AFI, Acid Fitness Islands genes).
Gene lists obtained by cluster or differential expression analysis were assessed for over-representation of KEGG (50) and Gene Ontology (51) functional terms using the open-source software DAVID (52,53) (http://david.abcc.ncifcrf.gov/). The GO analysis was performed with the lowest level of GO terms. In all cases, we used an FDR < 1% threshold to define a statistically significant enrichment.
Gene networks representing the neighbourhood of the two-component system regulators were inferred using the software application ARACNE (31,32) using the expression matrix representing the transcriptional state of 27 mutant strains and three replicates of the wild-type strain profiled at pH 7 and 5.5 (for a total of 60 arrays). ARACNE is an algorithm used for the identification of transcriptional interactions between gene products, by identifying statistical interactions based on the mutual information (32). Significant interactions were defined by a P-value threshold of P < 10−7 (corresponding to one over 2000 false positive connections). In order to eliminate non-direct interactions, we used the inequality principle as implemented in ARACNE with a DPI of 0.1 (32). The resulting networks were visualized using the Cytoscape software application (54).
To identify potential binding sites for OmpR, we have used the online tool Virtual Footprint (http://prodoric.tu-bs.de/vfp/) using the Prodoric library and performing the Regulon and the Promoter analysis (55). The PWM (Position Weight Matrix) considered were OmpC-box and OmpF-box of E. coli strain K-12.
Microarray data
Gene expression profiles data are accessible on the Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo/), with accession number GSE13361.
RESULTS
Escherichia coli adaptation to acid involves a rapid but transient transcriptional response
The approach we took in this study was to first characterize the dynamics of the transcriptional response of E. coli to acid adaptation. In order to do this, we monitored cultures that were kept in a balanced state of growth by removal of culture and replenishment with pre-warmed medium at regular intervals, a procedure that maintains cells in a constant transcriptional state (Supplementary Figure S1). The effect of acidification on gene expression was analysed using microarrays with RNA samples from six time points: prior to treatment, and 30 s, 5, 15, 25 and 60 min after exposure to pH 5.5. A high-level representation of the changes in the cells’ molecular state, performed using PCA, revealed that the process is defined by a rapid (30 s to 15 min) but transient response leading, after 1 h, to cells whose transcriptional state is similar to cells grown at pH 7 (Figure 1).
A more detailed analysis of the gene expression dynamics both by visual inspection of the data and using clustering revealed that the transcriptional events during this period of adaptation could be summarized by four main clusters of gene expression profiles (Figure 2). The first two clusters represented the earliest response to stimulation, including down- and up-regulated genes (Figure 2, clusters 1 and 2, respectively). In these two clusters, the largest changes in absolute value were detected 5 min after acid exposure. The second set of two clusters represented genes that are more gradually modulated down or up in response to pH 5.5 under these conditions (Figure 2, clusters 3 and 4, respectively) and where the largest change in absolute value was seen 25 min post-treatment.
We looked to see whether genes in each cluster were significantly enriched for any specific GO and KEGG functional terms (Figure 2). Genes represented in the significant terms were then mapped on the BW25113 genome and, as an additional level of quality control, the transcriptional response of genes in operons was analysed to check that all genes in a given operon showed that same transcriptional response (Supplementary Figures S3A–D and Figure S4: 17).
Consistent with our current understanding of acid adaptation, we observed the up-regulation of many of the genes associated with the glutamate, arginine and lysine-dependent acid response systems, and members of the AFI such as gadW, hdeA, hdeD, hdeB, yhiD, slp and yhiF. The exceptions found in this preliminary analysis were the genes coding for the isozyme GadA, and the GadX and the GadE regulators, since during early stationary phase they are supposed to be active, following the expression of RpoS (56). This apparent inconsistency may reflect a more complex regulatory system driving the transcriptional behaviour of the GAD genes. Genes coding for the FoF1 ATPase complex (thought to be a component of the amino acid-independent acid stress response system AR1) were also down-regulated. In addition, the transcriptional response to acid exposure involved the concomitant up-regulation of genes involved in anaerobic respiration (GO:0009061) and down-regulation of genes involved in aerobic respiration (GO:0009060). Genes involved in the regulation of the cell wall (GO:0005618) and translation (GO:0006412) were down-regulated whereas genes involved in membrane transport (ecd02010) were up-regulated in response to acid exposure (Table 1 and Figure 2). We also found that several genes coding for membrane proteins involved in osmoregulation were up-regulated (Supplementary Figures S6 and S10). More specifically, a number of osmoprotectant transporters involved in the response to hyperosmotic shock were transiently up-regulated. More specifically, a number of osmoprotectant transporters involved in the response to hyperosmotic shock were transiently up-regulated together with the genes mscL and mscS coding for the mechano-sensors involved in the response to hypo-osmotic shock.
Table 1.
Gene expression in response to acid adaptation
| Function | Complex | Genes | Regulation | P-value |
|---|---|---|---|---|
| Oxidative phosphorylation | NADH dehydrogenase | nuoA | ↓ | 1.18E-11 |
| nuoB | ↓ | |||
| nuoC | ↓ | |||
| nuoE | ↓ | |||
| nuoF | ↓ | |||
| nuoK | ↓ | |||
| FoF1 ATPase | atpB | ↓ | ||
| atpE | ↓ | |||
| atpH | ↓ | |||
| atpA | ↓ | |||
| Glycolysis | Galactose-1-epimerase | galE | ↓ | 3.58E-04 |
| galT | ↓ | |||
| galK | ↓ | |||
| galM | ↓ | |||
| Phosphoglycerate kinase | epd | ↓ | ||
| pgk | ↓ | |||
| Translation | Glycil-tRNA synthetase | glyS | ↓ | 3.07E-05 |
| glyQ | ↓ | |||
| Lysyl-tRNA synthetase | lysS | ↓ | ||
| dsbC | ↓ | |||
| Anaerobic respiration | Formate dehydrogenase | fdnG | ↑ | 1.20E-06 |
| fdnH | ↑ | |||
| fdnI | ↑ | |||
| Trimethylamine N-oxide reductase | torC | ↑ | ||
| torA | ↑ | |||
| torD | ↑ | |||
| Transport | Iron dicitrate ABC transporter | fecA | ↑ | 1.20E-06 |
| fecB | ↑ | |||
| fecC | ↑ | |||
| fecE | ↑ | |||
| Sulphonate/nitrate/taurine ABC transporter | ssuA | ↑ | ||
| ssuB | ↑ | |||
| ssuC | ↑ | |||
| ssuD | ↑ | |||
| Probable proton-driven drug efflux system | yjeP | ↑ | ||
| hsrA | ↑ |
The table list genes belonging to some of the pathways that are down (oxidative phosphorylation, glycolysis and translation) and up-regulated (anaerobic respiration and transport). The P-values obtained from DAVID functional annotation analysis are shown in the right side of the table alongside the direction of change.
As this initial analysis of the time course data was carried out using a single experiment only at each time point, we also compared three replicates of wild-type control cells with cells 15 min after acid exposure. Statistical analysis of these data identified 1871 differentially expressed genes. The result was largely overlapping with the results of the time course analysis (83% of the genes identified in the time course experiment were also identified by the single time point analysis, Supplementary Figures S5 and S6), showing our time course data to be robust.
Genes transcriptionally regulated by mild acid are generally required for surviving acid shock
Having performed an initial characterization of the E. coli transcriptional response during acid adaptation, we wished to see what fraction of the differentially expressed genes was required for survival during acid shock. In order to address this question, we selected 38 genes representative of the main functions modulated during acid adaptation (Supplementary Table S1) and tested the ability of strains where each of these genes was deleted to survive in strong acid conditions (pH 2.5). The mutants were assayed at different time points (5, 30, 60, 90, 120, 150 and 360 min) after acid exposure, and alive and dead cells were quantified by flow cytometry analysis. All tested mutants showed a detectable increase in sensitivity to low pH in the assay, though some effects were small (Figure 3, Supplementary Tables S2 and S3).
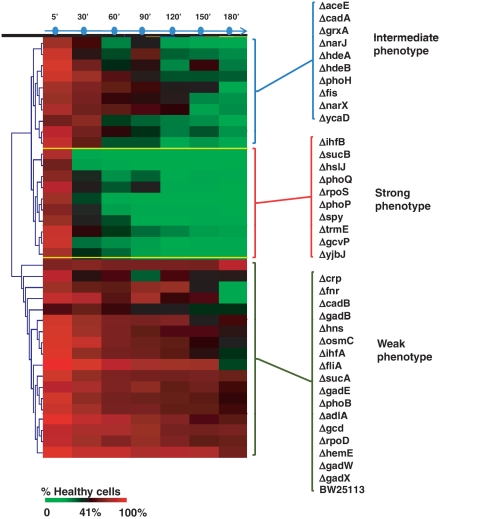
Figure 3.
Clustering of mutant strains according to their phenotypic response to acid shock. The figure represented in the format of a heatmap the percentage of healthy cells following exposure to LB at pH 2.5, without prior adaptation for the mutants we tested. Survival was measured at different time points (one sample every 30 min for 3 h). The colour code indicating the percentage of survival determined by flow cytometry analysis at each time point is below the heatmap. Hierarchical clustering of the survival time course data revealed three clusters of strains on the basis of their survival profiles. These were labelled as weak, intermediate and strong phenotypes.
In order to define the severity of the phenotype, we chose a threshold, which allowed us to cluster the mutants in three different groups as having strong, weak or intermediate phenotypes. We considered 30 min of exposure at pH 2.5 the parameter for the definition of the phenotype: strong phenotype mutants show <35–40% of healthy cells after 30 min at pH 2.5, intermediate phenotype show between 60% and 40% of healthy cells, while weak phenotype show >60% of healthy cells (Figure 3). The wild-type strain, included for comparison, shows the weakest phenotype, as expected, as this strain is quite acid resistant under the growth conditions of this assay.
Genes that when mutated gave a strong phenotype include transcriptional regulators (phoP, rpoS and ihfB), a glycine decarboxylase (gcvP), the 2-oxoglutarate dehydrogenase (sucB) and the two-component system sensor phoQ. Strains with an intermediate phenotype included mutants of two acid stress chaperones (hdeA and hdeB) and nitrate reduction (narJ and narX). Strains with a weak phenotype included mutants of several transcriptional regulators (crp, rpoD, hns, phoB, ihfA, fliA, fnr) and, interestingly, included three of the GAD system-specific regulators (gadE, gadW and gadX).
Defective expression of cell wall and energy metabolism genes correlates with increased sensitivity to acid
The experiments described above showed that the inactivation of genes that are transcriptionally regulated during adaptation often results in a significant loss of survival following acid shock. We reasoned that analysis of the transcriptional response to acid exposure in the different phenotypic groups would enable us to formulate hypotheses concerning the molecular pathways that are more important for survival of acid stress. We, therefore, subjected the same 38 mutants used above to a microarray analysis after acid adaptation and asked whether any component of the transcriptional response was predictive of loss of acid resistance. For practical purposes, this experiment was performed on a single time point (15 min after exposure at pH 5.5), which is where the highest change in gene expression in the early response clusters of the wild-type strain occurred (Figure 2). A PCA analysis confirmed that the acid adaptation transcriptional programme was defective in the mutant strains (Supplementary Figure S8).
In order to identify molecular pathways linked to the severity of the phenotype, we compared the gene expression profiles (expressed as a log-ratio between expression values at pH 5.5 and 7) between the wild-type (represented by four replicated experiments) and mutated strains that showed either an intermediate or a strong phenotype. We found 221 genes to be differentially expressed between the three experimental groups (FDR < 10%). Among the list of genes up-regulated in the wild-type strain, we found some of them involved in anaerobic respiration (hyfC, nrfF, menA), sugar transport (xylE, ulaA, ycjP, sgcC, malF, agaD, agaV), lipopolysaccharide biosynthesis (ylbH, rfaF, rfe, rfaJ, rhsA) and purine metabolism (yqeA, nudF, allB), whereas the list of down-regulated genes was enriched in aerobic respiration genes (including oxidative phosphorylation and TCA cycle) (ubiA, nuoF, acnB, sdhC, acnA, sdhD, ppc, nuoN, cyoA, cyoB, cyoD), DNA repair (uvrC, xthA, nei, exoX) and cell cycle (mrdB, ftsY, mukE, ftsA) (Figure 4).
Figure 4.
Differential gene expression in response to acid in the three phenotypic groups. The figure represents in a heatmap format the ratio of gene expression in acid-exposed cells respect to control cultures of the 221 differentially expressed genes between the wild-type and the mutants belonging to the intermediate (INT) and strong (STR) phenotype groups. The annotation on the right refers to functional groups of genes that were either down regulated (green) or up-regulated (red) in the wild-type strains at pH 5.5 relative to their expression at pH 7. These are shown in the first column of the heat map. In the second and third columns, the means of the log ratios of expression at pH 7 and 5.5 for these same genes are shown for the intermediate and strong groups.
We observed that the large majority of the genes predictive of phenotypic outcome (89%) were also differentially expressed in response to acid exposure (Figure 4). Remarkably, the ratios of the expression between cells grown at pH 5.5 and 7 were generally reversed in mutant strains that had an intermediate phenotype, while mutant strains with a strong phenotype were characterized by a general inability to regulate most of these genes (Figure 4).
Network inference analysis reveals new potential regulators of the global transcriptional response to acid stress
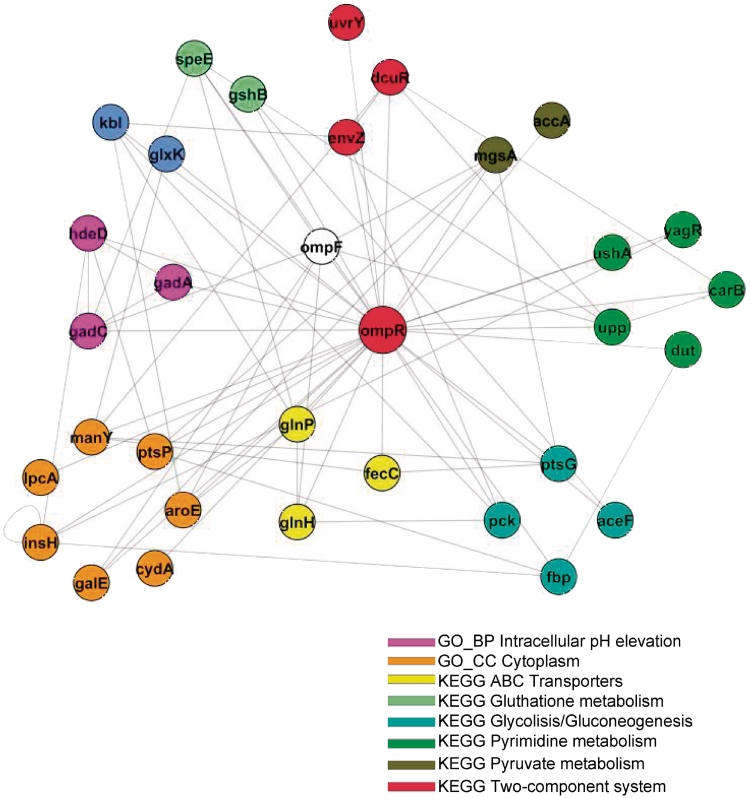
The results above identified molecular pathways whose levels of expression are linked to survival during acid shock. We therefore investigated which transcription factors might regulate the activity of these pathways. In order to focus on regulators whose activity may be directly influenced by environmental sensing we specifically looked for direct or indirect targets of the regulatory subunits of two-component systems. To achieve this, we used an unbiased approach that relies on a well-validated reverse engineering methodology (ARACNE). ARACNE uses a probabilistic measure of dependency (mutual information) to identify potential targets of a given transcription factor. It relies entirely on the analysis of a compendium of expression profiles representing different perturbations of a cell's homoeostatic state. Genes are inferred to be transcriptionally coupled when there is a statistically significant correlation between their levels of expression, across all samples in the compendium data set and after potential indirect connections are removed using the inequality principal criteria (32). Typically, large compendia of microarray data are used for the inference procedure. In this analysis, we chose to use the collection of 60 experiments representing expression profiles of genes in mutants and wild-type strains at pH 7 and 5.5, described in the previous paragraph. This had the advantage that it represented a collection of mutant strains relevant for acid adaptation while being sufficiently large to allow reliable inference (57). Since our objective was to identify regulators of the previously identified pathways, we ranked all the E. coli two-component system regulators on the basis of the number of connections they had with genes belonging to those pathways that we had shown to be predictive of phenotypic response and highly regulated during acid response. The ranking is shown in Table 2. The highest ranking gene by this analysis was OmpR, the regulatory subunit of the osmoregulator two-component system EnvZ/OmpR. OmpR was connected to genes involved in aerobic energy metabolism (pyruvate metabolism and glycolysis), signal transduction and transport, as well as some of the components of the GAD system (Figure 5 and Supplementary Table S5). NarP, the nitrate/nitrite response regulator, was connected to nucleotide and amino acid metabolism; the envelope stress response regulator BaeR was connected to genes involved in transport and metabolism. Most of the other two-component system regulatory subunits were poorly connected to genes in the pathways we had previously identified. On the basis of these observations, we proposed the novel hypothesis that OmpR may be a key regulator of acid response in E. coli BW25113 under the conditions of our study.
Table 2.
Potential two-component systems regulators targets as defined by the ARACNE analysis
| Gene | Two-component system | Targets based on MI | Function enriched | P-value |
|---|---|---|---|---|
| ompR | Omsoregulatory two-component system OmpR/EnvZ | 33 | GO_BP intracellular pH elevation (3 genes) | 4.50E-05 |
| GO_CC cytoplasm (7 genes) | 6.80E-03 | |||
| KEGG ABC transporters (3 genes) | 4.20E-02 | |||
| KEGG gluthatione metabolism (2 genes) | 3.90E-02 | |||
| KEGG glycolysis/gluconeogenesis(4 genes) | 3.00E-03 | |||
| KEGG pyrimidine metabolism (5 genes) | 3.60E-03 | |||
| KEGG pyruvate metabolism (2 genes) | 2.40E-03 | |||
| KEGG two-component system (3 genes) | 3.10E-03 | |||
| dcuR | Two-component system DcuR/DcuS, regulating anaerobic fumarate respiratory system | 27 | GO_BP ciliary or flagellar motility (3 genes) | 1.10E-03 |
| KEGG two-component system (7 genes) | 3.10E-05 | |||
| KEGG methane metabolism (3 genes) | 9.28E-05 | |||
| KEGG pyruvate metabolism (3 genes) | 3.46E-03 | |||
| baeR | Two-component regulatory system BaeS/BaeR | 43 | GO_BP transport (10 genes) | 1.25E-06 |
| KEGG ABC transporters (11 genes) | 3.21E-06 | |||
| KEGG galactose metabolism (5 genes) | 1.39E-05 | |||
| KEGG purine metabolism (3 genes) | 2.06E-03 | |||
| narL | NarX/NarL Two-component system, nitrate/nitrite dependent | 71 | GO_MF metal ion binding (4 genes) | 2.88E-03 |
| KEGG glycolysis/gluconeogenesis (3 genes) | 4.63E-04 | |||
| phoP | PhoP/PhoQ Two-component system, magnesium dependent | 48 | GO_BP oxidation reduction (3 genes) | 1.63E-03 |
| KEGG oxydative phosphorylation (3 genes) | 1.63E-03 | |||
| kdpE | Two-component regulatory system KdpD/KdpE involved in the regulation of the kdp operon | 31 | GO_BP nickel ion transport (2 genes) | 5.78E-04 |
| KEGG two-component system (6 genes) | 4.48E-04 | |||
| rstA | Two-component regulatory system RstA/RstB | 27 | GO_BP translation (4 genes) | 5.36E-03 |
| KEGG alanine, aspartate and glutamate metabolism (3 genes) | 9.81E-04 | |||
| KEGG Ribosome (3 genes) | 4.27E-05 | |||
| uvrY | Two-component regulatory system UvrY/BarA | 23 | GO_BP two-component signal transduction system (5 genes) | 2.48E-05 |
| GO_BP transcription (7 genes) | 1.45E-04 | |||
| KEGG two-component system (3 genes) | 4.27E-05 | |||
| narP | NarP/NarQ Two-component system, nitrate/nitrite dependent | 50 | GO_BP pyrimidine nucleotide biosynthetic process (6 genes) | 3.36E-10 |
| GO_BP iron–sulphur cluster assembly (6 genes) | 1.21E-07 | |||
| KEGG pyrimidine metabolism (7 genes) | 1.72E-06 | |||
| KEGG alanine, aspartate and glutamate metabolism (5 genes) | 1.57E-05 | |||
| rcsB | RcsB/RcsC regulates the expression of genes involved in colanic acid capsule synthesis | 36 | GO_BP transcription (9 genes) | 1.08E-04 |
| fimZ | involved in fimbrial expression | 34 | GO_BP transcription (7 genes) | 7.24E-04 |
| torR | TorR/TorS two-component system, responding to changes in the concentration of the TMAO | 20 | GO_BP metabolic process (5 genes) | 1.09E-02 |
| rssB | facilitates and regulates degradation of sigma S | 18 | GO_MF protein binding (8 genes) | 7.77E-03 |
| yfjR | Two-component system YfjR/YfhK | 18 | GO_BP transcription (5 genes) | 9.98E-03 |
| uhpA | UhpA/UhpB two-component system responding to external concentrations of glucose-6-phosphate | 13 | GO_CC integral to membrane (8 genes) | 4.33E-03 |
Regulators were ranked on the basis of how many connections were found to functions that were modulated during the acid adaptation time course (in red). P-values of the DAVID functional annotation analysis are shown.
Figure 5.
Network of regulatory interactions in the neighbourhood of OmpR, inferred using ARACNE. Nodes represent genes and edges represent inferred gene-to-gene connections. Genes are colour coded on the basis of their function.
Experimental confirmation that OmpR is a key regulator in the response to low pH
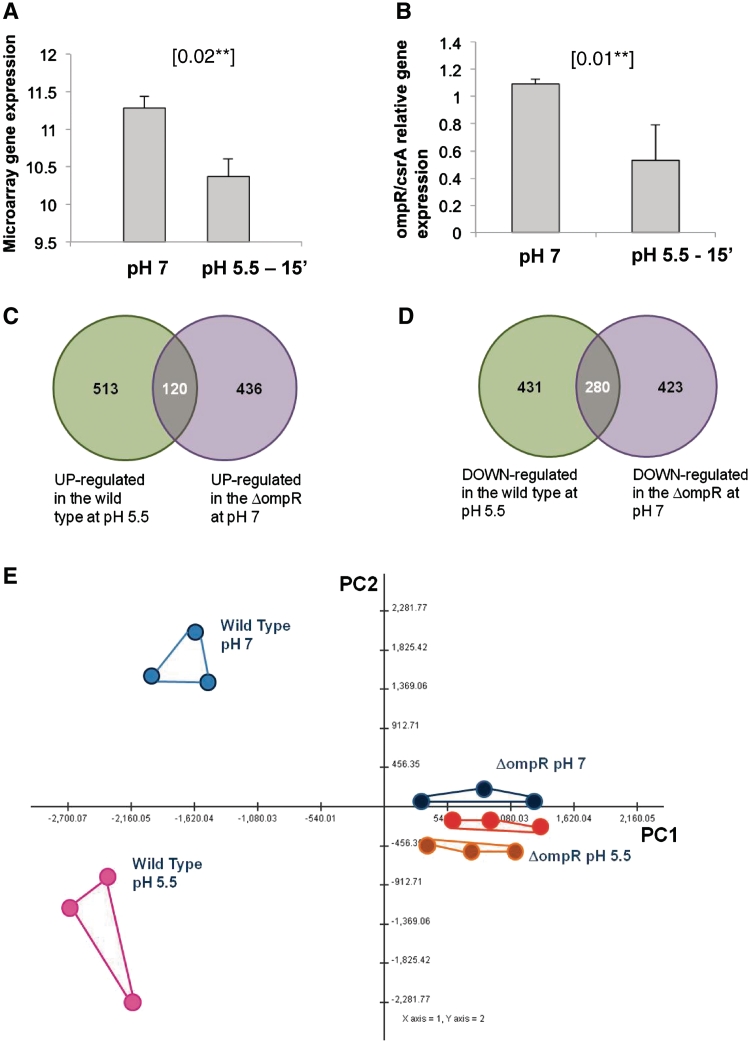
If our hypothesis is correct, we expect a mutant strain lacking ompR to display the following properties. First, the genes differentially regulated between ΔompR and the wild-type strain should significantly overlap with genes differentially regulated during acid adaptation in the wild-type. Second, this overlap should be consistent with the regulation of ompR itself during acid adaptation in the wild-type strain. For example, if ompR expression decreases on acid shock, then those genes that are normally repressed by OmpR should increase after acid shock, and show significant overlap with genes that are up-regulated if ompR is deleted. Third, the ΔompR mutant strain should be less able to initiate the normal transcriptional response to acid adaptation and, hence, should show a significant decrease in survival of acid shock relative to the wild-type strain. In order to test these predictions, we analysed the expression profile of a ΔompR mutant strain at pH 7 and 5.5, and performed a phenotypic characterization of this strain by flow cytometry after a direct challenge at pH 2.5. The results of this analysis closely matched our predictions. Genes differentially regulated between wild-type and the ΔompR mutant strain showed significant overlaps with genes differentially expressed in the wild-type strain during acid adaptation (Figure 6C and D). More specifically, 120 (23%) of the up-regulated genes in the wild-type during acid adaptation were also expressed more highly in the ΔompR mutant than in the wild-type at pH 7, while 280 genes (71%) genes down-regulated in the wild-type during acid adaptation were expressed more weakly in the ΔompR mutant than the wild-type at pH 7. The direction of the overlap was consistent with the observed down-regulation of ompR in response to acid exposure (Figure 6A and B). qPCR showed the down-regulation of ompR at pH 5.5 with values of slope and r2 for the efficiency and the accuracy of the measurements: csrA slope = −3.1 cycles/logdecade, r2 = 0.99; ompR slope = −3.01 cycles/logdecade, r2 = 0.99. Even more strikingly, there was no transcriptional response detectable to acid exposure in the ΔompR mutant either using a univariate statistical analysis approach (no differentially expressed genes were identified up to a FDR < 50% threshold) or using a multivariate exploratory analysis approach (Figure 6D). Flow cytometry analysis showed that the ΔompR mutant strain showed one of the strongest phenotypes detected in our study (Figure 7, Panel A). Mutant cells showed increased resistance to acid shock if pre-adapted to mild acid conditions, suggesting a small residual ability to mount an effective response, although they were still significantly less resistant than the unadapted wild-type strain. Complementation of the ΔompR mutant strain with a copy of the wild-type ompR gene expressed under its own promoter in the low-copy number plasmid pZC320 led to restoration of normal levels of acid resistance, whereas the vector alone had no effect [Figure 7, Panel B shows analysis by flow cytometry; all results were also confirmed with plate counts (data not shown)].
Figure 6.
Comparison of transcriptional responses of ΔompR and wild-type to acid stress of pH 5.5. Panels (A and B) show the expression levels of ompR at pH 7 and after 15 min exposure at pH 5.5, based on either microarray data (A) or qPCR data (B). Bars show standard deviations of three biological replicates. ompR is significantly down-regulated at pH 5.5 in both data sets (P-value 0.02 and 0.01, respectively, for microarrays and qPCR data, obtained with t-test). (C and D) show the extent of overlap between genes which are differentially up-regulated (C) or down-regulated (D) in the wild-type at pH 5.5, relative to expression at pH 7, and genes which are over-expressed (C) or under-expressed (D) in ΔompR at pH 7, relative to the wild-type at pH 7. (E) PCA plot of transcriptome changes in wild-type and the ΔompR mutant, analysed at pH 7 and 5.5. Blue dots, wild-type pH 7, pink dots: wild-type pH 5.5, black dots ΔompR at pH 7, red and orange dots ΔompR at pH 5.5 after 30 s and 15 min of exposure, respectively.
Figure 7.
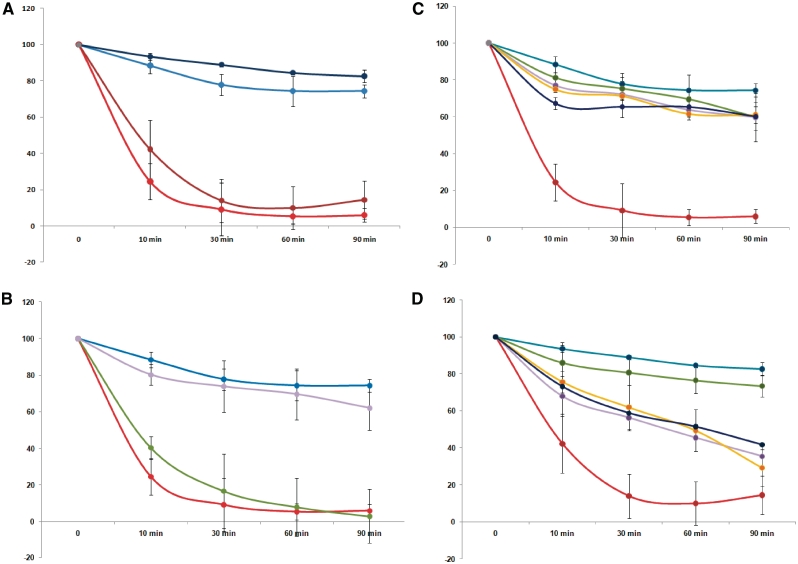
(A) Acid sensitive phenotype of the ΔompR strain. Survival of wild type and ΔompR strains in response to exposure to pH 2.5, either directly or after prior adaptation to pH 5.5, was measured by flow cytometry. Error bars show standard deviations of four independent biological replicates. The y axis shows % survival relative to viable cell numbers at t0. Wild type without adaptation, pale blue; wild-type with adaptation, dark blue; ΔompR without adaptation, red; ΔompR with adaptation, brown. (B) Complementation of the ΔompR strain restores acid resistance. The ΔompR mutant was complemented with the plasmid pZCompR, or as a control with the empty vector pZC320, and acid resistance measured at pH 2.5 without prior adaptation Error bars show standard deviation values of four biological replicates for WT and ΔompR and three biological replicates for the complemented strain. Wild type, pale blue; ΔompR without vector, red; ΔompR with empty vector, green, ΔompR with pZCompR, mauve. (C, D) Comparison of the ΔompR acid resistance phenotype with those of other mutations in key acid response genes. Survival curves of the following mutants after exposure to pH 2.5, without (A) or with (B) prior adaptation at pH 5.5 for one hour: ΔompR (red), ΔgadE (yellow), ΔgadC (purple), ΔadiC (green) and ΔrpoS (dark blue) compared to the wild type (blue). Error bars show standard deviation values of four independent biological replicates for wild-type and ΔompR and three biological replicates for the other mutant strains.
Comparison between effects of acid exposure in different mutant strains confirms the importance of OmpR
To compare the effects of different mutations on the acid resistant phenotype with that of the ΔompR mutation,we did flow cytometry experiments on BW25113 strains deleted for gadE, gadC, adiC and rpoS. GadE is a central regulator of the GAD system, GadC is the GABA/glutamate antiporter needed for function of the AR2 system, AdiC is the arginine/agmatine antiporter needed for function of the AR3 system, and RpoS is a sigma factor of RNA polymerase involved in the acid resistant phenotype. Under our conditions, we confirmed that the ΔompR strain showed the strongest phenotype of all these mutants when cells were not adapted with a prior shock at pH 5.5 (Figure 7, Panel C). After 10 min of exposure, <30% of the cell in ΔompR were still alive, while in the other strains, >70% of the cells were healthy at this stage. Strains carrying mutations in either of the two amino acid antiporters showed a weak phenotype. Different effects were seen when cells were induced at pH 5.5, before exposure to acid shock at pH 2.5 (Figure 7, Panel D). All mutants in this case (apart from the adiC mutant) showed large reductions in viability compared to the wild-type strains; however, the ΔompR strain still had the most severe phenotype.
DISCUSSION
Our work has shown that the ability of E. coli BW25113 to survive in strong acid conditions is dependent on an OmpR-dependent transcriptional programme that involves the regulation of energy metabolism. Mutant strains affecting the classic acid resistance genes generally have a weaker phenotype than a ΔompR strain, suggesting that this novel response programme is the key to acid adaptation.
A rapid shift between the expression of genes involved in aerobic and anaerobic energy metabolism is a key landmark of acid adaptation
Several lines of evidence that emerge from this work support the view that acid exposure induces a shift between aerobic and anaerobic metabolism and that this is a strong requirement for survival during acid shock. First of all, E. coli BW25113 cells under the conditions of our experiment express high levels of mRNA for enzymes involved in aerobic metabolism and lower levels of genes involved in anaerobic energy metabolism. A shift to low pH culture conditions induces a reversal of this pattern (Supplementary Figure S4: 1–7). The functional analysis revealed a trend in the modulation of the genes belonging to the anaerobic respiration pathway (according to GO terms), which involves the down-regulation of the fumarate-dependant respiration and the up-regulation of nitrate and formate respiratory enzymes (Table 3). Enterobacteria are predominantly fermentative anaerobes that generate acids. The dominant metabolism of E. coli in an anaerobic environment is based on the mixed acid fermentation, and on the adaptation to readily decrease external pH, for example, by inducing the synthesis of formate hydrogen lyase pathway. The metabolic switches imply that acid triggers a shift from aerobic respiration driven by the tricarboxylic acid cycle to adaptation to oxygen-limited growth. This suggests that E. coli has evolved the ability to prepare for anaerobic lifestyle by a pre-emptive induction of some anaerobic pathways prior to oxygen starvation. The comparison between the wild-type and mutant strains (with intermediate and severe phenotypes) revealed that failure to invert the expression of aerobic and anaerobic metabolism genes correlates with a strongly acid sensitive phenotype. Moreover, many of the strains which show a strong phenotype are mutated in genes involved in energy metabolism (Figure 4 and Supplementary Figure S9: 1–2). This indicates that under the conditions described here, effective proton scavenging alone may not be sufficient for survival unless the expression of bioenergetics genes is also modulated during adaptation to low pH conditions. We have seen that the genes transcriptionally controlled by the pH change are essential for the entire process previously described. This implies that the gene products are part of an integrated adaptive process in which transcription regulation depends on the completion of the process itself. There are currently few insights into the mechanism underlying this response. Suggestions can be made if an fnr mutant may be defective in adaptation to low pH. Although FNR is an iron–sulphur protein that is inactivated by oxygen (58,59), in cultures of moderately high density such as those used in the current experiments, FNR is partially active despite vigorous aeration. It would also be interesting to see whether FNR is critical for adaptation to acid of cultures at a much lower bacterial density.
Table 3.
Regulation of the genes involved in the anaerobic respiration pathway, according to GO terms
| DOWN-regulated | UP-regulated | ||
|---|---|---|---|
| aceF | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | aceE | Pyruvate dehydrogenase E1 component |
| acnA | Aconitate hydratase 1 | fdnH | Formate dehydrogenase, nitrate-inducible, iron-sulphur subunit |
| acnB | Aconitate hydratase 2 | fdnI | Formate dehydrogenase, nitrate-inducible, cytochrome b556(fdn) subunit |
| dcuR | Transcriptional regulatory protein dcuR | fdoI | Formate dehydrogenase, cytochrome b556(fdo) subunit |
| dmsA | Anaerobic dimethyl sulfoxide reductase chain A | glpA | Anaerobic glycerol-3-phosphate dehydrogenase subunit A |
| dmsB | Anaerobic dimethyl sulfoxide reductase chain B | glpB | Anaerobic glycerol-3-phosphate dehydrogenase subunit B |
| fdhE | Protein fdhE | glpC | Anaerobic glycerol-3-phosphate dehydrogenase subunit C |
| fdnG | Formate dehydrogenase, nitrate-inducible, major subunit | hyaA | Hydrogenase-1 small chain |
| fdoG | Formate dehydrogenase-O major subunit | hyaB | Hydrogenase-1 large chain |
| fnr | Fumarate and nitrate reduction regulatory protein | hyaE | Hydrogenase-1 operon protein hyaE |
| frdA | Fumarate reductase flavoprotein subunit | hyaF | Hydrogenase-1 operon protein hyaF |
| frdB | Fumarate reductase iron–sulphur subunit | hycE | Formate hydrogenlyase subunit 5 |
| frdC | Fumarate reductase subunit C | hycF | Formate hydrogenlyase subunit 6 |
| frdD | Fumarate reductase subunit D | hyfD | Hydrogenase-4 component D |
| fumC | Fumarate hydratase class II | hyfE | Hydrogenase-4 component E |
| glpE | Thiosulfate sulphurtransferase glpE | hyfF | Hydrogenase-4 component F |
| glpR | Glycerol-3-phosphate regulon repressor | hyfG | Hydrogenase-4 component G |
| gltA | Citrate synthase | hyfR | Hydrogenase-4 transcriptional activator |
| hybB | Probable Ni/Fe-hydrogenase 2 b-type cytochrome subunit | menA | 1,4-dihydroxy-2-naphthoate octaprenyltransferase |
| hybC | Hydrogenase-2 large chain | napD | Protein napD |
| hybD | Hydrogenase 2 maturation protease | napF | Ferredoxin-type protein napF |
| hybF | Probable hydrogenase nickel incorporation protein hybF | narG | Respiratory nitrate reductase 1 alpha chain |
| hybO | Hydrogenase-2 small chain | narH | Respiratory nitrate reductase 1 beta chain |
| hypD | Hydrogenase isoenzymes formation protein hypD | narI | Respiratory nitrate reductase 1 gamma chain |
| hypE | Hydrogenase isoenzymes formation protein hypE | narJ | Respiratory nitrate reductase 1 delta chain |
| hypF | Carbamoyltransferase hypF | narL | Nitrate/nitrite response regulator protein narL |
| icd | Isocitrate dehydrogenase [NADP] | narX | Nitrate/nitrite sensor protein narX |
| lldD | l-lactate dehydrogenase [cytochrome] | narY | Respiratory nitrate reductase 2 beta chain |
| lpd | Dihydrolipoyl dehydrogenase | narZ | Respiratory nitrate reductase 2 alpha chain |
| mdh | Malate dehydrogenase | ndh | NADH dehydrogenase |
| menC | o-succinylbenzoate synthase | nikE | Nickel import ATP-binding protein nikE |
| mltD | Membrane-bound lytic murein transglycosylase D | nirB | Nitrite reductase [NAD(P)H] large subunit |
| napC | Cytochrome c-type protein napC | nrfA | Cytochrome c-552 |
| narW | Respiratory nitrate reductase 2 delta chain | nrfE | Cytochrome c-type biogenesis protein nrfE |
| nrfD | Protein nrfD | pflC | Pyruvate formate-lyase 2-activating enzyme |
| nuoB | NADH-quinone oxidoreductase subunit B | pflD | Formate acetyltransferase 2 |
| nuoC | NADH-quinone oxidoreductase subunit C/D | torA | Trimethylamine-N-oxide reductase 1 |
| nuoE | NADH-quinone oxidoreductase subunit E | torC | Cytochrome c-type protein torC |
| nuoF | NADH-quinone oxidoreductase subunit F | torT | Periplasmic protein torT |
| nuoG | NADH-quinone oxidoreductase; NADH-quinone oxidoreductase subunit G | ugpA | sn-glycerol-3-phosphate transport system permease protein ugpA |
| nuoH | NADH-quinone oxidoreductase subunit H | ugpC | sn-glycerol-3-phosphate import ATP-binding protein ugpC |
| nuoJ | NADH-quinone oxidoreductase subunit J | ugpE | sn-glycerol-3-phosphate transport system permease protein ugpE |
| nuoK | NADH-quinone oxidoreductase subunit K | ybiY | Putative pyruvate formate-lyase 3-activating enzyme |
| nuoL | NADH-quinone oxidoreductase subunit L | ||
| nuoN | NADH-quinone oxidoreductase subunit N | ||
| pykA | Pyruvate kinase II; Pyruvate kinase | ||
| tdcE | Keto-acid formate acetyltransferase | ||
| torS | Sensor protein torS; Sensor protein | ||
| ynfH | Anaerobic dimethyl sulphoxide reductase chain ynfH | ||
The list includes genes down-regulated in response to acid exposure (left) and up-regulated (right).
OmpR: an important acid response regulator
Our study provides strong evidence for a role of OmpR as a regulator of the transcriptional response to acid adaptation in BW25113. Although OmpR has not been directly implicated hitherto in acid resistance in E. coli, it has been shown than an ompR UPEC mutant shows reduced survival in the mouse urinary tract, and that the growth defect seen in this mutant in high salt is enhanced at low pH (5.5); lethal pH was not tested in this experiment (60). OmpR has been shown to regulate the stationary phase acid inducible response in Salmonella Typhimurium (61) potentially by counteracting H-NS-mediated repression (61,62), and an OmpR-like regulator (HP0166) has also been implicated in the acid response of H. pylori (63) suggesting a broader role for regulators of this type in acid stress responses.
Gene expression in the ompR mutant partially mimics the response to acid of the parent strain (Figure 6) but mutant cells are unable to mount any response to acid, suggesting that modulation of OmpR is required for an effective response to acid. This result, coupled with the acid sensitivity of the ΔompR mutant, are consistent with a model where OmpR is required for the expression of cellular components, or for the establishment of a particular cellular state, which is needed for the cells to be able to respond to acid stress in a way that enhances their survival. In the absence of OmpR, this state no longer exists and so the ΔompR mutant fails to respond to acidification and shows enhanced acid sensitivity. Cells lacking this important regulator are even less able to survive in extreme acid conditions than those carrying mutations in other important genes implicated in acid resistance. The key role of OmpR in regulating adaptation to low pH was unexpected and has not featured in an extensive literature on the EnvZ–OmpR two-component system. Key questions for future research to answer are whether EnvZ mediates the response and, if so, to what chemical signal EnvZ responds. In a previous study on Shigella, FNR, the dual transcriptional regulator of the switch between aerobic and anaerobic metabolism, was found to regulate the length of type III secretion system needles, required for secretion of the invasion plasmid antigen. Exposure to oxygen at the surface of the gastrointestinal mucosa inactivates FNR, reversing the block on invasion antigen secretion and hence priming the bacteria for the attack (58,59). Perhaps the OmpR-mediated response to acid fulfils a similar role. Moreover, a deeper analysis of FNR targets revealed a down-regulation of the functions modulated by FNR, which we found important for our work (Supplementary Table S6).
Whether OmpR exerts its function directly or by modulating the activity of other regulators remains an important question. We found that some of the genes downstream from OmpR (as defined by correlation and KO analysis) have OmpR-binding sites in their promoter regions (Supplementary Figure S7), suggesting that at least in some cases OmpR may directly activate genes involved in acid response.
The relevance of the osmotic stress response to acid adaptation
The novel role we have found for OmpR suggests a link between osmotic and acid stress responses. A potential connection between the activation of anaerobic metabolism (an important feature of acid adaptation in this strain) and osmotic stress has been previously described (64), where it was linked to changes in DNA topology. Many genes involved in the response to osmotic shock are also modulated in response to acid in our experiments (Supplementary Figures S6 and S10). Most of the channels involved in the transport and production of osmoprotectants were down-regulated at pH 5.5. However, the H+/proline symporter (65), the choline transporter (66) and two K+ channels (67,68) (Supplementary Figure S10) were up-regulated after acid exposure, a response that makes adaptive sense in light of the finding that K+ and proline have a beneficial effect on pH homoeostasis in E. coli (69). It may also be the case that the import of amino acids into the cytoplasm, as part of the acid adaptation response, could drive the cells to a weak condition of hypo-osmolarity. Consistent with this hypothesis, the mechanosensor channels, MscS (70) and MscL (71), with the acquaporine AqpZ (72) and the K+ mechanosensor (73) were up-regulated in the first 15 min after acid exposure. Substantial changes are known to take place in the balance of ions across the membrane on acid shock in E. coli, leading to reversal of membrane polarity, and in this context it is not surprising that a regulator that itself responds indirectly to changed osmolarity is crucial in regulating this response.
A major two-component system involved in regulating the acid stress response, the EvgAS system, did not emerge from this study. This is unsurprising as it is known that the expression of the evgAS operon is little affected by acid and so it would not have been detected as being important using the methods described here. Furthermore, the evgAS system is mainly thought to be important in regulating the gad system, but this system has a limited role in the conditions described here, as can be seen by the fact that a gadE deletion only confers a mild phenotype.
Different E. coli strains show a wide range of resistance to acid. It is not yet clear whether OmpR expression can explain some of the variation in this naturally occurring resistance. It may be that the relative importance of OmpR may depend on the strain being used, and that BW25133 is particularly dependent on OmpR. We are in the process of testing this hypothesis.
Overall, three aspects of the physiological response of E. coli K-12 to mild acid stress require further analysis and explanation. First, the response regulator OmpR, which normally is thought of as responding to osmotic stress, is essential for adaptation to low pH and for priming survival of more severe acid stress. Second, many genes previously identified as components of OmpR regulon as well as additional previously unrecognized members, are essential for adaptation. Third, mild acid stress induces various metabolic switches, for example, from glycolysis to gluconeogenesis and fatty acid synthesis, and from energy generation powered by the TCA cycle to expression of FNR-regulated genes associated with anaerobic respiration (Supplementary Table S6, enriched functions of FNR target genes in response to acid exposure in our time course data).
CONCLUSIONS
We have taken a broadly applicable, systems-level approach to defining the response to acid of E. coli BW25133. This approach has revealed several entirely novel features of this well-studied response, including the importance of a shift in the metabolic state of the cell, and a central role for the OmpR regulator, a regulator that was already know to play an important role in modulating E. coli gene expression under standard conditions (74). The next steps in this study will be to see how broadly applicable these findings are across different strains, including pathogens and different growth conditions, to dissect the molecular and physiological mechanisms that underlie the changes we have detected, and to understand in more detail their adaptive value for the cell.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) (BBC5151041). A.S. is a recipient of a Darwin Trust of Edinburgh PhD fellowship, N.D. and P.A. are recipients of a BBSRC PhD studentships. Funding for open access charge: BBSRC, UK Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Professor Simon Andrews from the University of Reading, for permissions to use the Keio collection library. We are grateful to Elsa Boudadi from the University of Birmingham for help with the qPCR experiments.
REFERENCES
- 1.Giannella RA, Broitman SA, Zamcheck N. Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro. Gut. 1972;13:251–256. doi: 10.1136/gut.13.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhagwat AA, Tan J, Sharma M, Kothary M, Low S, Tall BD, Bhagwat M. Functional heterogeneity of RpoS in stress tolerance of enterohemorrhagic Escherichia coli strains. Appl. Environ. Microbiol. 2006;72:4978–4986. doi: 10.1128/AEM.02842-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin MM, Datta AR. Acid tolerance of enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 1995;61:1669–1672. doi: 10.1128/aem.61.4.1669-1672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conner DE, Kotrola JS. Growth and survival of Escherichia coli O157:H7 under acidic conditions. Appl. Environ. Microbiol. 1995;61:382–385. doi: 10.1128/aem.61.1.382-385.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 6.Richard HT, Foster JW. Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 2003;52:167–186. doi: 10.1016/s0065-2164(03)01007-4. [DOI] [PubMed] [Google Scholar]
- 7.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J. Bacteriol. 2004;186:6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin-Galiano AJ, Ferrandiz MJ, de la Campa AG. The promoter of the operon encoding the F0F1 ATPase of Streptococcus pneumoniae is inducible by pH. Mol. Microbiol. 2001;41:1327–1338. doi: 10.1046/j.1365-2958.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- 10.Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J. Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Biase D, Tramonti A, Bossa F, Visca P. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 1999;32:1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- 12.Gorden J, Small PL. Acid resistance in enteric bacteria. Infect. Immun. 1993;61:364–367. doi: 10.1128/iai.61.1.364-367.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucker DL, Tucker N, Conway T. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 2002;184:6551–6558. doi: 10.1128/JB.184.23.6551-6558.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hommais F, Krin E, Coppee JY, Lacroix C, Yeramian E, Danchin A, Bertin P. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology. 2004;150:61–72. doi: 10.1099/mic.0.26659-0. [DOI] [PubMed] [Google Scholar]
- 16.Mates AK, Sayed AK, Foster JW. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J. Bacteriol. 2007;189:2759–2768. doi: 10.1128/JB.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuda N, Church GM. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 2002;184:6225–6234. doi: 10.1128/JB.184.22.6225-6234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tramonti A, Visca P, De Canio M, Falconi M, De Biase D. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 2002;184:2603–2613. doi: 10.1128/JB.184.10.2603-2613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tramonti A, De Canio M, Delany I, Scarlato V, De Biase D. Mechanisms of transcription activation exerted by GadX and GadW at the gadA and gadBC gene promoters of the glutamate-based acid resistance system in Escherichia coli. J. Bacteriol. 2006;188:8118–8127. doi: 10.1128/JB.01044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayed AK, Odom C, Foster JW. The Escherichia coli AraC-family regulators GadX and GadW activate gadE, the central activator of glutamate-dependent acid resistance. Microbiology. 2007;153:2584–2592. doi: 10.1099/mic.0.2007/007005-0. [DOI] [PubMed] [Google Scholar]
- 21.Eguchi Y, Itou J, Yamane M, Demizu R, Yamato F, Okada A, Mori H, Kato A, Utsumi R. B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and PhoQ/PhoP in Escherichia coli. Proc. Natl Acad. Sci. USA. 2007;104:18712–18717. doi: 10.1073/pnas.0705768104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castanie-Cornet MP, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 2010;38:3546–3554. doi: 10.1093/nar/gkq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster JW, Hall HK. Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 1990;172:771–778. doi: 10.1128/jb.172.2.771-778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boot IR, Cash P, O'Byrne C. Sensing and adapting to acid stress. Antonie Van Leeuwenhoek. 2002;81:33–42. doi: 10.1023/a:1020565206835. [DOI] [PubMed] [Google Scholar]
- 25.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, Selbig J, Willmitzer L. Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 6:364. doi: 10.1038/msb.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 2002;184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 2005;187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes ET, Wilks JC, Sanfilippo P, Yohannes E, Tate DP, Jones BD, Radmacher MD, BonDurant SS, Slonczewski JL. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 2006;6:89. doi: 10.1186/1471-2180-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kannan G, Wilks JC, Fitzgerald DM, Jones BD, Bondurant SS, Slonczewski JL. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol. 2008;8:37. doi: 10.1186/1471-2180-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margolin AA, Wang K, Lim WK, Kustagi M, Nemenman I, Califano A. Reverse engineering cellular networks. Nat. Protoc. 2006;1:662–671. doi: 10.1038/nprot.2006.106. [DOI] [PubMed] [Google Scholar]
- 32.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl. 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2006;2:2006.0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachmann BJ. Linkage map of Escherichia coli K-12, edition 8. Microbiol. Rev. 1990;54:130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hewitt CJ, Nebe-von Caron G, Nienow AW, McFarlane CM. The use of multi-parameter flow cytometry to compare the physiological response of Escherichia coli W3110 to glucose limitation during batch, fed-batch and continuous culture cultivations. J. Biotechnol. 1999;75:251–264. doi: 10.1016/s0168-1656(99)00168-6. [DOI] [PubMed] [Google Scholar]
- 38.Shapiro HM, Nebe-von-Caron G. Multiparameter flow cytometry of bacteria. Methods Mol. Biol. 2004;263:33–44. doi: 10.1385/1-59259-773-4:033. [DOI] [PubMed] [Google Scholar]
- 39.Raymond M, Rousset F. An exact test for population differentiation. Hoboken, NJ: Wiley; 1995. [DOI] [PubMed] [Google Scholar]
- 40.Shi J, Biek DP. A versatile low-copy-number cloning vector derived from plasmid F. Gene. 1995;164:55–58. doi: 10.1016/0378-1119(95)00419-7. [DOI] [PubMed] [Google Scholar]
- 41.Burton NA, Johnson MD, Antczak P, Robinson A, Lund PA. Novel aspects of the acid response network of E. coli K-12 are revealed by a study of transcriptional dynamics. J. Mol. Biol. 401:726–742. doi: 10.1016/j.jmb.2010.06.054. [DOI] [PubMed] [Google Scholar]
- 42.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 43.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 44.Herrero J, Valencia A, Dopazo J. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics. 2001;17:126–136. doi: 10.1093/bioinformatics/17.2.126. [DOI] [PubMed] [Google Scholar]
- 45.Raychaudhuri S, Stuart JM, Altman RB. Principal components analysis to summarize microarray experiments: application to sporulation time series. Pac. Symp. Biocomput. 2000:455–466. doi: 10.1142/9789814447331_0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bar-Joseph Z, Gifford DK, Jaakkola TS. Fast optimal leaf ordering for hierarchical clustering. Bioinformatics. 2001;17(Suppl. 1):S22–S29. doi: 10.1093/bioinformatics/17.suppl_1.s22. [DOI] [PubMed] [Google Scholar]
- 47.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 48.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 49.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogata H, Goto S, Fujibuchi W, Kanehisa M. Computation with the KEGG pathway database. Biosystems. 1998;47:119–128. doi: 10.1016/s0303-2647(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 51.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 53.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Munch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- 56.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daub CO, Steuer R, Selbig J, Kloska S. Estimating mutual information using B-spline functions–an improved similarity measure for analysing gene expression data. BMC Bioinformatics. 2004;5:118. doi: 10.1186/1471-2105-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.George SM, Richardson LC, Pol IE, Peck MW. Effect of oxygen concentration and redox potential on recovery of sublethally heat-damaged cells of Escherichia coli O157:H7, Salmonella enteritidis and Listeria monocytogenes. J. Appl. Microbiol. 1998;84:903–909. doi: 10.1046/j.1365-2672.1998.00424.x. [DOI] [PubMed] [Google Scholar]
- 59.Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prevost MC, Sansonetti P, Tang CM. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwan WR. Survival of uropathogenic Escherichia coli in the murine urinary tract is dependent on OmpR. Microbiology. 2009;155:1832–1839. doi: 10.1099/mic.0.026187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bang IS, Kim BH, Foster JW, Park YK. OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar typhimurium. J. Bacteriol. 2000;182:2245–2252. doi: 10.1128/jb.182.8.2245-2252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bang IS, Audia JP, Park YK, Foster JW. Autoinduction of the ompR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol. Microbiol. 2002;44:1235–1250. doi: 10.1046/j.1365-2958.2002.02937.x. [DOI] [PubMed] [Google Scholar]
- 63.Bury-Mone S, Thiberge JM, Contreras M, Maitournam A, Labigne A, De Reuse H. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 2004;53:623–638. doi: 10.1111/j.1365-2958.2004.04137.x. [DOI] [PubMed] [Google Scholar]
- 64.Ni Bhriain N, Dorman CJ, Higgins CF. An overlap between osmotic and anaerobic stress responses: a potential role for DNA supercoiling in the coordinate regulation of gene expression. Mol. Microbiol. 1989;3:933–942. doi: 10.1111/j.1365-2958.1989.tb00243.x. [DOI] [PubMed] [Google Scholar]
- 65.Culham DE, Lasby B, Marangoni AG, Milner JL, Steer BA, van Nues RW, Wood JM. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J. Mol. Biol. 1993;229:268–276. doi: 10.1006/jmbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- 66.Lamark T, Kaasen I, Eshoo MW, Falkenberg P, McDougall J, Strom AR. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 1991;5:1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 67.Bossemeyer D, Borchard A, Dosch DC, Helmer GC, Epstein W, Booth IR, Bakker EP. K+-transport protein TrkA of Escherichia coli is a peripheral membrane protein that requires other trk gene products for attachment to the cytoplasmic membrane. J. Biol. Chem. 1989;264:16403–16410. [PubMed] [Google Scholar]
- 68.Bossemeyer D, Schlosser A, Bakker EP. Specific cesium transport via the Escherichia coli Kup (TrkD) K+ uptake system. J. Bacteriol. 1989;171:2219–2221. doi: 10.1128/jb.171.4.2219-2221.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kitko RD, Wilks JC, Garduque GM, Slonczewski JL. Osmolytes contribute to pH homeostasis of Escherichia coli. PLoS ONE. 2010;5:e10078. doi: 10.1371/journal.pone.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akitake B, Anishkin A, Sukharev S. The “dashpot” mechanism of stretch-dependent gating in MscS. J. Gen. Physiol. 2005;125:143–154. doi: 10.1085/jgp.200409198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sukharev SI, Martinac B, Arshavsky VY, Kung C. Two types of mechanosensitive channels in the Escherichia coli cell envelope: solubilization and functional reconstitution. Biophys. J. 1993;65:177–183. doi: 10.1016/S0006-3495(93)81044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Borgnia MJ, Kozono D, Calamita G, Maloney PC, Agre P. Functional reconstitution and characterization of AqpZ, the E. coli water channel protein. J. Mol. Biol. 1999;291:1169–1179. doi: 10.1006/jmbi.1999.3032. [DOI] [PubMed] [Google Scholar]
- 73.Booth IR, Epstein W, Giffard PM, Rowland GC. Roles of the trkB and trkC gene products of Escherichia coli in K+ transport. Biochimie. 1985;67:83–89. doi: 10.1016/s0300-9084(85)80233-9. [DOI] [PubMed] [Google Scholar]
- 74.Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.