Abstract
Various environmental oxidative stresses are sensed by redox-sensitive regulators through cysteine thiol oxidation or modification. A few zinc-containing anti-sigma (ZAS) factors in actinomycetes have been reported to respond sensitively to thiol oxidation, among which RsrA from Streptomyces coelicolor is best characterized. It forms disulfide bonds upon oxidation and releases bound SigR to activate thiol oxidative stress response genes. Even though numerous ZAS proteins exist in bacteria, features that confer redox sensitivity to a subset of these have been uncharacterized. In this study, we identified seven additional redox-sensitive ZAS factors from actinomycetes. Comparison with redox-insensitive ZAS revealed characteristic sequence patterns. Domain swapping demonstrated the significance of the region K33FEHH37FEEC41SPC44LEK47 that encompass the conserved HX3CX2C (HCC) motif. Mutational effect of each residue on diamide responsive induction of SigR target genes in vivo demonstrated that several residues, especially those that flank two cysteines (E39, E40, L45, E46), contribute to redox sensitivity. These residues are well conserved among redox-sensitive ZAS factors, and hence are proposed as redox-determinants in sensitive ZAS. H37A, C41A, C44A and F38A mutations, in contrast, compromised SigR-binding activity significantly, apparently affecting structural integrity of RsrA. The residue pattern around HCC motif could therefore serve as an indicator to predict redox-sensitive ZAS factors from sequence information.
INTRODUCTION
Bacteria respond to environmental changes primarily by changing transcription profiles in gene expression. An efficient way of changing transcriptome globally is to use alternate sigma factors, whose activities are elaborately controlled via various sensing mechanisms including anti-sigma factors (1,2). Among σ70 family of sigma factors, which contain conserved DNA-binding domains for recognizing −35 and −10 regions of promoters, group 4 or extracytoplasmic function (ECF) sigma factors, are the most abundant and diverse subclass (3–6). They constitute ∼60% of all σ70 family of sigma factors predicted from bacterial genomes. About one-third of their genes lie next to genes for anti-sigma factors containing structurally conserved sigma-binding (anti-sigma) domain (3). A prototypical example of the group 4 sigma–anti-sigma pair is RpoE-RseA of Escherichia coli (Eco) that respond to a variety of cell envelope stresses (7,8).
Among group 4 anti-sigma factors, RsrA from Streptomyces coelicolor (Sco) that binds σR (SigR) and regulates response to thiol oxidative stresses is the first reported ZAS (zinc-containing anti-sigma) factor (9,10). ChrR from Rhodobacter sphaeroides (Rsp) that binds σE and regulates response to singlet oxygen generated during photosynthesis also contains zinc (11). These two, along with many others, share a conserved sequence motif of HX3CX2C (HCC) in the anti-sigma domain (ASD), which can be called as a ZAS motif. About 40% of ECF-linked anti-sigma factors are predicted to possess this ZAS motif (3). Examples of the anti-sigma factors with ZAS motif include SigH-binding RshA of Mycobacterium tuberculosis (Mtu) that responds to thiol oxidative stress (12), SigW-binding RsiW of Bacillus subtilis (Bsu) that responds to cell envelope stresses (13,14), SigU-binding RsuA of S. coelicolor that regulates protein secretion and cell differentiation (15,16), SigL-binding RslA of M. tuberculosis (17), and a RpoE-binding ZAS factor of Neisseria meningitides (18).
In S. coelicolor, RsrA responds to thiol oxidants such as diamide by forming disulfide bond primarily between C11 and C44 residues conserved among ZAS members (19,20). Mutational studies revealed that zinc is bound through the two conserved cysteine residues in the ZAS motif (C41 and C44), and possibly through additional conserved residues H37 and C11 (10,20,21) or less conserved residues C3 and H7 (19). Zinc release accompanies disulfide bond formation, which causes a drastic conformational change to release bound SigR (9,20) to transcribe its target genes to cope with thiol oxidative stress (22–24).
Among verified or predicted ZAS, only ScoRsrA and MtuRshA have been shown experimentally to be sensitive to thiol oxidation by diamide (9,12). There have been some indications that SigH of Corynebacterium glutamicum could be regulated by its cognate anti-sigma factor in a similar way as ScoSigR or MtuSigH (25,26). However, experimental evidence for redox-sensitive regulation of its anti-sigma factor has been lacking. Another ZAS from M. tuberculosis, RslA, which regulates SigL has been proposed to be redox-sensitive on the basis of disulfide bond formation in vitro by treating 10 mM H2O2 (27). However, since SigL does not respond to oxidative stress in vivo (17), it may not respond as sensitively to thiol oxidative stress in vivo as ScoRsrA and MtuRshA do. YlaD, which binds its cognate sigma factor YlaC in B. subtilis, has been controversial in terms of redox sensitivity (28,29). However, since its inactivation was observed only under high concentration of H2O2 (2.5 mM) in vitro (29), it can be regarded relatively insensitive to thiol oxidation. Other ZAS factors such as RspChrR, BsuRsiW, and ScoRsuA have all been reported to be insensitive to thiol oxidants (11,13–15).
What makes certain ZAS factors sensitive to redox changes? Before we proceed to find clues to this question, we need more ZAS factors at hand that are sensitive to oxidation. In this study, we identified seven additional redox-sensitive ZAS proteins among close homologs of ScoRsrA in Actinomycetes. Comparing their sequences with the known redox-sensitive and insensitive ZAS proteins, and through swapping and alanine-scanning mutagenesis, we define signatures around the ZAS motif that contribute to redox sensitivity.
MATERIALS AND METHODS
Bacterial strains and culture conditions
Streptomyces coelicolor A3(2) strain M145 (wild-type) and the mutants were grown in YEME liquid medium containing 5 mM MgCl2·6H2O and 10% sucrose at 30°C by inoculating spore suspension (30). Other actinomycetes obtained either from the stock center at the Institute of Microbiology, SNU or from individual researchers were grown to prepare DNA as described below. Nocardia farcinica (ATCC 3318) was grown in GY medium (1% glucose, 1% yeast extract, pH 7.0) at 30°C. Corynebacterium glutamicum (ATCC 13032), Arthrobacter aurescens (IFO 12136) and Brevibacterium linens (ATCC 9172) were grown in brain heart infusion broth (Difco) at 30°C whereas C. diphtheriae (ATCC 11913) was grown at 37°C in the same medium. Thermobifida fusca (ATCC 27730) was grown in GYM medium (0.4% glucose, 0.4% yeast extract, 1% malt extract, 0.2% CaCO3, pH 7.2) at 50°C. M. smegmatis (IFO 3083) was grown in nutrient broth (Difco) at 37°C. Rhodococcus jostii RHA1 (from Dr William W. Mohn, University of British Columbia) was cultured in LB (1% tryptone, 0.5% yeast extract and 1% NaCl) at 30°C. Cells were grown by shaking at 200 rpm and harvested at the early stationary phase. Cultured cells of Frankia alni ACN14, donated by Dr Beth C. Mullin (University of Tennessee) were used directly to extract chromosomal DNA. The chomosomal DNA of M. tuberculosis H37Rv was a kind gift from Dr M.Y. Hahn (Yonsei University). Bacillus subtilis strains used in this work were provided by Dr Thomas Wiegert (University of Bayreuth). Bacillus subtilis and E. coli strains were cultured in LB according to standard procedures (14). The strains and plasmids used in this study were summarized in the Supplementary Table S1.
Homolog prediction and sequence analysis
BlastP algorithm was used to predict sequence homologs of ScoRsrA from NCBI nr database (ftp://ftp.ncbi.nih.gov/blast/db/). Multiple sequence alignment and generation of phylogenetic tree were done by using MEGA4 software (31). A probable structure of ScoRsrA was created based on the determined structure of R. sphaeroides ChrR (3; PDB ID:2Z2S) by using Modeller program (32), and was visualized through PyMOL (33). Sequence logos were generated by using TEXshade (34,35).
Replacement of the rsrA gene in S. coelicolor with selected homologs from other actinomycetes
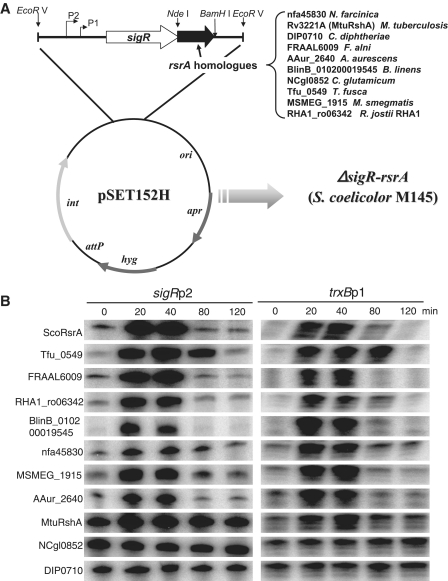
Chromosomal DNAs were prepared from various actinomycetes according to standard procedures (30), and were subjected to PCR to amplify rsrA ortholog genes. The primer pairs amplified the entire open reading frames with NdeI and BamHI sites at the start codon and at immediately downstream of the stop codon, respectively (Supplementary Table S2). The NdeI/BamHI-digested PCR products were cloned into pUC19-based recombinant plasmid (pUC19sigRrsrA-NB) that harbors S. coelicolor sigR-rsrA operon to replace the rsrA gene. The resulting hybrid sigR operon was cut with HindIII/BamHI and recloned into the EcoRV site of pSET152H that is capable of conjugation (36) (Figure 2A). The final pSET152H-based recombinant plasmids where the rsrA gene was replaced with nfa45830 (N. farcinica), Rv3221A (sigH, M. tuberculosis), DIP0710 (C. diphtheria), FRAAL6009 (F. alni), AAur_2640 (A. aurescens), BlinB_010200019545 (B. linens), NCgl0852 (C. glutamicum), Tfu_0549 (T. fusca), MSMEG_1915 (M. smegmatis) and RHA1_ro06342 (R. jostii), were confirmed by sequencing and introduced into the ΔsigR rsrA mutant of S. coelicolor through conjugation. The exconjugants that contain the recombinant plasmid in the chromosome were selected and confirmed by nucleotide sequencing.
Figure 2.
Determination of redox-senstitivity for ten RsrA homologs. (A) Cloning strategy. Genes for ten RsrA homologs were cloned downstream of the sigR gene in place of the rsrA gene of S. coelicolor as described in the text. The final pSET152H-based recombinant plasmids were introduced into the ΔsigR rsrA mutant of S. coelicolor M145 through conjugation. The resulting exconjugants contain the hybrid sigR operon in the chromosome with replaced rsrA homolog from Nocardia farcinica (nfa45830), M. tuberculosis H37Rv (Rv3221A; rshA), C. diphtheriae NCTC13129 (DIP0710), Frankia alni ACN14a (FRAAL6009), Brevibacterium linens (BlinB_010200019545), C. glutamicum ATCC13032 (NCgl0852), Thermobifida fusca YX (Tfu_0549), M. smegmatis MC2155 (MSMEG_1915), or Rhodococcus jostii RHA1 (RHA1_ro06342). (B) Diamide-sensitive induction of SigR target genes monitored by S1 mapping. The recombinant S. coelicolor strains that harbor different rsrA homologs were grown to OD600 of ∼0.3 in YEME and treated with 0.5 mM diamide for 20, 40, 80 and 120 min before cell harvest. Transcripts from SigR-dependent sigRp2 and trxBp1 promoters were analyzed by S1 mapping. The quantified values from more than three independent experiments were presented in Supplementary Figure S1.
Swapping of HCC region between ScoRsrA and BsuRsiW
The nucleotide sequences corresponding to 15 amino acids from K33 to K47 (KFEHHFEECSPCLEK) of ScoRsrA and those from V26 to H40 (VLNEHLETCEKCRKH) of BsuRsiW, harboring the conserved HCC motif (bold letters), were swapped with each other in the rsrA and rsiWΔ2 (deleted of trans-membrane and extracellular domains) genes of S. coelicolor and B. subtilis. For this purpose, a modified overlapping PCR was done (37). For swapped RsrA (S-RsrA), the primer pair S-RsrAP1 (5′tcg cg cat atg agc tgc gga3′) and S-RsrAP2 (5′CTC GCA TGT CTC CAG ATG TTC ATT TAA TAC cac gca gtc cga gtc cgg3′) were used to create the upstream half, and the pair S-RsrAP3 (5′CTG GAG ACA TGC GAG AAA TGC AGA AAG CAT tac ggg ctg gag cag gcc3′) and S-RsrAP4 (5′gac gct cag gga tcc tca gga ctc3′) were used to create the downstream half, which overlap by 15 complementary nucleotides. The capital letters indicate swapped sequence. The second PCR from partially hybridized half templates were done with the primer pair S-RsrAP1 and S-RsrAP4 to create the entire open reading frame for S-RsrA. For swapped and trunctated RsiW (ST-RsiW), the primer pair ST-RsiWP1 (5′agg tga gga tcc atg agc tgt3′) and ST-RsiWP2 (5′CGA GCA CTC CTC GAA GTG GTG CTC GAA CTT gtg ttc atc ttt tgg aag3′) were used to generate the upstream half, and the primers ST-RsiWP3 (5′TTC GAG GAG TGC TCG CCC TGC CTG GAG AAG ttt tac gag atg gag aaa3′) and ST-RsiWP4 (5′tgc gca tgc tca atg ggt tct gaa cca tct3′) were used to generate the downstream half, which overlap by 15 complementary nucleotides. The second PCR was done with ST-RsiWP1 and ST-RsiWP4. The final PCR product for S-RsrA was digested with NdeI and BamHI, and cloned into the the NdeI/BamHI site of pUC19sigRrsrA-NB to substitute the rsrA gene, followed by recloning into pSET152H and introduction to the ΔsigRrsrA mutant of S. coelicolor as described above. The final PCR product for ST-RsiW was inserted into the HindIII/SphI site of pALrsiW (14) to replace the wild-type rsiW gene. The resulting pAL-ST-rsiW plasmid was introduced into the B. subtilis 1012-1 strain, which is resistant to neomycin and spectinomycin resistant (NeoR and SpcR), to replace the SpcR gene in the lacA locus with the erythromycin resistance (ErmR) gene (14) (Supplementary Table S1). Transformants (NeoR, ErmR and SpcS) were selected to isolate 1012-ST-rsiW strain. To knock out the wild-type rsiW gene in the 1012-ST-rsiW strain, the rsiW::SpcR chromosome was introduced into the strain. The ST-rsiW strain was selected by antibiotics (NeoR, ErmR and SpcR) and confirmed by PCR and DNA sequencing. Consequently, the ST-rsiW gene at the lacA locus is under the control of an IPTG-inducible promoter (14). The recombinant B. subtilis strains were grown in LB with appropriate antibiotics, and 0.1-mM IPTG was added to induce RsiW variants. For oxidative stress, LB-grown cultures were treated with 1 mM diamide at OD600 of 0.7 for varying lengths of time before cell harvest.
Alanine-scanning mutagenesis
Each residue from K33 to K47 in ScoRsrA was replaced with alanine by site-directed mutagenesis, using alanine-scanning primers (Supplementary Table S2) and pUC19-sigRrsrA, according to the protocol provided by GENEART® site-directed mutagenesis system (Invitrogen). The mutated rsrA genes were confirmed by sequencing. The HindIII/BamHI fragments from the resulting plasmids were cloned into the EcoRV site of pSET152H, and the final recombinant plasmids were introduced into the ΔsigR rsrA mutant of S. coelicolor through conjugation. The desired exconjugants were selected and confirmed by DNA sequencing.
S1 nuclease mapping
Exponentially growing cells in YEME media were treated with different concentrations of diamide for varying lengths of time before cell harvest. RNA preparation and S1 nuclease mapping were done according to the standard procedure (30). Specific probes for SigR-responsive transcripts from the sigR and trxB genes were generated as described previously (36). For each sample, 25 μg RNA was hybridized at 50°C with gene-specific probes labeled with [γ-32P]-ATP. Following S1 nuclease treatment, the protected DNA probes were loaded on 5% polyacrylamide gel containing 7 M urea, and the radioactive signals were detected and quantified by BAS-2500 system (Fuji).
Determination of redox sensitivity of RsrA mutants
The amount of S1 mapped transcripts (response) from SigR target genes following treatment with increasing amounts of diamide (dose) for 10 min was quantified and plotted against diamide concentration. The resulting dose-response curve was fit to Lineweaver-Burk equation, in an analogous way as used in enzyme kinetics or pharmacokinetics to determine Km or LD50, respectively, (38). The concentration (induction dose) of diamide that allowed half-maximal induction of SigR regulon, termed IDIF50, for each variant of ScoRsrA was determined, and was regarded as an index to reflect redox sensitivity of each RsrA variant.
RESULTS
Excavating new redox-sensitive ZAS factors
In order to find more ZAS factors that are sensitive to thiol oxidation, close homologs of ScoRsrA in NCBI nr database (as of 8 November 2008) were searched by BlastP. Only the ASD corresponding to 103 amino acids of ScoRsrA was compared. Using a cutoff E-value of 0.3, 114 candidates were retrieved (See Supplementary Table S3 for sequence information). Most of them are from actinobacteria, except four; Rmet_0596 (Ralstonia metallidurans CH34, Betaproteobacteria), CHU_0600 (Cytophaga hutchinsonii ATCC 33406, Bacteroidetes), Ccel_0804 (Clostridium cellulolyticum H10, Firmicutes) and Ctha_1264 (Chloroherpeton thalassium ATCC 35110, Chlorobi). The list included reported ZAS factors such as RshA from M. tuberculosis (12,17,27) and a predicted ZAS RshA from C. glutamicum (25). Except for NCgl0852 from C. glutamicum, all ZAS genes are adjacent to ECF sigma factor genes. In M. tuberculosis and M. bovis strains, genes for ZAS (RshA) and sigma factor (SigH) are separated by a hypothetical ORF.
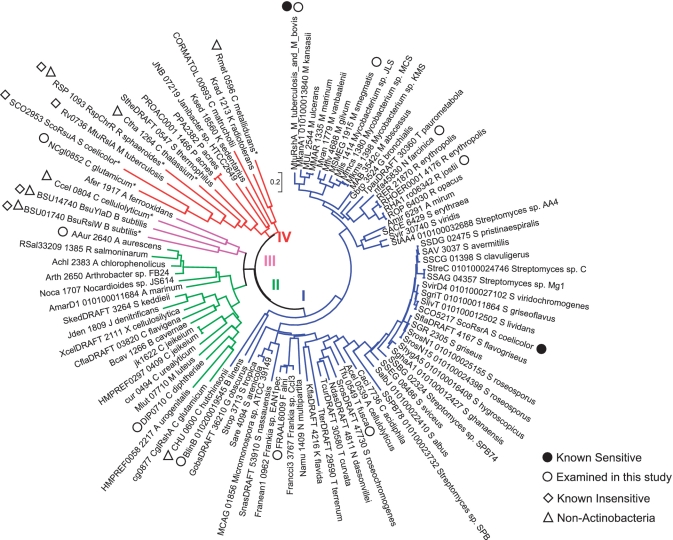
We constructed a phylogenetic tree using a neighbor-joining algorithm provided by MEGA4 program (Figure 1). Four reported redox-insensitive ZAS members were included in the phylogenetic analysis for comparison. These are RspChrR, BsuYlaD, BsuRsiW and ScoRsuA. Only the N-terminal ASD domain was included in multiple sequence alignment and phylogenetic tree construction. The phylogenetic tree demonstrated that the majority of the retrieved homologs cluster closely with ScoRsrA (Group I), next to the second cluster (Group II), which harbors CglRshA from C. glutamicum (25). Known insensitive ZAS genes were clustered within Groups III and IV. For experimental analysis, we selected nine candidates from dispersed branches of group I and group II clusters (Figure 1, open circles); Rv3221A (MtuRshA), MSMEG_1915 (M. smegmatis), BlinB_010200019545 (Brevibacterium linens), nfa45830 (Nocardia farcinica), RHA1_ro06382 (Rhodococcus jostii), FRAAL6009 (Frankia alni), Tfu_0549 (Thermobifida fusca), DIP0710 (C. diphtheria) and AAur_2640 (Arthrobacter aurescens). A candidate from a branch in Group IV, NCgl0852 (C. glutamicum), was also selected for experimental analysis. The corresponding genes were synthesized through PCR amplification of chromosomal DNA as described in ‘Materials and Methods’ section. We cloned these genes in pSET152H to replace the rsrA gene in the sigR-rsrA operon of S. coelicolor as diagramed in Figure 2A. The resulting hybrid operons were introduced into the chromosome of ΔsigRrsrA strain through the att site. If the product of the cloned gene behaves similarly to ScoRsrA, it is expected to bind SigR in S. coelicolor and keep it inactive under non-stressed condition, but will release SigR to transcribe its target genes under oxidative stress conditions. We analyzed transcripts from SigR-dependent sigRp2 and trxBp1 promoters by S1 mapping in various recombinant S. coelicolor strains before and after treatment with 0.5-mM diamide for varying lengths of time.
Figure 1.
Phylogenetic relatedness of ScoRsrA homologs. A neighbor-joining tree of RsrA homologs were constructed by using MEGA4 with default parameters. In addition to 104 homologs retrieved by BlastP, ZAS factors which have been reported to be insensitive to oxidation (marked with diamonds) were included in the tree. Only the ASD that corresponds to ScoRsrA in each entry was compared. Sequence information for all entries is provided in the Supplementary Table S3. Reported redox-senstive RsrA homologs were marked with close circles. MtuRshA_M_tuberculosis_and_M_bovis represents 25 identical sequences from M. tuberculosis or M. bovis strains. The 10 homologs whose redox-sensitivity was experimentally examined in this study were marked with open circles. Homologs from non-actinobacteria were marked with open triangles.
Figure 2B shows representative S1 results, which demonstrate that several homologs behave similarly as ScoRsrA and thus deserve to be called its orthologs. For example, Tfu_0549, FRAAL6009, RHA1_ro06342, BlinB_010200019545, nfa45830, MSMEG_1915 and AAur_2640 kept ScoSigR sufficiently inactive under non-stressed conditions (0 min), and transiently allowed SigR activation as ScoRsrA did. MtuRshA (Rv3221A) was less efficient in sequestering ScoSigR under non-stressed condition, resulting in higher basal-level expression. However, it responded to diamide induction as expected (12). On the other hand, those from Corynebacterium spp. (NCgl0852 and DIP0710) appeared inactive in binding SigR, since the basal-level expression of both sigRp2 and trxBp1 transcripts were constitutively high regardless of diamide treatment. Therefore, it is not possible to assess their redox sensitivity by our analyses. Considering a difference in GC content between Corynebacterium (54%) and S. coelicolor (72%) genomes, there also exist a possibility that not enough corynebacterial ZAS proteins are expressed due to codon bias and other post-transcriptional defects. We quantified S1 results from more than three independent experiments for each recombinant strain, and presented diamide sensitivity of each homolog as fold-induction values compared with the unstressed level (Supplementary Figure S1). This analysis, therefore, allowed us to identify seven new redox-sensitive ZAS factors, which appear to be functional orthologs of ScoRsrA in binding sigma factor SigR and responding to thiol oxidative stress.
Comparison of ASDs in redox-sensitive versus -insensitive ZAS factors
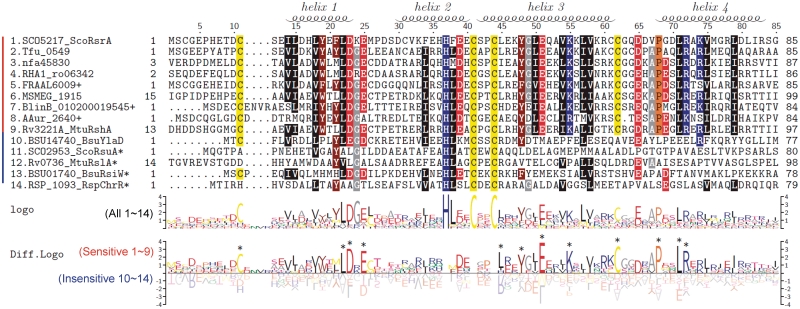
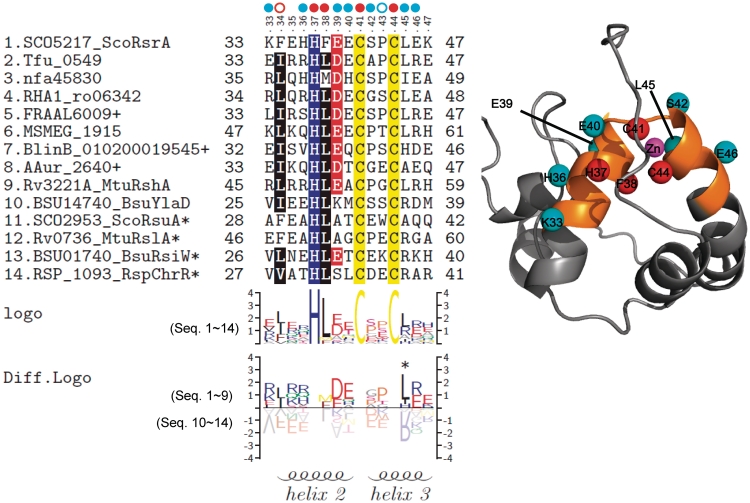
Multiple sequence alignment of ASDs from nine redox-sensitive and five insensitive ZAS factors reveal residues that are conserved across all ZAS and those that are restricted to redox-sensitive ZAS from actinomycetes (Figure 3). In order to delineate broadly functional residues from subfamily-specific residues in a more systematic way, we compared differentially conserved sequence patterns by constructing sequence logos through TEXshade package (34,35). Figure 3 demonstrated conserved sequence logo of all 14 ZAS members, and a differentially conserved logo of experimentally validated sensitive ZAS (nos 1–9) and insensitive ZAS (nos 10–14). Residues around the HCC motif (from K33 to E46 in ScoRsrA) show some differentially conserved pattern in sensitive versus insensitive ZAS proteins. There are also highly conserved residues among sensitive ZAS such as L22, D23, E25, Y48, E51, K55, C62, P68, L71 and R72. However, from sequence gazing, it is not possible to distinguish residues that are conserved across actinomycetes from those that may contribute to redox sensitivity. We therefore proceeded to examine the contribution of the region that surrounds the conserved ZAS motif to determining redox sensitivity.
Figure 3.
Multiple sequence alignment and sequence logo of ScoRsrA homologs. Mulitple sequence alignment was carried out by using MEGA4, followed by manual curation. Sequence logos were produced by using TEXshade. The secondary structure prediction is based on the structural information of RspChrR (PDB;2Z2S). For alignment, we trimmed both N-terminal and C-terminal residues that extend beyond the ScoRsrA sequence. The nucleotide sequencing identified some sequence differences from the public database, and we used modified sequences for BlinB_010200019545, AAur_2640 and FRAAL6009 (+; Supplementary Table S3). Asterisk indicates entries that contain C-terminal domains in addition to ASD.
Evaluation of the HCC region for determining redox sensitivity by domain swapping
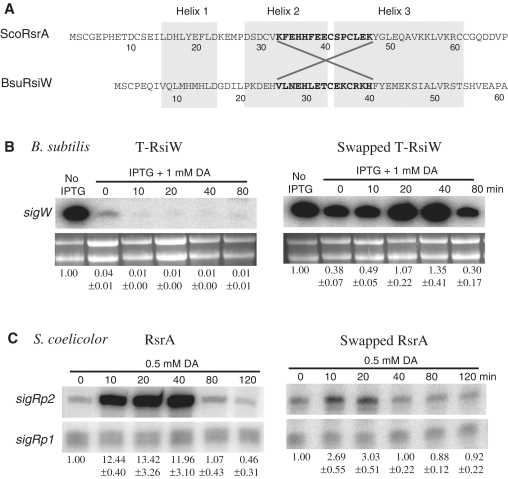
We created recombinant proteins by exchanging residues from K33 to K47 in ScoRsrA with the corresponding residues (V26 to H40) in RsiW from B. subtilis (Figure 4A). The truncated version of RsiW (T-RsiW) that contains only the N-terminal 87 amino acid of ASD without trans-membrane and extracellular domains was modified to make the recombinant. The secondary structure of the swapped product (ST-RsiW) was predicted unchanged by Jnet program (39). The recombinant B. subtilis strains that harbor either T-rsiW or ST-rsiW genes in place of the wild-type rsiW were examined to determine whether diamide can induce SigW-specific gene expression. Production of ST-RsiW or T-RsiW was controlled by IPTG (14), and the amount of auto-regulated sigW transcripts following diamide treatment was examined by S1 mapping.
Figure 4.
Swapping of HCC region between ScoRsrA and BsuRsiW. (A) Amino acid sequences and secondary structure predictions were shown for B. subtilis RsiW (BsuRsiW) and ScoRsrA. The swapped regions (K33 to K47 in ScoRsrA and V26 to H40 in BsuRsiW) were indicated. (B) The S1 mapping results of swapped ScoRsrA and T-RsiW (truncated soluble form of BsuRsiW). In B. subtilis, the Tw32Δ2 (T-RsiW) and swapped T-RsiW (ST-RsiW) strains were grown to OD600 of 0.7 in LB and harvested at 0, 10, 20, 40, 80 min after diamide (1 mM) treatment. Transcripts from sigW promoters were analyzed by S1 mapping. Quantified values of the sigW transcript bands were normalized to ribosomal RNAs in each sample, and the relative expression values from three independent experiments were presented in comparison with the level in the absence of RsiW induction by IPTG. (C) In S. coelicolor, strains that contain wild-type rsrA (MK2) or swapped rsrA (S-RsrA) in the same genetic background (Supplementary Table S1) were grown to OD600 of ∼0.3 in YEME, and were harvested at 0, 10, 20, 40, 80, 120 min after treatment with 0.5 mM diamide. Transcripts from sigRp2 promoter were analyzed by S1 mapping. Quantified value of the SigR-dependent sigRp2 signal was normalized to the constitutive sigRp1 level in each sample. Relative expression values from more than three independent experiments were presented in comparison with the un-induced level.
Figure 4B demonstrates that when T-RsiW is induced, sigW expression is drastically reduced, confirming the ability of T-RsiW to bind and sequester SigW. Treatment with diamide did not activate SigW at all, and the sigW gene expression was even lowered compared with the unstressed level. Production of ST-RsiW reduced the sigW gene expression by ∼60%, indicating some partial binding of SigW by ST-RsiW. When diamide was treated, the expression level increased up to about 4-fold after 40 min of treatment, and then decreased to the basal level at 80 min. This result demonstrates that the HCC region of ScoRsrA conferred redox sensitivity to the insensitive RsiW.
We then examined the converse swapping in S. coelicolor. In the presence of wild-type RsrA, SigR-target transcripts (sigRp2) increased dramatically after diamide treatment, by 12- to 13-fold as expected (Figure 4C). However, in the presence of S-RsrA, the diamide induction was significantly reduced to only ∼3-fold level. The low basal level of sigRp2 transcript in untreated sample (0 min) indicates that S-RsrA binds SigR nearly as well as the wild-type. This result again clearly indicates that the HCC region of RsrA is a critical determinant of redox sensitivity. The residual low sensitivity of S-RsrA appears to reflect contribution from other regions in conferring redox sensitivity.
Determination of residues that contribute to redox sensitivity in the HCC region
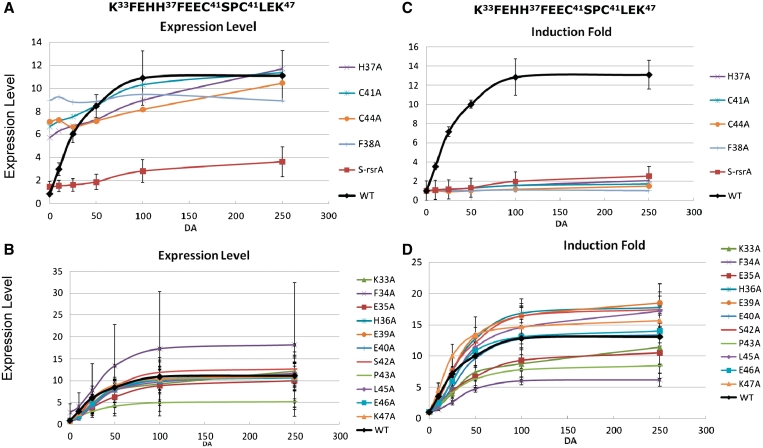
In order to find residue(s) that confer ScoRsrA, its characteristic redox sensitivity, we changed each of the 15 residues in the swapped HCC region (from K33 to K47) to alanine, and introduced the mutated rsrA gene and the upstream sigR to ΔsigR-rsrA strain. As a way to assess sensitivity to diamide, we treated each cell samples with increasing amounts of diamide from 0 to 0.25 mM for 10 min, and then monitored the level of SigR-dependent transcripts (sigRp2) by S1 mapping. The quantified results were plotted either as expression levels or induction fold against diamide concentrations. Figure 5 exhibits profiles of the sigRp2 expression in the mutants, in comparison with that in the wild-type (black curve). It was evident from the graph that some mutations affected SigR-specific expression drastically in such a way to elevate the un-stressed level and lower fold-induction accordingly (Figure 5A and C). These mutations are H37A, C41A, C44A and F38A. These residues are those that are very well conserved in all ZAS, including F38 which shares similar hydrophobicity with the conserved leucine at this position (Figure 3). Since the highly elevated level of basal expression reflects free dissociated SigR, these mutations must have damaged the integrity of the overall structure of RsrA. The swapped S-RsrA was also examined by this assay, and demonstrated very low induction, consistent with the observation in Figure 4C.
Figure 5.
Redox sensitivies of Ala-scanned ScoRsrA mutants. Each of the 15 residues from K33 to K47 in ScoRsrA was changed to alanine in recombinant S. coelicolor strains that harbor mutated rsrA in sigR-rsrA operon inserted at the att site of the ΔsigR-rsrA strain. Cells were grown to OD600 of ∼0.3 in YEME and treated with varying concentrations of diamide (0, 10, 25, 50, 100, 250 μM) for 10 min. Following S1 mapping, the normalized sigRp2 signal values were plotted either as expression levels (A and B) or induction fold (C and D) against diamide concentrations. For expression values, the uninduced basal level of the wild-type was set as 1.0. For induction folds, the uninduced levels of all strains were set as 1.0. Mutants that constitutively induced SigR-specific expression were presented separately (panels A and C) from those that affected primarily sensitivity to diamide (panels B and D). Quantified values for induction folds were presented in Table 1.
The rest of the mutants exhibited relatively subtle effects (Figure 5B). Under non-stressed condition, they showed low basal-level expression of sigRp2 comparable with the wild-type, indicating that the mutations did not damage the structural integrity or the activity of RsrA to bind SigR. Regarding diamide sensitivity, some behaved almost like the wild-type whereas the others showed less sensitive induction. F34A produced widely varying results. In order to normalize different levels of basal expression and maximum induction level, we re-plotted the expression data of Figure 5A and B to present changes in induction fold for each mutant (Figure 5C and D). The graphs in Figure 5B and D can be regarded as dose-dependent response curves typically observed in enzyme- or pharmaco-kinetic analyses. We therefore applied Lineweaver–Burk equations to fit the data and calculated the concentration of diamide that caused 50% maximal induction for each mutant, defined as IDIF50 (induction dose at 50% maximum induction fold) or IDEL50 (induction dose at 50% maximum expression level). IDEL50 and IDIF50 values were very close to each other. The quantified values for the basal level sigRp2 expression and IDIF50 were summarized in Table 1. Compared with the wild-type (IDIF50 of 39.5 µM diamide), several mutants showed significant increases in IDIF50 values. For example, K33A, H36A, E39A, E40A, S42A, L45A and E46A caused elevation of IDIF-50 by more than 2-fold, implying that these residues contribute to redox sensitivity. Relatively larger effects were observed in L45A, E40A, E46A and E39A mutants by 7.7, 4.8, 4.0 and 3.2-fold increases in IDIF50, respectively. These residues flank the two conserved cysteines (C41 and C44) and show pronounced conservation among sensitive ZAS members.
Table 1.
SigR-binding activity and diamide sensitivity of RsrA variants
| RsrA | BLa | IDIF50 (µM)b | Relative IDIF50c | RsrA | BL | IDIF50 (µM) | Relative IDIF50 |
|---|---|---|---|---|---|---|---|
| WT | 0.8 ± 0.1 | 39.5 ± 3.5 | 1.0 | C41A | 6.7 | ND | ND |
| K33A | 1.1 ± 0.1 | 79.6 ± 7.2 | 2.0 | S42A | 0.7 ± 0.1 | 110.3 ± 21.9 | 2.8 |
| F34A | 2.8 ± 1.9 | 49.9 ± 6.8 | 1.3 | P43A | 0.4 ± 0.3 | 45.5 ± 12.3 | 1.2 |
| E35A | 0.9 ± 0.5 | 54.8 ± 38.3 | 1.4 | C44A | 7.1 | ND | ND |
| H36A | 0.6 ± 0.2 | 103.8 ± 14.0 | 2.6 | L45A | 0.7 ± 0.3 | 303.3 ± 86.7 | 7.7 |
| H37A | 5.7 | ND | ND | E46A | 0.8 ± 0.3 | 158.6 ± 34.1 | 4.0 |
| F38A | 8.0 ± 3.1 | ND | ND | K47A | 0.7 ± 0.4 | 38.5 ± 16.8 | 1.0 |
| E39A | 0.6 ± 0.2 | 128.3 ± 42.4 | 3.2 | S-RsrA | 1.4 ± 0.5 | 9.1 ± 0.6 | 0.2 |
| E40A | 0.9 ± 0.3 | 191.4 ± 146.6 | 4.8 |
aThe basal-level (BL) expression of sigRp2 was normalized in each sample to the level of constitutive sigRp1 expression in the absence of diamide. For each sample, the value was presented as an average ± standard deviation from more than three independent experiments except for H37A, H41A and H44A.
bThe diamide concentration in micromolar (induction dose; ID) needed to induce sigRp2 to the level corresponding to half-maximum induction fold (IF50). Each value represents an average ± standard deviation from more than three independent experiments. ND, not-determined.
cRelative IDIF50 is in comparison with the wild-type value set as 1.0.
DISCUSSION
Extended list of redox-sensitive ZAS
Among 9 RsrA homologs selected for experimental analysis from dispersed branches of a phylogenetic tree composed of 114 homologs of ScoRsrA, we identified seven new redox-sensitive ZAS proteins from M. smegmatis, B. linens, N. farcinica, R. jostii, F. alni and T. fusca. Two candidates from C. glutamicum and C. diphtheria were not eligible for our assay, since they appear incapable of binding ScoSigR under unstressed conditions, resulting in constitutive expression of SigR target genes. Considering the difference in GC content, there is also a possibility that not enough proteins were expressed. The high success rate of finding sensitive ZAS leads to a prediction that the majority of the retrieved homologs, especially those of groups I and II (Figure 1), are likely to be functional orthologs of ScoRsrA, regulating SigR-like sigma factor encoded from a neighboring gene, in response to thiol oxidative stresses. Since our assay was restricted by the ScoSigR-binding ability of the homolog, only close homologs that can bind ScoRsrA can be evaluated for their redox sensitivity. If a more generalized assay that utilizes cognate sigma factor were available, it is expected that a broader spectrum of sensitive ZAS proteins could be evaluated.
Contribution of zinc-binding domain to determine redox sensitivity
Sequence comparison revealed a number of conserved residues among sensitive ZAS (Figure 3). However, since all the sensitive ZAS proteins are from actinomycetes, it is not possible to distinguish the residues that contribute to redox sensitivity from those conserved among actinomycetous RsrA homologs. Swapping of zinc-binding regions between sensitive (ScoRsrA) and insensitive (BsuRsiW) ZAS demonstrated that the 15 amino acid stretch centered around the HCC motif contributed significantly to determining sensitivity. Estimation of redox sensitivity of each Ala-substituted mutant of this region revealed the importance of residues surrounding the HCC motif (Figure 5 and Table 1). In Figure 6, we presented the position of these residues in a 3D-structural model of ScoRsrA based on the reported structure of RspChrR (3).
Figure 6.
The conserved sequence pattern and the position of the residues near the HCC motif in 3D structural model of ScoRsrA. Multiple sequence alignment and sequence logos for the 15 amino acid region examined for mutational analyses were presented as in Figure 3. A 3D structural model was produced through homology modeling, using RspChrR structure (PDB; 2Z2S) as a template. Residues that affected redox-sensitivity (IDIF50) by more than 2-fold were indicated with cyan circles, whereas those that affected structural integrity of ScoRsrA were indicated with red ones. Mutations that partially affected ScoRsrA structure by decreasing interaction with SigR (F34, red open circle) or increasing interaction with SigR (P43, blue open circle) were also indicated. In the structure model, Cβ atoms of effective residues were highlighted as spheres with indicated colors. The mutated 15 amino acid region in 3D structure was highlighted in orange color.
Mutation of the universally conserved residues across ZAS (H37, C41 and C44), which are ligands of zinc in RspChrR (3), and are reported to serve a similar role in ScoRsrA (20,21), all caused constitutive expression of the SigR target gene. Another highly conserved hydrophobic residue F38 (leucine in most other cases) showed similar effect. Based on biochemical analyses of SigR binding and disulfide bond formation, loss of zinc in ScoRsrA is known to decrease its binding affinity to ScoSigR (19) and to render RsrA more labile to oxidation (20). For ScoRsrA, whose structural information is not yet available, the zinc-binding residues are not well resolved. The extended X-ray absorption fine structure (EXAFS) spectroscopy of a mutant form of RsrA, where non-conserved cysteines (C3, C31, C61, C62) were all substituted with alanines, suggested that the bound zinc closely interacts with C11, H37, C41 and C44, in a way similarly observed in the structure of RspChrR as observed previously (10,21). Assessment of zinc content in various ScoRsrA mutants suggested the possibility of C3 and H7 in the extended N-terminal loop, in addition to C41 and C44, to participate in zinc binding (19). Even though the H37-corresponding residue in ChrR structure (H31) serves as a zinc ligand, its mutation did not affect anti-sigma activity of ChrR, implying that it may contribute marginally to zinc-binding affinity (3). Similarly weak interaction between H37 and zinc has been reported (19,21). Therefore, the conserved H37 may contribute further in determining ScoRsrA structure beyond serving as a zinc-binding ligand. Based on these studies, it is highly likely that C41 and C44 are involved in zinc binding. However, identification of additional zinc ligands in ScoRsrA requires further studies. Whether zinc binds to more than two alternative sites, and whether these alternative forms are at equilibrium in the cell remain open for deeper investigation. More systematic analyses combined with structural studies are in need to get better understanding of zinc-binding in ScoRsrA.
The residues, whose mutation enhanced IDIF50 by more than 2-fold, were marked in Figure 6 (sky blue dots; K33, H36, E39, E40, S42, L45, E46). The most pronounced effect was observed for L45A, E40A, E46A and E39A, which increased IDIF50 by 7.7-, 4.8-, 4.0- and 3.2-fold, respectively. These four residues immediately flank the two conserved cysteines (C41 and C44), and show pronounced differential conservation among sensitive ZAS members (Figure 6). In this respect, the presence of negatively charged residues (DE) preceding C41, and a hydrophobic (L) and a charged residue (R/E) following C44, could be a critical determinant of redox sensitivity. The P43A mutation did not change sensitivity significantly, but lowered the basal level by about 2-fold (Table 1). This suggests that the mutation could have caused tighter binding of SigR under unstressed condition.
Factors that contribute to the reactivity of zinc-coordinated protein thiols
RsrA is an example of redox sensors containing reactive zinc-cysteine centers (40,41). Protein thiols with low pKa value get easily deprotonated, and exist as an anionic thiolate form. Zinc can lower pKa of the protein thiol through stabilizing the thiolate form, and thus is capable of increasing the reactivity of cysteine thiols toward electrophilic oxidants and alkylating compounds (42). However, the actual reactivity of the zinc-thiol (or thiolate) center also depends on its structural environment that includes hydrogen bonding, dielectric properties, electrostatic screening, and protein packing (42–44). For example, an increase in the steric and electrostatic shielding of anionic zinc cores in zinc fingers is correlated with a decrease in their reactivity (43).
Previous observation that de-metallated RsrA becomes more vulnerable to oxidation in vitro can be interpreted to suggest that (i) zinc binding in RsrA protects thiolates, especially that of C41, from labile oxidation, and/or (ii) the reactive thiolate in apo-RsrA is more surface-exposed due to structural relaxation compared with the zinc-bound form. In this respect, the binding affinity as well as the flexibility in choosing ligands of zinc are likely to affect the reactivity of active-site cysteine (C41 in ScoRsrA), which most likely is located in the middle of the loop between helixes 2 and 3 of the ASD (3). Therefore, one can hypothesize that the zinc binding to (H)CC motif in sensitive ZAS could be weaker and/or more flexible relative to that in insensitive ZAS, due to the specified residue environment of the motif. Further comparative studies of sensitive versus insensitive ZAS are in need to find whether any differences in zinc-binding properties, if present, contribute to determine redox sensitivity.
An extended component of structural environment involves protein-protein interaction with interacting partners. Studies on the sequence variation of dipeptide sequence in the CXXC motifs of thioredoxin and related proteins revealed that cysteine reactivity depends not only on pKa and redox potential values, but also on interacting partners (45–47). Since nearly all RsrA in the reduced cytoplasm is thought to exist as a bound complex with SigR, the interacting SigR environment around the reactive zinc–thiol center can also contribute to redox sensitivity. The HCC motif, which spans from the C-terminal end of helix 2 to the N-terminal end of helix 3, is modeled to be relatively exposed to the surface, away from the interacting sigma factor (3). However, the remaining residues of helixes 2 and 3, especially helix 3, are thought to interact heavily with the bound sigma factor. In the absence of structural information on SigR-RsrA complex, it is not possible to delineate residues that interact with SigR. A structural model based on the template of RpoE-ChrR complex structure suggests that S42 and E46 in helix 3 are positioned toward the SigR-interacting face (C.O. Seok, personal communication). However, the relatively distant sequence relatedness between ScoSigR and RspRpoE hinders precise modeling. To assess the contribution from the interacting sigma factor, precise structural information on SigR-RsrA interaction is in need.
Extensive studies on finding determinants of reactivity in protein thiols of thioredoxin family demonstrated the contribution of dipeptide (XX) residues in the CXXC motif in determining pKa, redox potential, and possibly interaction with partner proteins (47,48). A sensitive motif for cysteine S-nitrosylation in proteins that are nitrosylated by S-nitroso-glutathione has also been proposed (49). In this study, a new type of redox-sensitive motif in zinc-bound ASD has been suggested. Considering the growing list of redox-regulated proteins, where disulfide bonds serve as switches for modulating various functional aspects such as localization, interaction and stability, we expect this study to add a new insight in finding novel redox-sensitive proteins (41). Further biochemical and structural analyses will reveal the mechanism behind the redox-determining function of specific residues in this class of proteins, and give insight in understanding the mechanism behind redox-sensing in other zinc proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NRL of Molecular Microbiology to JH Roe (NRF-2009-0079278); Post-doctoral and doctoral fellowships, respectively, from the second-stage BK21 Program for Life Sciences at SNU (to Y.-G.J. and J.-S.Y). Funding for open access charge: Office of Research Affairs, Seoul National University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Drs William Mohn, Beth Mullin, Thomas Wiegert and Mi-Young Hahn for providing strains, plasmids, or chromosomal DNA.
REFERENCES
- 1.Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr. Opin. Microbiol. 2008;11:121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helmann JD. Regulation by alternative sigma factors. In: Storz G, Hengge R, editors. Bacterial Stress Responses. 2nd edn. Washington: ASM press; 2010. pp. 31–41. [Google Scholar]
- 3.Campbell EA, Greenwell R, Anthony JR, Wang S, Lim L, Das K, Sofia HJ, Donohue TJ, Darst SA. A conserved structural module regulates transcriptional responses to diverse stress signals in bacteria. Mol. Cell. 2007;27:793–805. doi: 10.1016/j.molcel.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 5.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 6.Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayden JD, Ades SE. The extracytoplasmic stress factor, sigmaE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One. 2008;3:e1573. doi: 10.1371/journal.pone.0001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ades SE, Hayden JD, Laubacher ME. Envelope stress. In: Storz G, Hengge R, editors. Bacterial Stress Responses. 2nd edn. Washington: ASM press; 2010. pp. 115–131. [Google Scholar]
- 9.Kang JG, Paget MS, Seok YJ, Hahn MY, Bae JB, Hahn JS, Kleanthous C, Buttner MJ, Roe JH. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 1999;18:4292–4298. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paget MS, Bae JB, Hahn MY, Li W, Kleanthous C, Roe JH, Buttner MJ. Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch. Mol. Microbiol. 2001;39:1036–1047. doi: 10.1046/j.1365-2958.2001.02298.x. [DOI] [PubMed] [Google Scholar]
- 11.Anthony JR, Warczak KL, Donohue TJ. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc. Natl Acad. Sci. USA. 2005;102:6502–6507. doi: 10.1073/pnas.0502225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song T, Dove SL, Lee KH, Husson RN. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 2003;50:949–959. doi: 10.1046/j.1365-2958.2003.03739.x. [DOI] [PubMed] [Google Scholar]
- 13.Cao M, Wang T, Ye R, Helmann JD. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol. Microbiol. 2002;45:1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x. [DOI] [PubMed] [Google Scholar]
- 14.Schobel S, Zellmeier S, Schumann W, Wiegert T. The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 2004;52:1091–1105. doi: 10.1111/j.1365-2958.2004.04031.x. [DOI] [PubMed] [Google Scholar]
- 15.Gehring AM, Yoo NJ, Losick R. RNA polymerase sigma factor that blocks morphological differentiation by Streptomyces coelicolor. J. Bacteriol. 2001;183:5991–5996. doi: 10.1128/JB.183.20.5991-5996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon ND, Ottaviano GL, Connell SE, Tobkin GV, Son CH, Shterental S, Gehring AM. Secreted-protein response to sigmaU activity in Streptomyces coelicolor. J. Bacteriol. 2008;190:894–904. doi: 10.1128/JB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn MY, Raman S, Anaya M, Husson RN. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J. Bacteriol. 2005;187:7062–7071. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopman CT, Speijer D, van der Ende A, Pannekoek Y. Identification of a novel anti-sigmaE factor in Neisseria meningitidis. BMC Microbiol. 2010;10:164. doi: 10.1186/1471-2180-10-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae JB, Park JH, Hahn MY, Kim MS, Roe JH. Redox-dependent changes in RsrA, an anti-sigma factor in Streptomyces coelicolor: zinc release and disulfide bond formation. J. Mol. Biol. 2004;335:425–435. doi: 10.1016/j.jmb.2003.10.065. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Bottrill AR, Bibb MJ, Buttner MJ, Paget MS, Kleanthous C. The Role of zinc in the disulphide stress-regulated anti-sigma factor RsrA from Streptomyces coelicolor. J. Mol. Biol. 2003;333:461–472. doi: 10.1016/j.jmb.2003.08.038. [DOI] [PubMed] [Google Scholar]
- 21.Zdanowski K, Doughty P, Jakimowicz P, O'Hara L, Buttner MJ, Paget MS, Kleanthous C. Assignment of the zinc ligands in RsrA, a redox-sensing ZAS protein from Streptomyces coelicolor. Biochemistry. 2006;45:8294–8300. doi: 10.1021/bi060711v. [DOI] [PubMed] [Google Scholar]
- 22.Kallifidas D, Thomas D, Doughty P, Paget MS. The sigmaR regulon of Streptomyces coelicolor A32 reveals a key role in protein quality control during disulphide stress. Microbiology. 2010;156:1661–1672. doi: 10.1099/mic.0.037804-0. [DOI] [PubMed] [Google Scholar]
- 23.Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigmaR regulon. Mol. Microbiol. 2001;42:1007–1020. doi: 10.1046/j.1365-2958.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- 24.Park JH, Roe JH. Mycothiol regulates and is regulated by a thiol-specific antisigma factor RsrA and sigma(R) in Streptomyces coelicolor. Mol. Microbiol. 2008;68:861–870. doi: 10.1111/j.1365-2958.2008.06191.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim TH, Kim HJ, Park JS, Kim Y, Kim P, Lee HS. Functional analysis of sigH expression in Corynebacterium glutamicum. Biochem. Biophys. Res. Commun. 2005;331:1542–1547. doi: 10.1016/j.bbrc.2005.04.073. [DOI] [PubMed] [Google Scholar]
- 26.Nakunst D, Larisch C, Huser AT, Tauch A, Puhler A, Kalinowski J. The extracytoplasmic function-type sigma factor SigM of Corynebacterium glutamicum ATCC 13032 is involved in transcription of disulfide stress-related genes. J. Bacteriol. 2007;189:4696–4707. doi: 10.1128/JB.00382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thakur KG, Praveena T, Gopal B. Structural and biochemical bases for the redox sensitivity of Mycobacterium tuberculosis RslA. J. Mol. Biol. 2010;397:1199–1208. doi: 10.1016/j.jmb.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto T, Nakanishi K, Asai K, Sadaie Y. Transcriptional analysis of the ylaABCD operon of Bacillus subtilis encoding a sigma factor of extracytoplasmic function family. Genes Genet. Syst. 2005;80:385–393. doi: 10.1266/ggs.80.385. [DOI] [PubMed] [Google Scholar]
- 29.Ryu HB, Shin I, Yim HS, Kang SO. YlaC is an extracytoplasmic function (ECF) sigma factor contributing to hydrogen peroxide resistance in Bacillus subtilis. J. Microbiol. 2006;44:206–216. [PubMed] [Google Scholar]
- 30.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich Research Park, Colney, Norwich NR4 7UH, UK: John Innes Foundation; 2000. [Google Scholar]
- 31.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 32.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 33.Schrodinger LLC. The PyMOL Molecular Graphics System. 2008 Version 1.1r1 from http://www.pymol.org. [Google Scholar]
- 34.Beitz E. TEXshade: shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics. 2000;16:135–139. doi: 10.1093/bioinformatics/16.2.135. [DOI] [PubMed] [Google Scholar]
- 35.Schuster-Bockler B, Schultz J, Rahmann S. HMM Logos for visualization of protein families. BMC Bioinformatics. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim MS, Hahn MY, Cho Y, Cho SN, Roe JH. Positive and negative feedback regulatory loops of thiol-oxidative stress response mediated by an unstable isoform of sigmaR in actinomycetes. Mol. Microbiol. 2009;73:815–825. doi: 10.1111/j.1365-2958.2009.06824.x. [DOI] [PubMed] [Google Scholar]
- 37.Liu Q, Thorland EC, Heit JA, Sommer SS. Overlapping PCR for bidirectional PCR amplification of specific alleles: a rapid one-tube method for simultaneously differentiating homozygotes and heterozygotes. Genome Res. 1997;7:389–398. doi: 10.1101/gr.7.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell. Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 39.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 40.Ilbert M, Graf PC, Jakob U. Zinc center as redox switch–new function for an old motif. Antioxid. Redox. Signal. 2006;8:835–846. doi: 10.1089/ars.2006.8.835. [DOI] [PubMed] [Google Scholar]
- 41.Wouters MA, Fan SW, Haworth NL. Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxid. Redox. Signal. 2010;12:53–91. doi: 10.1089/ars.2009.2510. [DOI] [PubMed] [Google Scholar]
- 42.Maret W. Zinc coordination environments in proteins as redox sensors and signal transducers. Antioxid. Redox. Signal. 2006;8:1419–1441. doi: 10.1089/ars.2006.8.1419. [DOI] [PubMed] [Google Scholar]
- 43.Maynard AT, Covell DG. Reactivity of zinc finger cores: analysis of protein packing and electrostatic screening. J. Am. Chem. Soc. 2001;123:1047–1058. doi: 10.1021/ja0011616. [DOI] [PubMed] [Google Scholar]
- 44.Smith JN, Shirin Z, Carrano CJ. Control of thiolate nucleophilicity and specificity in zinc metalloproteins by hydrogen bonding: lessons from model compound studies. J. Am. Chem. Soc. 2003;125:868–869. doi: 10.1021/ja029418i. [DOI] [PubMed] [Google Scholar]
- 45.Lin TY. Protein-protein interaction as a powering source of oxidoreductive reactivity. Mol. Biosyst. 2010;6:1454–1462. doi: 10.1039/b927132e. [DOI] [PubMed] [Google Scholar]
- 46.Lin TY, Chen TS. A positive charge at position 33 of thioredoxin primarily affects its interaction with other proteins but not redox potential. Biochemistry. 2004;43:945–952. doi: 10.1021/bi0355138. [DOI] [PubMed] [Google Scholar]
- 47.Quan S, Schneider I, Pan J, Von Hacht A, Bardwell JC. The CXXC motif is more than a redox rheostat. J. Biol. Chem. 2007;282:28823–28833. doi: 10.1074/jbc.M705291200. [DOI] [PubMed] [Google Scholar]
- 48.Chivers PT, Prehoda KE, Raines RT. The CXXC motif: a rheostat in the active site. Biochemistry. 1997;36:4061–4066. doi: 10.1021/bi9628580. [DOI] [PubMed] [Google Scholar]
- 49.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell. Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.