Abstract
Planarians can regenerate any missing body part, requiring mechanisms for the production of organ systems in the adult, including their prominent tubule-based filtration excretory system called protonephridia. Here, we identify a set of genes, Six1/2-2, POU2/3, hunchback, Eya and Sall, that encode transcription regulatory proteins that are required for planarian protonephridia regeneration. During regeneration, planarian stem cells are induced to form a cell population in regeneration blastemas expressing Six1/2-2, POU2/3, Eya, Sall and Osr that is required for excretory system formation. POU2/3 and Six1/2-2 are essential for these precursor cells to form. Eya, Six1/2-2, Sall, Osr and POU2/3-related genes are required for vertebrate kidney development. We determined that planarian and vertebrate excretory cells express homologous proteins involved in reabsorption and waste modification. Furthermore, we identified novel nephridia genes. Our results identify a transcriptional program and cellular mechanisms for the regeneration of an excretory organ and suggest that metazoan excretory systems are regulated by genetic programs that share a common evolutionary origin.
Keywords: Kidney, Neoblast, Planarian, Protonephridia, Regeneration, Stem cell
INTRODUCTION
The planarian Schmidtea mediterranea can regenerate from small body fragments (Reddien and Sánchez Alvarado, 2004). This robust regenerative capacity, together with emerging molecular tools for planarian gene function studies, make these animals a powerful system for studying the regeneration of cell types, tissues and even entire organ systems. In addition to uncovering regenerative mechanisms, studying the regeneration of planarian cell types could also identify broadly used developmental mechanisms.
Here, we investigate the genetic mechanisms underlying regeneration of the planarian protonephridial system, which is involved in waste excretion and osmoregulation. Excretory systems consist of a filtration surface and a tubule, which modifies the ultrafiltrate by reabsorption and secretion and connects to the animal exterior (Ruppert and Smith, 1988; Bartolomaeus and Ax, 1992; Ruppert, 1994). Protonephridia consist of blind tubules ending in a terminal cell (Ruppert and Smith, 1988; Bartolomaeus and Ax, 1992), in which beating cilia generate negative pressure allowing filtration from the extracellular space into the tubule lumen through membrane fenestrations (Ruppert and Smith, 1988; Bartolomaeus and Ax, 1992). Metanephridia involve specialized epithelial cells called podocytes, which filter pressurized fluid from circulatory systems into a tubule (Quaggin and Kreidberg, 2008). Within the Bilateria, most deuterostomes have metanephridia, whereas both protonephridia and metanephridia are found within protostomes (Bartolomaeus and Ax, 1992) (see Fig. S1 in the supplementary material). In addition to morphological differences at the filtration site, the two nephridial systems can also have different germ layer origins; most invertebrate nephridia (protonephridia and metanephridia) have an ectoderm origin, whereas metanephridia in vertebrates have a mesoderm origin. These characteristics, together with the presence of both systems in different developmental stages of some invertebrates, have generated disparate conclusions regarding the homology of nephridia (Wilson and Webster, 1974; Bartolomaeus and Ax, 1992; Ruppert, 1994). Therefore, despite the importance and ubiquity of excretory systems, their evolutionary origin remains unclear.
The vertebrate kidney is the most extensively studied metanephridial system (Dressler, 2006; Dressler, 2009; Costantini and Kopan, 2010). In the mouse, nephrons are the basic functional units of kidneys and develop from the intermediate mesoderm, and numerous genes involved in their development have been characterized (Dressler, 2009). Protonephridia are mostly found in small adult animals or larvae of several phyla, including Nermertea, Gastrotricha, Platyhelminthes, Mollusca and Annelida (Ruppert and Smith, 1988; Bartolomaeus and Ax, 1992). The two classical protostome model organisms, Caenorhabditis elegans and Drosophila melanogaster, have specialized excretory systems with only one excretory cell in the former (Nelson et al., 1983) and a separation of ultrafiltration from excretion in the latter (Denholm and Skaer, 2009). Therefore, despite morphological and ultrastructural studies of protonephridial systems in several organisms, our understanding of the regulatory genes required for protonephridia formation is limited.
Most animals share a discrete and relatively small number of organ systems, such as eyes, muscle, gut, heart and an excretory system. Characterization of regulatory programs required for the formation of these major tissues/organs is a central challenge in developmental biology. The identity or existence of a common genetic regulatory program for excretory systems has not been established. Here, we identify a regeneration program for the planarian tubule-based filtration excretory system. Our results suggest that there existed a genetic program for nephridia formation in the common ancestor of the Bilateria with components still responsible for the formation and function of varied excretory systems in extant bilaterians.
MATERIALS AND METHODS
RNAi experiments
RT-PCR and RACE were used to amplify and determine gene sequences. Genes were cloned into pPR244 for RNA interference (RNAi) as described (Reddien et al., 2005a). cDNAs used to generate RNA probes were: SAAH-aaa15g04 (carbonic anhydrase VII, CA), H.85.1h (alkaline phosphatase), H.2.8b (rootletin), H14.9d (EF-hand domain) and SAAH-aaa96a06 (ring finger). Six1/2-2, POU2/3, Sall and hunchback RNAi was performed by feeding. Bacterial culture (10 ml) was pelleted and resuspended in 30 μl of liver. Animals were fed on days 0, 4 and 7. For regeneration, animals were amputated at day 8 and scored at day 15 (Six1/2-2 RNAi), or fed and cut again at day 16 and scored at day 23 (POU2/3 and hunchback RNAi). For intact animals undergoing tissue turnover, feedings were on days 0, 4 and 7 for Six1/2-2 with an additional feeding on day 14 for POU2/3 and hunchback, seven additional feedings for Eya, and four extra feedings for Sall. For RNAi by injection, dsRNA from in vitro transcription reactions (Promega) was injected on days 0, 1 and 2, with animals amputated on day 2. Six days later, animals were injected and amputated again, with fixation six days thereafter.
In situ hybridization and immunolabeling
In situ hybridization and immunolabeling were performed as described (Pearson et al., 2009; Glazer et al., 2010). Numbers of carbonic anhydrase (CA)-expressing cells and tubule branches in clusters were counted in tails of intact animals and in the anterior and posterior blastemas of regenerating trunk pieces. X1 cells were isolated as described (Scimone et al., 2010), adhered to poly-l-lysine-coated coverslips and fixed in 4% paraformaldehyde for 20 minutes. Images were obtained using a Zeiss AxioImager with Apotome or a Zeiss LSM710 confocal microscope.
Transmission electron microscopy
Animals were fixed in 2.5% glutaraldehyde, 3% paraformaldehyde with 5% sucrose in 0.1 M sodium cacodylate buffer (pH 7.4) for one hour, then post-fixed in 1% OsO4 in veronal-acetate buffer. Animals were stained overnight with 0.5% uranyl acetate in veronal-acetate buffer (pH 6.0), dehydrated, and embedded in Spurr's resin. Sections were cut on a Reichert Ultracut E microtome with a Diatome diamond knife at a thickness setting of 50 nm then stained with 2% uranyl acetate and lead citrate. The sections were examined using a FEI Tecnai spirit at 80 KV and photographed with an AMT CCD camera.
Microarray experiments
RNA from Six1/2-2(RNAi) and POU2/3(RNAi) animals was harvested with Trizol at days 8 and 15 after RNAi initiation for the former and days 15 and 21 for the latter. Three biological replicates were used. Cy3- and Cy5-labeled cRNA was prepared using a QuickAmp Labeling Kit (Agilent) starting with 1 μg total RNA. Agilent custom planarian 4×44,000 expression arrays were hybridized according the manufacturer's instructions and scanned using an Agilent DNA microarray scanner. Array images were quantified and statistical significance of differential expression was calculated using Agilent's Feature Extraction Image Analysis software with the default two-color gene expression protocol. Agilent two-color arrays were within-array normalized by loess, followed by between-array quantile normalization of average intensities across channels (Aquantile). Differential expression analysis was performed using a moderated t-test, as implemented in the limma package of Bioconductor, with P-value correction by false discovery rate. Microarray data are available at http://www.ncbi.nlm.nih.gov with accession GSE31618.
RESULTS
Six1/2-2, POU2/3 and hunchback are required for excretory system function
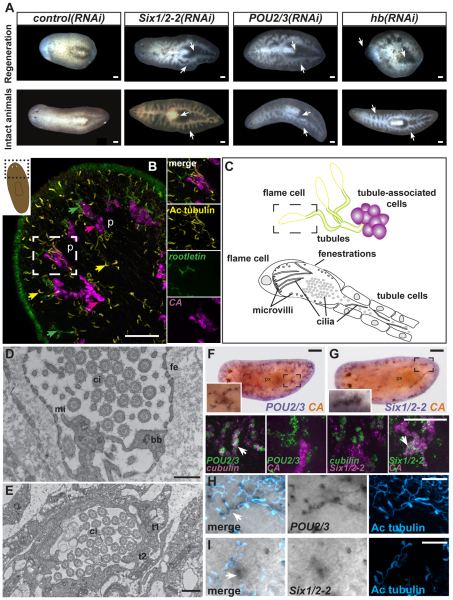
To study organ system regeneration we sought genes in the planarian S. mediterranea that are required for organ functions using RNAi screening. Inhibition of three genes resulted in a similar phenotype that suggested a defect in excretion/osmoregulation [this work and the work of Reddien et al. (Reddien et al., 2005a)]. Smed-Six1/2-2 (abbreviated to Six1/2-2) encodes a homeodomain-containing protein homologous to the Drosophila sine oculis gene (see Fig. S2A in the supplementary material), Smed-POU2/3 (abbreviated to POU2/3) encodes a representative of the POU family of homeobox genes (see Fig. S2B in the supplementary material) and Smed-hunchback (abbreviated to hunchback) encodes a zinc finger protein homologous to the Drosophila hunchback gene (see Fig. S2C in the supplementary material). Following RNAi of Six1/2-2, POU2/3 or hunchback, animals regenerated but displayed blisters, were bloated with fluid, and lysed (Fig. 1A, upper row) (Reddien et al., 2005a). Planarians maintain their adult tissues through the action of neoblasts, proliferative cells that replace aged adult cells (Reddien and Sánchez Alvarado, 2004). Inhibition of Six1/2-2, POU2/3 or hunchback during normal tissue turnover in intact animals also caused bloating, blistering and lysis (Fig. 1A, lower row). Protonephridia contain cilia (Ruppert and Smith, 1988; Glazer et al., 2010) and inhibition of genes required for cilia biology can cause similar defects to those observed in Six1/2-2, POU2/3 and hunchback RNAi animals, indicating that protonephridia dysfunction might underlie this phenotype (Reddien et al., 2005a; Glazer et al., 2010).
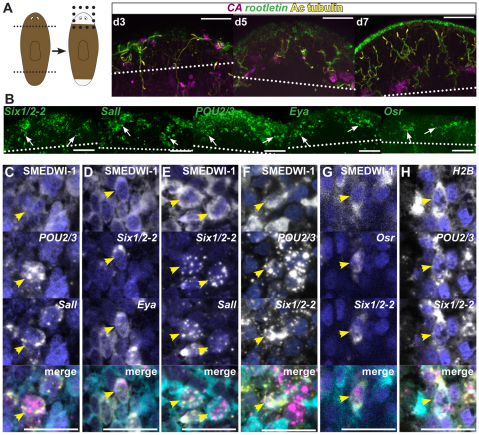
Fig. 1.
POU2/3, Six1/2-2 and hunchback are required for excretory function and are expressed in planarian protonephridia. (A) POU2/3, Six1/2-2 and hunchback RNAi animals bloated during regeneration (upper) and adult life without amputation (lower). Bloating initiated 12-15 days (Six1/2-2) or 21-30 days (POU2/3 and hunchback) after RNAi initiation (>50 animals each condition, 100% penetrance). Arrows indicate bloated regions. Animal translucence, with gut branches visible, is caused by fluid accumulation. Anterior, left. Scale bars: 0.1 mm. (B) Fluorescence in situ hybridization (FISH) and immunolabeling of a wild-type animal using carbonic anhydrase VII (CA, magenta) and rootletin (green) RNA probes and an anti-acetylated tubulin antibody (Ac tubulin, yellow). The cartoon depicts the region shown. p, photoreceptors. Arrowheads indicate examples of protonephridial cell types (magenta, tubule-associated cells; green, tubule cells; and yellow, flame cells). White dotted box indicates region shown in insets on right, showing single fluorescence channels of a cluster of tubule and tubule-associated cells. Scale bar: 0.1 mm. (C) Schematic of planarian protonephridial cell types: flame (yellow), tubule (yellow and green) and tubule-associated (magenta) cells. Dotted box indicates area magnified and shown in detail below with a flame cell and tubule cells. (D) Transmission electron microscopy (TEM) of a flame cell showing fenestrations (fe), microvilli (mi), cilia (ci) and basal bodies (bb). Scale bar: 500 nm. (E) TEM of two interdigitated tubule cells (t1 and t2). Scale bar: 500 nm. (F,G) Double colorimetric in situ hybridization of a wild-type animal using a CA (red, INT/BCIP) and POU2/3 or Six1/2-2 (purple, BCIP/NBT) RNA probes (F,G upper panels). Anterior, left. p, photoreceptors; px, pharynx. Scale bars: 0.1 mm. Insets: higher magnification of boxed area. POU2/3 and CA expression do not colocalize, whereas Six1/2-2 and CA expression do colocalize (>10 animals were similar). Lower panels: FISH of a wild-type day 6 regenerating blastema using RNA probes for POU2/3 or Six1/2-2 and for tubules (cubilin) and tubule-associated cells (CA). Arrowheads identify double-labeled cells. (H,I) Labeling of wild-type animals using POU2/3 (H) and Six1/2-2 (I) RNA probes and an anti-acetylated tubulin antibody (blue). POU2/3 expression colocalizes with the acetylated-tubulin pattern, whereas Six1/2-2 expression does not. Scale bars: 0.05 mm.
POU2/3 is expressed in tubule cells and Six1/2-2 is expressed in tubule-associated cells
Flatworm excretory systems have been described in many species using vital dye labeling, and light and electron microscopy (Reisinger, 1922; McKanna, 1968a; McKanna, 1968b). To image S. mediterranea protonephridia, we combined fluorescence in situ hybridization and immunolabeling using Smed-carbonic anhydrase VII (Smed-CA; abbreviated to CA) and Smed-rootletin (abbreviated to rootletin) RNA probes, which were previously shown to label the excretory system (Sánchez Alvarado et al., 2002; Glazer et al., 2010), and an antibody that recognizes acetylated tubulin and labels planarian protonephridia cilia (Glazer et al., 2010). CA encodes an enzyme required for the interconversion of carbon dioxide and bicarbonate to maintain acid-base balance (Purkerson and Schwartz, 2007) and rootletin encodes a ciliary rootlet component (Yang et al., 2002). The S. mediterranea excretory system includes at least three cell types: a ciliated terminal cell (anti-acetylated tubulin+ and often called a `flame cell' because its beating cilia resemble a flame), a ciliated tubule cell type (labeled with the anti-acetylated tubulin antibody and the rootletin RNA probe), and clusters of CA-expressing cells located in close proximity to the tubules (Fig. 1B,C; see Fig. S3A in the supplementary material) (see note added in proof). In some flatworms, there exist tubule-associated cells, with a suggested excretory role, called paranephrocytes (Hertel, 1993). Because of the uncertain relationship between paranephrocytes and the CA-expressing cells, we refer to the CA-expressing cells as tubule-associated cells.
In transmission electron micrographs, S. mediterranea flame cells were subepidermal (see Fig. S3B in the supplementary material) with numerous cilia, membrane fenestrations and peripheral microvilli (Fig. 1D; see Fig. S3B,C in the supplementary material). Cilia beating generates negative pressure, allowing filtration from the extracellular space into the flame cell through the fenestrations (McKanna, 1968a; Ruppert and Smith, 1988). The filtrate travels into the tubule lumen (see Movie 1 in the supplementary material) to the animal exterior. The tubule lumen was formed by two interdigitating, ciliated cells that lack fenestrations or microvilli (Fig. 1E).
Six1/2-2 and POU2/3 were expressed in a pattern similar to the distribution of the excretory system (Fig. 1F-I). hunchback was expressed broadly, indicating that it probably plays a role in multiple tissues (see Fig. S3D in the supplementary material). POU2/3-expressing cells were co-labeled with the anti-acetylated tubulin antibody and the cubilin RNA probe (cubilin is specifically expressed in tubule cells, see below) but not with the CA RNA probe (Fig. 1F,H). By contrast, Six1/2-2-expressing cells co-expressed CA but were not labeled with the anti-acetylated tubulin antibody or the cubilin RNA probe (Fig. 1G,I). These expression patterns were observed in intact animals and in day 6 regenerating blastemas (Fig. 1F,G lower panels; see Fig. S3E in the supplementary material). RNAi reduced detectable corresponding mRNAs (see Fig. S3F,G in the supplementary material). We conclude that Six1/2-2 is expressed in tubule-associated cells and POU2/3 is expressed in tubule cells of the planarian protonephridia.
Six1/2-2, POU2/3 and hunchback are required for formation and maintenance of protonephridia
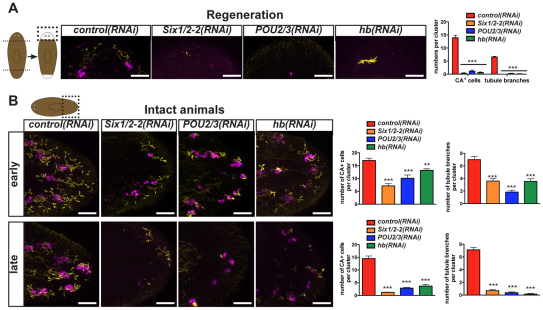
To assess the roles of Six1/2-2, POU2/3 and hunchback in planarian protonephridia regeneration, RNAi animals were amputated to remove heads and tails, and regenerating blastemas (unpigmented outgrowths at wound sites) were analyzed. Six1/2-2, POU2/3 and hunchback were essential for formation of the entire protonephridial system during regeneration; too few flame (acetylated tubulin+), tubule (acetylated tubulin+) or tubule-associated (CA+) cells were regenerated when any of these genes was inactivated (Fig. 2A). Inhibition of Six1/2-2, POU2/3 and hunchback specifically affected the nephridial system, with other ciliated cells remaining unaffected (see Fig. S4 in the supplementary material). The requirement for Six1/2-2, POU2/3 and hunchback for protonephridia regeneration provides an explanation for the animal bloating, blistering and lysis phenotype observed following RNAi of these genes.
Fig. 2.
POU2/3, hunchback and Six1/2-2 are required for protonephridia regeneration and maintenance. (A) Six1/2-2, POU2/3 and hunchback RNAi animals failed to regenerate protonephridia. Left, cartoon of amputation and regeneration; blastemas are white. Dotted box indicates the area shown in images. Numbers of tubule branches and tubule-associated cells per cluster were determined in regenerating blastemas (day 7 shown). (B) Fluorescence in situ hybridization and immunolabeling of control, POU2/3, hunchback or Six1/2-2 intact RNAi animals using a CA (magenta) RNA probe and an anti-acetylated tubulin antibody (yellow). Animals were analyzed at early (day 10 for Six1/2-2 and day 18 for POU2/3 and hunchback) and late (day 14 for Six1/2-2 and day 26 for POU2/3 and hunchback) time points following RNAi initiation. Cartoon indicates the region displayed in images and quantified in graphs. Anterior, left. Scale bars: 0.05 mm. Graphs display numbers of CA+ cells and tubule branches (Ac tubulin+) per cluster in tail regions. Data are mean ± s.e.m. (>6 animals per group). One-way ANOVA tests were performed, followed by a Dunnet post-hoc test, comparing different groups relative to the control. ***P<0.0001.
RNAi animals undergoing tissue turnover (without amputation) were examined to assess gene requirements in protonephridia maintenance. Tubule cells (acetylated tubulin+), which normally express POU2/3, were gradually lost following POU2/3 RNAi; tubule-associated cells (CA+) were also lost, but at a slower rate (Fig. 2B). hunchback RNAi resulted in a similar pattern of cell loss (Fig. 2B). Conversely, tubule-associated cells, which normally express Six1/2-2, were rapidly lost in Six1/2-2(RNAi) animals, with loss of tubule cells occurring more slowly (Fig. 2B). Six1/2-2, POU2/3 and hunchback are, therefore, required for protonephridia maintenance. Requirements for mature cell viability and/or production of new differentiated cells during natural tissue turnover could contribute to these phenotypes. In the case of Six1/2-2 RNAi, tubule-associated cells are lost faster than their normal tissue-turnover rate [e.g. irradiated animals lacking neoblasts do not show signs of rapid protonephridia loss (Reddien et al., 2005b)], indicating a requirement for maintenance of existing cells for (at least) this gene. In POU2/3 and hunchback RNAi animals at early time points, flame cells aggregated (Fig. 2B); lack of acetylated tubulin+ signal for tubules in these RNAi animals is, therefore, suggestive of tubule loss rather than simply cilia loss. Additional markers (see below) indicate further that it is not only cilia that are lost from protonephridia in these RNAi animals. Together, these RNAi data demonstrate a requirement for Six1/2-2, POU2/3 and hunchback in both regeneration and maintenance of planarian protonephridia.
Genes associated with protein clearance and ion transport are expressed in planarian protonephridia
Vertebrate homologs of Smed-Six1/2-2 and Smed-POU2/3 are essential for normal kidney development in mice and zebrafish (Hauptmann and Gerster, 2000; Nakai et al., 2003; Xu et al., 2003; Self et al., 2006; Kobayashi et al., 2008; Dressler, 2009). This raises the possibility that animal excretory systems utilize regulatory programs with a common evolutionary origin for their development. If the similarities between planarian protonephridia and the vertebrate kidney reflect homology, then homologs of other vertebrate genes involved in kidney function should be expressed in the planarian nephridia. In order to investigate this possibility, microarray experiments with Six1/2-2 and POU2/3 RNAi animals were performed (Fig. 3A). Because protonephridial cells became depleted over time in intact Six1/2-2 and POU2/3 RNAi animals, we reasoned that RNA from these animals should be depleted of mRNAs expressed specifically in protonephridia. Numerous genes displayed decreased expression in RNAi animals compared with the control. For example, expression of 736 and 351 genes was reduced to less than 50% of the control expression level in Six1/2-2(RNAi) (day 15 following RNAi) and POU2/3(RNAi) (day 21 post-RNAi) animals, respectively, with 208 of these genes in both lists.
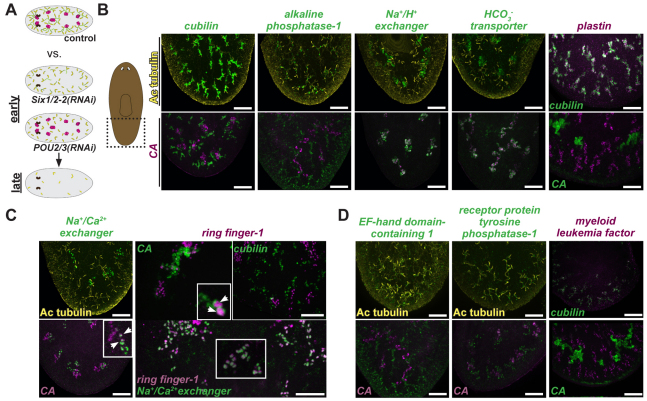
Fig. 3.
Microarray experiments identify homologs of vertebrate kidney genes expressed in S. mediterranea protonephridia. (A) Schematic of microarray experiments. RNA was collected as tubule-associated cells (purple) and tubule cells (green) were lost from Six1/2-2 or POU2/3 RNAi animals. (B-D) Cartoon depicts region imaged. Fluorescence in situ hybridization and immunolabeling of a wild-type animal (tail region shown) using different candidate genes, carbonic anhydrase (CA) and cubilin RNA probes, and an anti-acetylated tubulin antibody (Ac tubulin). Anterior, up. Scale bars: 0.05 mm. >10 animals per group were similar. Insets with higher magnification show colocalization (arrowheads).
Several genes with significant loss of expression in Six1/2-2 or POU2/3 RNAi animals encode proteins associated with ion transport and protein clearance, and other genes encode hydrolytic enzymes and enzymes involved in the maintenance of acid-base balance (see Tables S1 and S2 in the supplementary material). These genes encode alkaline phosphatase (Smed-alkaline phosphatase-1), carbonic anhydrase (Smed-carbonic anhydrase, described above), cubilin (Smed-cubilin), plastin (Smed-plastin) and solute carrier proteins (Smed-HCO3– transporter, Smed-Na+/H+ exchanger and Smed-Na+/Ca2+ exchanger; see below for more information on Smed-Na+/Ca2+ exchanger), and were expressed specifically in tubule or tubule-associated cells (Fig. 3B). These predicted proteins are homologous to proteins present in the vertebrate kidney (Christensen and Birn, 2002; Delanote et al., 2005; Purkerson and Schwartz, 2007; El-Sheikh et al., 2008; Lisowska-Myjak, 2010). In conclusion, planarian protonephridia and the vertebrate kidney share similarities in addition to developmental roles for Six1/2 and POU2/3 genes: they both possess a cell type for fluid filtration (flame cells and podocytes), and the planarian tubule/tubule-associated cells and the vertebrate kidney tubule cells utilize a similar set of excretory fluid-processing proteins.
Identification of novel excretory system genes and a second tubule-associated cell type
Two genes identified in the microarray experiments, Smed-Na+/Ca2+ exchanger (encoding a solute carrier protein) and Smed-ring finger-1 (abbreviated to Smed-RNF-1; encoding a novel ring finger protein), were co-expressed in cells associated spatially with protonephridia. Expression of the Smed-Na+/Ca2+ exchanger gene was absent in regenerating Six1/2-2 and POU2/3 RNAi animals (see Fig. S5A in the supplementary material). Some CA+ tubule-associated cells also expressed Smed-Na+/Ca2+ exchanger; however, most did not (Fig. 3C). These data suggest that a second tubule-associated cell type exists in the planarian protonephridia (Fig. 3C).
Additional genes identified from the microarray experiments have homologs in vertebrates, but do not have well-characterized roles in excretory systems. A myelodysplasia/myeloid leukemia factor homologous gene, Smed-myeloid leukemia factor (Smed-MLF), was expressed in tubule cells (Fig. 3D). Human MLF1 was identified from a translocation associated with myodysplastic syndrome and acute myeloid leukemia (Hitzler et al., 1999) and its biochemical function is poorly understood. Smed-receptor protein tyrosine phosphatase-1 (Smed-RPTP-1; encoding a receptor tyrosine phosphatase) and Smed-EF-hand domain-containing 1 (Smed-EFHC1; encoding an EF-hand containing-1-like protein) were also expressed in tubule cells (Fig. 3D). Mutations in vertebrate EFHC1 have been demonstrated in some forms of human epilepsy and might be associated with cilia biology (Suzuki et al., 2004; Ikeda et al., 2005). The identification of Smed-RNF-1, Smed-MLF, Smed-RPTP-1 and Smed-EFHC-1 highlights how the planarian protonephridia can be utilized to identify novel excretory organ genes.
Eya and Sall are required for protonephridia formation and maintenance and Osr is expressed in tubule cells
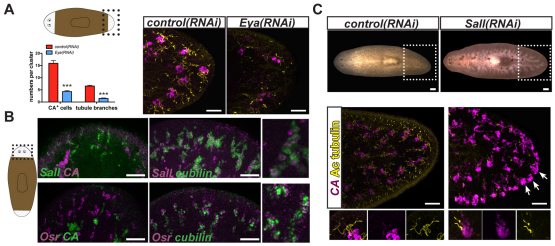
The microarray experiments described above also identified homologs of transcriptional regulators. RNAi of an eyes absent gene, Smed-Eya (abbreviated to Eya; see Fig. S2D in the supplementary material), decreased numbers of tubule and tubule-associated cells in blastemas (Fig. 4A). Moreover, intact animals under Eya RNAi conditions lost flame, tubule and tubule-associated cells (see Fig. S5B in the supplementary material). No Eya expression in mature protonephridia cells was detected; however, Eya is expressed in eye and parenchymal cells in planarians (Lapan and Reddien, 2011; Mannini et al., 2004) and Eya expression was detected during protonephridia regeneration (see below). Eya knockout mice lack kidneys (Xu et al., 1999). Eya acts together with sine oculis during eye formation in D. melanogaster as well as in the organogenesis of several other tissues (Bonini et al., 1993; Cheyette et al., 1994; Li et al., 2003). Our results suggest that Smed-Six1/2-2 and Smed-Eya might act together in planarian protonephridia regeneration.
Fig. 4.
Eya and Sall are required for protonephridia regeneration and Osr is expressed in tubule cells. (A) Eya(RNAi) animals failed to regenerate protonephridia. Cartoon depicts region analyzed. Left: numbers of tubule branches (Ac tubulin+) and tubule-associated cells (CA+) per cluster in control and Eya RNAi animals six days following amputation, 15 days following RNAi initiation. Data are mean ± s.e.m. (>12 animals per group). ***P<0.0001 determined by Student's t-test. Right: fluorescence in situ hybridization and immunolabeling of a regenerating control or Eya RNAi animal (tail blastema shown; anterior, left) using the CA (magenta) RNA probe and an anti-acetylated tubulin antibody (yellow). Scale bars: 0.05 mm. (B) Sall and Osr are expressed in tubule cells. Cartoon depicts blastema region imaged. Anterior, up. Scale bars: 0.05 mm. Insets on right show colocalization of transcription regulatory gene RNA (magenta) and cubilin (green). (C) Top: Sall(RNAi) animals bloated during tissue turnover in intact animals (17/20 animals bloated between days 43 and 46 after initial RNAi). Dotted box indicates the area depicted below. Middle: A bloated Sall(RNAi) animal with reduced tubule (Ac tubulin+, yellow) and increased tubule-associated cell numbers (CA+, magenta) is shown. Arrows indicate example clusters of ectopic tubule-associated cells at the animal periphery. Bottom: Higher magnification images of control and Sall(RNAi) clusters. Anterior, left. Scale bars: 0.05 mm.
A second candidate regulatory gene, Smed-Sall (abbreviated to Sall) (see Fig. S2E in the supplementary material), which encodes a transcription factor similar to Drosophila Spalt (Salm – FlyBase), was expressed in tubule cells in day six regenerating blastemas (Fig. 4B). Sall(RNAi) intact and regenerating animals bloated (Fig. 4C; see Fig. S5C in the supplementary material). Bloated Sall(RNAi) animals showed a severe reduction of tubule cells with an attendant increased number of CA-expressing cells (Fig. 4C). One possible explanation for this phenotype is that Sall might act in tubule cell formation/maintenance to repress the tubule-associated cell fate. Sall1-deficient mice do not form kidneys (Nishinakamura et al., 2001).
Smed-Osr (abbreviated to Osr) is homologous to the Drosophila odd skipped gene (see Fig. S2F in the supplementary material) and was also expressed in the planarian protonephridial tubule cells (Fig. 4B). A vertebrate Osr family gene, Osr1, is essential for nephron progenitor development (Mugford et al., 2008) and osr1 inactivation causes loss of kidney progenitors in zebrafish and a subsequent absence of kidney tubules (Tena et al., 2007; Mudumana et al., 2008). Similarly, Osr1-knockout mice do not form the metanephric mesenchyme and, therefore, lack kidneys (Wang et al., 2005; James et al., 2006). We did not detect a phenotype for Smed-Osr(RNAi) animals, but the expression of Osr in protonephridia and roles for vertebrate Osr1 in kidney development indicate further similarity between transcription factors involved in animal excretory systems. The microarray results expand the number of identified transcriptional regulatory factors associated with protonephridia regeneration to six: Six1/2-2, POU2/3, hunchback, Eya, Sall and Osr.
Blastema cells express Six1/2-2, Eya, POU2/3, Sall and Osr
Although Six1/2-2 and POU2/3 expression are restricted to specific nephridial cell types, formation of all protonephridial cell types was severely impaired during regeneration in Six1/2-2, POU2/3 and Eya RNAi animals (described above). We therefore studied the roles of these genes in regeneration. In the wild type, two symmetrical cell clusters, containing tightly associated tubule cells (rootletin- and acetylated tubulin-positive) and tubule-associated cells (CA+), formed at the edge of day three blastemas, indicating that both cell types form in blastemas at a similar time and location (Fig. 5A, left panel). This tight association persisted at day five of regeneration (Fig. 5A, middle panel); by day seven (Fig. 5A, right panel) the tubule-associated cells were found in a more internal location, distant from the animal periphery and similar to their normal location. Numerous cells that highly expressed Six1/2-2, POU2/3, Eya, Osr or Sall were present in day three blastemas (Fig. 5B; see Fig. S6A in the supplementary material). Many of these cells expressed more than one transcription regulatory gene (Fig. 5C-G; see Fig. S6A in the supplementary material). We examined the possibility that these cells include regenerative precursors of the protonephridia.
Fig. 5.
Six1/2-2, Eya, POU2/3, Sall and Osr define a population of blastema cells. (A-H) Fluorescence in situ hybridization and immunolabeling of wild-type animals using carbonic anhydrase (CA), rootletin, histone H2B (H2B), Six1/2-2, POU2/3, Eya, Osr and Sall RNA probes, and an anti-acetylated tubulin (Ac tubulin) or SMEDWI-1 antibody. Dotted lines indicate blastema boundary. (A) Formation of tubule and tubule-associated cells in blastemas of wild-type animals at day three (d3), five (d5) and seven (d7) of regeneration (>10 animals/time point were similar). Cartoon depicts amputation and regeneration; blastemas are white. Dotted box indicates the area depicted. Anterior, up. Scale bars: 0.05 mm. (B) Large numbers of cells expressing Six1/2-2, POU2/3, Eya, Osr and Sall were present in day three blastemas of wild-type animals (>30 animals were similar). Arrowheads indicate example single cells. Anterior, up. Scale bars: 0.05 mm. (C-G) Co-labeling of Sall and POU2/3 (C), Six1/2-2 and Eya (D), Six1/2-2 and Sall (E), Six1/2-2 and POU2/3 (F) and Six1/2-2 and Osr (G) RNA probes with the SMEDWI-1 antibody in wild-type, three day blastemas (>10 animals were similar). Arrowheads show gene co-expression and SMEDWI-1-protein presence. (H) Co-expression of Six1/2-2 and POU2/3 with the S-phase marker histone H2B in a wild-type, regenerating blastema (day three, >20 animals were similar). Arrowheads show cells co-expressing the three genes. Scale bars: 0.02 mm.
Regeneration in planarians requires dividing adult cells called neoblasts (Reddien and Sánchez Alvarado, 2004). Some neoblasts, called cNeoblasts, are adult, pluripotent stem cells (Wagner et al., 2011). Neoblasts express smedwi-1 (Reddien et al., 2005b) and respond to wounds to produce differentiating cells (Newmark and Sánchez Alvarado, 2000; Wenemoser and Reddien, 2010). Once neoblasts stop dividing and initiate differentiation, smedwi-1 mRNA disappears; SMEDWI-1 protein, however, persists in differentiating neoblast progeny cells for several days (Guo et al., 2006; Scimone et al., 2010). Therefore, an antibody recognizing SMEDWI-1 labels neoblasts and newly made neoblast progeny, including for differentiating cells of the nervous system and intestine (Wagner et al., 2011). We used this SMEDWI-1 antibody to describe a population of blastema cells with extensive co-expression of the protonephridia transcription factors. Most POU2/3+ cells in day three blastemas co-expressed Sall (107/120 POU2/3+ cells were Sall+) (see Fig. S6A in the supplementary material) and frequently had detectable SMEDWI-1 protein (55/70 POU2/3+/Sall+ cells were SMEDWI-1+), indicating that these cells are newly made neoblast progeny cells (Fig. 5C). In day six blastemas, both POU2/3 and Sall were expressed in tubule cells (Fig. 4C; see Fig. S3E in the supplementary material); therefore, day three blastema cells co-expressing POU2/3 and Sall could represent precursor cells of protonephridial tubules (see below). Several Six1/2-2+ cells co-expressed Eya (146/301 Six1/2-2+ cells were Eya+) and these cells frequently had SMEDWI-1 (97/104 Six1/2-2+/Eya+ cells were SMEDWI-1+) (Fig. 5D). Surprisingly, considering the different expression sites for Six1/2-2 and Sall in day six blastemas (Fig. 4C; see Fig. S3E in the supplementary material), many Six1/2-2+/Sall+ double-positive cells existed in day three blastemas (85/91 Six1/2-2+ cells were Sall+) (see Fig. S6A in the supplementary material). Most of these cells also possessed SMEDWI-1 protein (52/65 Six1/2-2+/Sall+ cells were SMEDWI-1+) (Fig. 5E). Furthermore, we found colocalization of Six1/2-2 and POU2/3 mRNAs themselves in day three blastemas (187/275 Six1/2-2+ cells were POU2/3+) (see Fig. S6A in the supplementary material). Many Six1/2-2+/POU2/3+ cells also possessed SMEDWI-1 (36/57 Six1/2-2+/POU2/3+ cells were SMEDWI-1+) (Fig. 5F). Cells also co-expressed Six1/2-2 and Osr in day three blastemas (see Fig. S6A in the supplementary material) and some of these cells were SMEDWI-1+ (Fig. 5G). We also found cells that co-express Osr and POU2/3 or Osr and Sall, together with SMEDWI-1 (see Fig. S6B in the supplementary material).
The significant co-expression of these transcription factor genes (pair-wise tests) indicates the existence of blastema cells that co-express Six1/2-2, Eya, POU2/3 and Sall, with some cells also expressing Osr. For example, the likelihood that Sall+/POU2/3+ and Six1/2-2+/POU2/3+ populations overlap in day three blastemas is significant (Fisher's test, P<0.0001); similarly, the likelihood that Six1/2-2+/POU2/3+ and Six1/2-2+/Eya+ cell populations overlap is significant (Fisher's test, P=0.0029). These cells are newly produced (SMEDWI+) and present in early blastemas following amputation. Cells co-expressing the transcription factors POU2/3 and Six1/2-2 or Six1/2-2 and Sall were not detected in day three blastemas of animals that were lethally irradiated the day after amputation (see Fig. S6C in the supplementary material), further indicating that these cells represent a transient cell population detected prominently during regeneration.
We next assessed whether expression of nephridia transcription factors initiates within neoblasts. Cells that co-express each transcription factor tested and the neoblast marker smedwi-1 were indeed present (see Fig. S6D in the supplementary material). Moreover, cells co-expressing POU2/3, Six1/2-2 and the S-phase marker histone H2B (H2B) (which labels neoblasts, as the only dividing somatic cell population) were observed in day three blastemas (Fig. 5H). These results suggest that initial specification of candidate nephridial precursor cells occurs within neoblasts. Similarly, eye precursor cells originate within neoblasts during regeneration (Lapan and Reddien, 2011). Examination of individual neoblasts isolated by flow cytometry (neoblasts can be isolated by DNA labeling because of their greater than 2N DNA content and are called X1 cells), has previously revealed gene expression heterogeneity in dividing planarian cells (Hayashi et al., 2010), and we detected some X1 cells expressing POU2/3 from day three blastemas (see Fig. S6E in the supplementary material). Whereas some neoblasts are pluripotent (Wagner et al., 2011), these eye data (Lapan and Reddien, 2011) and nephridia data together demonstrate that gene expression heterogeneity of neoblasts exists in vivo with some neoblasts expressing lineage-specific genes. It is unknown whether these nephridia transcription factor+ neoblasts will divide to expand in number or directly differentiate.
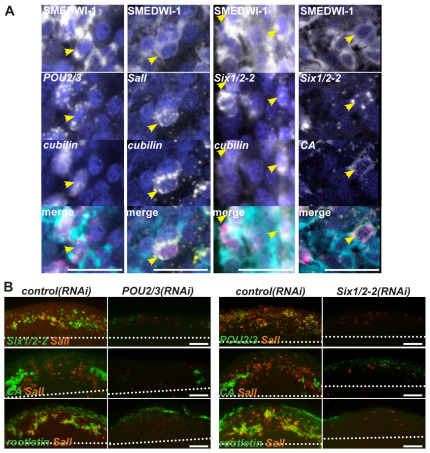
Next, we reasoned that if blastema cells expressing Sall, Six1/2-2, Eya, Osr and POU2/3 are indeed precursor cells for protonephridia, there might exist cells in intermediate stages between precursors and differentiated cells that co-express both the transcription factors and the differentiated markers. We found that some of the cells co-expressing POU2/3 and SMEDWI-1, Six1/2-2 and SMEDWI-1, or Sall and SMEDWI-1 also expressed low levels of cubilin (Fig. 6A), which is specifically expressed in protonephridial tubule cells (Fig. 3B). Similarly, some cells co-expressing Six1/2-2 and SMEDWI-1 also expressed low levels of CA, the tubule-associated cell marker (Fig. 6A). These results further support the hypothesis that Sall, Six1/2-2, POU2/3, and by association (because of co-expression data above) Eya and Osr, label blastema cells that, as a population, will produce tubule and tubule-associated cells.
Fig. 6.
Differentiation and formation of nephridia precursors. (A) Fluorescence in situ hybridization (FISH) and immunolabeling using carbonic anhydrase (CA), cubilin, Six1/2-2, POU2/3 and Sall RNA probes and the anti-SMEDWI-1 antibody in wild-type, three-day regenerating blastemas. Arrowheads show cells co-expressing a transcription factor, a differentiated marker and the SMEDWI-1 protein (>10 animals were similar). Scale bars: 0.02 mm. (B) Six1/2-2 and POU2/3 RNAi animals failed to generate nephridia precursors. FISH with control, Six1/2-2 or POU2/3 RNAi animals (>20 animals were similar). Dotted lines indicate blastema boundary. Anterior, up. Scale bars: 0.05 mm.
Six1/2-2 and POU2/3 RNAi animals fail to generate precursor cells
If the population of Six1/2-2+, POU2/3+, Eya+, Osr+ and Sall+ day 3 blastema cells does indeed include precursors for tubule and tubule-associated cells, ablation of this cell population should block both differentiation pathways. We therefore assessed the presence of the candidate nephridial precursors in Six1/2-2 and POU2/3 RNAi animals. As expected, no tubule (rootletin+) or tubule-associated cells (CA+) were found in Six1/2-2 and POU2/3 RNAi regeneration blastemas (Fig. 6B). Furthermore, no POU2/3+/Sall+ blastema cells were found in Six1/2-2(RNAi) animals and no Six1/2-2+/Sall+ blastema cells were found in POU2/3(RNAi) animals (Fig. 6B). In fact, few blastema cells expressing Six1/2-2 or POU2/3 were present at all following RNAi of either gene. These results support a model in which Six1/2-2 and POU2/3 functions are essential for the generation of a population of nephridial precursor cells, with some cells of this population differentiating into tubule cells and others differentiating into tubule-associated cells. The absence of such a population of precursors would explain the failure to regenerate both tubule and tubule-associated cells in these RNAi animals.
DISCUSSION
Six1/2-2, POU2/3, hunchback, Eya, Sall and Osr are involved in regeneration of an excretory system
The ability to regenerate missing body parts is a phenomenon that is broadly distributed in multicellular organisms. However, the mechanisms underlying this phenomenon are not well understood. Because planarians can regenerate all adult organs and their genes can be efficiently inhibited by RNAi, planarians can be utilized to identify mechanisms controlling regeneration of numerous cell types. We investigated genes consist of in regeneration of the planarian excretory system, called protonephridia. The S. mediterranea protonephridia consist of numerous tubules, two types of tubule-associated cells, and ciliated terminal cells. Because this organ system is dispersed throughout planarian bodies, most injuries will remove or damage regions of the protonephridia. Adult planarians must, therefore, possess mechanisms for making each protonephridial cell type and for integrating new cells with the remaining tubule system.
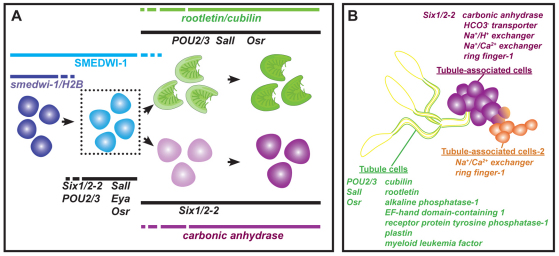
We identified multiple genes (Six1/2-2, POU2/3, hunchback, Eya, Sall and Osr) encoding conserved transcription regulatory proteins that are involved in protonephridia regeneration. Six1/2-2, POU2/3, hunchback and Eya were required for regeneration of all protonephridial cells, and Sall was required for regeneration of the proper ratio of tubule to tubule-associated cells. During regeneration, a population of blastema cells co-expressed many of these transcription regulatory genes. These blastema cells were newly produced neoblast progeny (SMEDWI protein+ and irradiation-sensitive). Some transcription regulatory gene+/SMEDWI-1+ blastema cells co-expressed differentiated nephridial markers. Finally, RNAi of Six1/2-2 or POU2/3 eliminated this population of blastema cells and blocked regeneration of differentiated protonephridial cells. Based on these observations, we propose a regenerative model for protonephridia (Fig. 7). First, protonephridia transcription factors are expressed together in neoblasts (some Six1/2-2 and POU2/3 double-positive cells were co-labeled with smedwi or H2B mRNA). Next, neoblast progeny (SMEDWI+) that co-express Six1/2-2, POU2/3, Eya, Sall and Osr in the blastema are produced (Fig. 7A), requiring Six1/2-2 and POU2/3 for formation. Cells from within this population produce terminally differentiated cells, either a tubule cell expressing POU2/3, Osr and Sall and the markers rootletin and cubilin, or a tubule-associated cell expressing Six1/2-2 and CA (Fig. 7A). These precursor cells are described as a population and could, therefore, include multipotent nephridial precursors or could possess distinct lineage-restricted cells with one subset of cells making tubules and a different subset making tubule-associated cells. Because of the lack of a planarian flame cell marker, there is no direct evidence that the described blastema cells can produce cells that differentiate into flame cells. However, flame cells are not regenerated in either Six1/2-2 or POU2/3 RNAi animals. During regeneration, newly formed cells in the blastema combine with remaining pre-existing protonephridia cells in the remainder of the body to reconstitute the excretory system.
Fig. 7.
A genetic and cellular program for regeneration of the planarian protonephridia. (A) Model for protonephridia regeneration: neoblasts (dark blue, dividing cell expressing smedwi-1 and histone H2B) produce a transient population of nephridia precursor cells (light blue) that co-expresses Six1/2-2, Eya, Osr, POU2/3, Sall and SMEDWI-1. Some cells expressing Six1/2-2, POU2/3 and Sall were found to express neoblast-specific H2B (S-phase marker) and/or smedwi-1 mRNA. Dotted box indicates that data cannot distinguish whether this precursor population is homogeneous or heterogeneous in potential. Precursor cells differentiate either as a tubule cell (green), maintaining expression of POU2/3, Sall and Osr and expressing rootletin and cubilin, or as a tubule-associated cell (magenta), maintaining Six1/2-2 expression and expressing CA. (B) Schematic of the planarian protonephridial system. Genes expressed in either tubule (green and yellow), tubule-associated cells (magenta) or tubule-associated cells-2 (orange) are listed.
In vertebrates, the existence of adult renal progenitors is controversial (Anglani et al., 2010; Guo and Cantley, 2010), and no adult, multipotent renal stem cell has been isolated (Guo and Cantley, 2010). However, cells with long-term self-renewal capacity in vitro and expression of renal transcription factors such as Cited1, Wt1, Osr1, Six1, Six2, Sall, Eya and Hoxa11 have been identified in the embryonic metanephric mesenchyme (Osafune et al., 2006; Guo and Cantley, 2010; Lusis et al., 2010). In addition, single cells expressing Sall isolated from the embryonic metanephric mesenchyme in mice formed colonies in vitro and differentiated into cells expressing podocyte, proximal tubule, distal tubule and Henle's loop markers (Osafune et al., 2006). Moreover, in vivo studies found that most of the mouse kidney cells arise from an Osr1-expressing population of cells (James et al., 2006; Mugford et al., 2008) and that a cap mesenchyme Six2+ population represents a multipotent progenitor cell population with the ability to differentiate into an epithelial glomerular and tubular cell (Boyle et al., 2008; Kobayashi et al., 2008). Therefore, dividing mesenchymal cells can promote nephron development in vertebrates and parenchymal proliferating cells (neoblasts) promote protonephridia regeneration in planarians, with these developmental and regenerative processes involving several similar transcription factors.
A common transcriptional regulatory program for nephridia formation in bilaterians
Diverse animals utilize organs for similar tasks. The discovery that similar developmental genes are utilized in organisms across the Bilateria (the bilaterally symmetrical animals) for the development of similar organs, such as Pax6/eyeless for the eye (Quiring et al., 1994), MyoD for muscle (Weintraub et al., 1991; Chen et al., 1992), Gata and Forkhead/HNF3 for the gut (Ang et al., 1993; Rehorn et al., 1996; Zhu et al., 1997) and tinman/Nkx2-5 for the heart (Evans et al., 1995), has raised the possibility that certain organ regulatory programs existed in the common ancestor of the Bilateria. However, no conserved gene regulatory paradigm has yet been established for animal excretory systems.
In vertebrates, the POU3-family member Brn-1 (Pou3f3) is expressed in the developing kidney and is required for kidney tubule formation (Hauptmann and Gerster, 2000; Nakai et al., 2003). Six1, Eya1, Osr1 and Sall are expressed in the metanephric mesenchyme and are essential for early mammalian kidney development. Mice deficient for each of these genes are similar; they die prenatally or at birth and lack kidneys owing to a failure of ureteric bud outgrowth and metanephric induction resulting in mesenchyme apoptosis (Abdelhak et al., 1997; Xu et al., 1999; Nishinakamura et al., 2001; Xu et al., 2003; Sajithlal et al., 2005; Wang et al., 2005). Moreover, Six2-deficient mice also show severe kidney hypoplasia because Six2 is required for function of a nephron-progenitor population (Self et al., 2006; Kobayashi et al., 2008). The identification of a molecular program including POU2/3, Six1/2, Eya, Sall and Osr family genes involved in the formation of excretory systems in both planarians (protostomes) and vertebrates (deuterostomes) suggests that a similar set of genes might have specified excretory system cell development in the common ancestor of bilaterally symmetrical animals.
Similarity between characters of different animal groups could reflect homology or could occur by independent evolution (Abouheif et al., 1997). If the roles for POU2/3, Six1/2, Eya, Sall and Osr family genes in planarian and vertebrate excretory systems reflect homology, cell types expressing or requiring these genes for their development should utilize similar proteins for their differentiated functions. We found several genes encoding proteins involved in protein clearance, acid-base balance and ion transport expressed in the planarian protonephridia that are indeed similar to vertebrate kidney genes (Fig. 7B). A further prediction of the homology hypothesis is that additional similarities would exist for excretory systems in other organisms. Metanephridial and protonephridial excretory systems are found in the three superphyla comprising bilaterians: deuterostomes, and the two groups among protostomes, the Ecdysozoa and the Lophotrochozoa (see Fig. S1 in the supplementary material). Future molecular analyses in additional organisms will, therefore, be an important direction for assessing potential homologous roles for the gene families described here in excretory system development. The specialized ecdysozoan excretory systems studied in C. elegans and Drosophila might reflect evolutionarily derived characters from the ancestral condition for the Ecdysozoa. Nonetheless, the recent discovery that orthologous genes are involved in nephrocyte slit formation in Drosophila and mouse podocytes points to potential evolutionary conservation of the functional morphology between waste filtration cells in ecdysozoans and deuterostomes (Weavers et al., 2009). Furthermore, a C. elegans POU2/3 family gene, ceh-6, is essential for the function of the only excretory cell in this animal (Burglin and Ruvkun, 2001). Therefore, the hypothesis of a homologous role in excretory systems for POU2/3 genes has support from all three bilaterian superphyla.
Some gene classes that we identified to be expressed in or to be important for formation of planarian nephridia have no known roles in the vertebrate kidney, raising the possibility that a similar role for orthologous genes might exist in other organisms. These include a hunchback gene, an MLF gene, a conserved Ring finger-encoding gene, an EF-hand domain-containing-1 gene, and a receptor protein tyrosine phosphatase-1 gene. Study of planarians can thus identify previously uncharacterized excretory system proteins.
Our analysis of planarian protonephridia identifies genes promoting regeneration of a filtration-based tubule excretory system. Identification of central regulatory programs for cell types, and distinguishing the core factors from those with species-specific function, is facilitated by comparing functional data from different organisms. Comparing data from planarians and vertebrates indicates that Six1/2-2, Eya, POU2/3, Sall and Osr, among the many factors possible, are candidate central regulators of animal excretory cell types. This could indicate candidate genes to utilize for future attempts at reprogramming-based regenerative medicine applications for kidney diseases. We propose that nephridia were present in the bilaterian ancestor with molecular mechanisms underlying nephridia formation conserved over millions of years of bilaterian evolution.
Note added in proof
While this manuscript was in production, Rink et al. (Rink et al., 2011) published work demonstrating a role for an EGFR ortholog in tubule branching in planarian nephridia. This manuscript is also a resource for anatomical markers and features of protonephridia.
Supplementary Material
Acknowledgments
We thank E. Alvarez-Saavedra and members of the Reddien laboratory for discussions; Sylvain Lapan for sharing methods and advice for detecting precursor cells; Danielle Wenemoser for help with the X1 in situ experiment; Irving Wang for illustrations; and N. Watson at the Whitehead, W. M. Keck Microscopy Facility for electron microscopy.
Footnotes
Funding
M.L.S. and M.S. were supported by Jane Coffin Childs Fellowships. P.W.R. is an early career scientist of the Howard Hughes Medical Institute. We acknowledge support by NIH R01GM080639 and Keck Foundation support, and a Cabot career development professorship. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.068098/-/DC1
References
- Abdelhak S., Kalatzis V., Heilig R., Compain S., Samson D., Vincent C., Weil D., Cruaud C., Sahly I., Leibovici M., et al. (1997). A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat. Genet. 15, 157-164 [DOI] [PubMed] [Google Scholar]
- Abouheif E., Akam M., Dickinson W. J., Holland P. W., Meyer A., Patel N. H., Raff R. A., Roth V. L., Wray G. A. (1997). Homology and developmental genes. Trends Genet. 13, 432-433 [DOI] [PubMed] [Google Scholar]
- Ang S. L., Wierda A., Wong D., Stevens K. A., Cascio S., Rossant J., Zaret K. S. (1993). The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development 119, 1301-1315 [DOI] [PubMed] [Google Scholar]
- Anglani F., Mezzabotta F., Ceol M., Cristofaro R., Del Prete D., D’Angelo A. (2010). The regenerative potential of the kidney: what can we learn from developmental biology? Stem Cell Rev. 6, 650-657 [DOI] [PubMed] [Google Scholar]
- Bartolomaeus T., Ax P. (1992). Protonephridia and Metanephridia – their relation within the Bilateria. J. Zool. Syst. Evol. Res. 30, 21-45 [Google Scholar]
- Bonini N. M., Leiserson W. M., Benzer S. (1993). The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72, 379-395 [DOI] [PubMed] [Google Scholar]
- Boyle S., Misfeldt A., Chandler K. J., Deal K. K., Southard-Smith E. M., Mortlock D. P., Baldwin H. S., de Caestecker M. (2008). Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev. Biol. 313, 234-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin T. R., Ruvkun G. (2001). Regulation of ectodermal and excretory function by the C. elegans POU homeobox gene ceh-6. Development 128, 779-790 [DOI] [PubMed] [Google Scholar]
- Chen L., Krause M., Draper B., Weintraub H., Fire A. (1992). Body-wall muscle formation in Caenorhabditis elegans embryos that lack the MyoD homolog hlh-1. Science 256, 240-243 [DOI] [PubMed] [Google Scholar]
- Cheyette B. N., Green P. J., Martin K., Garren H., Hartenstein V., Zipursky S. L. (1994). The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12, 977-996 [DOI] [PubMed] [Google Scholar]
- Christensen E. I., Birn H. (2002). Megalin and cubilin: multifunctional endocytic receptors. Nat. Rev. Mol. Cell Biol. 3, 256-266 [DOI] [PubMed] [Google Scholar]
- Costantini F., Kopan R. (2010). Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev. Cell 18, 698-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanote V., Vandekerckhove J., Gettemans J. (2005). Plastins: versatile modulators of actin organization in (patho)physiological cellular processes. Acta Pharmacol. Sinica 26, 769-779 [DOI] [PubMed] [Google Scholar]
- Denholm B., Skaer H. (2009). Bringing together components of the fly renal system. Curr. Opin. Genet. Dev. 19, 526-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler G. R. (2006). The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 22, 509-529 [DOI] [PubMed] [Google Scholar]
- Dressler G. R. (2009). Advances in early kidney specification, development and patterning. Development 136, 3863-3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sheikh A. A., Masereeuw R., Russel F. G. (2008). Mechanisms of renal anionic drug transport. Eur. J. Pharmacol. 585, 245-255 [DOI] [PubMed] [Google Scholar]
- Evans S. M., Yan W., Murillo M. P., Ponce J., Papalopulu N. (1995). tinman, a Drosophila homeobox gene required for heart and visceral mesoderm specification, may be represented by a family of genes in vertebrates: XNkx-2.3, a second vertebrate homologue of tinman. Development 121, 3889-3899 [DOI] [PubMed] [Google Scholar]
- Glazer A. M., Wilkinson A. W., Backer C. B., Lapan S. W., Gutzman J. H., Cheeseman I. M., Reddien P. W. (2010). The Zn finger protein Iguana impacts Hedgehog signaling by promoting ciliogenesis. Dev. Biol. 337, 148-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. K., Cantley L. G. (2010). Cellular maintenance and repair of the kidney. Annu. Rev. Physiol. 72, 357-376 [DOI] [PubMed] [Google Scholar]
- Guo T., Peters A. H., Newmark P. A. (2006). A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 11, 159-169 [DOI] [PubMed] [Google Scholar]
- Hauptmann G., Gerster T. (2000). Combinatorial expression of zebrafish Brn-1- and Brn-2-related POU genes in the embryonic brain, pronephric primordium, and pharyngeal arches. Dev. Dyn. 218, 345-358 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Shibata N., Okumura R., Kudome T., Nishimura O., Tarui H., Agata K. (2010). Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its ‘index sorting’ function for stem cell research. Dev. Growth Differ. 52, 131-144 [DOI] [PubMed] [Google Scholar]
- Hertel L. A. (1993). Excretion and osmoregulation in the flatworms. Trans. Am. Microsc. Soc. 112, 10-17 [Google Scholar]
- Hitzler J. K., Witte D. P., Jenkins N. A., Copeland N. G., Gilbert D. J., Naeve C. W., Look A. T., Morris S. W. (1999). cDNA cloning, expression pattern, and chromosomal localization of Mlf1, murine homologue of a gene involved in myelodysplasia and acute myeloid leukemia. Am. J. Pathol. 155, 53-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T., Ikeda K., Enomoto M., Park M. K., Hirono M., Kamiya R. (2005). The mouse ortholog of EFHC1 implicated in juvenile myoclonic epilepsy is an axonemal protein widely conserved among organisms with motile cilia and flagella. FEBS Lett. 579, 819-822 [DOI] [PubMed] [Google Scholar]
- James R. G., Kamei C. N., Wang Q., Jiang R., Schultheiss T. M. (2006). Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development 133, 2995-3004 [DOI] [PubMed] [Google Scholar]
- Kobayashi A., Valerius M. T., Mugford J. W., Carroll T. J., Self M., Oliver G., McMahon A. P. (2008). Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3, 169-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan S. W., Reddien P. W. (2011). dlx and sp6-9 control optic cup regeneration in a prototypic eye. PLoS Genetics 7, e1002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Oghi K. A., Zhang J., Krones A., Bush K. T., Glass C. K., Nigam S. K., Aggarwal A. K., Maas R., Rose D. W., et al. (2003). Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426, 247-254 [DOI] [PubMed] [Google Scholar]
- Lisowska-Myjak B. (2010). Serum and urinary biomarkers of acute kidney injury. Blood Purif. 29, 357-365 [DOI] [PubMed] [Google Scholar]
- Lusis M., Li J., Ineson J., Christensen M. E., Rice A., Little M. H. (2010). Isolation of clonogenic, long-term self renewing embryonic renal stem cells. Stem Cell Res. 5, 23-39 [DOI] [PubMed] [Google Scholar]
- Mannini L., Rossi L., Deri P., Gremigni V., Salvetti A., Salo E., Batistoni R. (2004). Djeyes absent (Djeya) controls prototypic planarian eye regeneration by cooperating with the transcription factor Djsix-1. Dev. Biol. 269, 346-359 [DOI] [PubMed] [Google Scholar]
- McKanna J. A. (1968a). Fine structure of the protonephridial system in Planaria. I. Flame cells. Z. Zellforsch. Mikrosk. Anat. 92, 509-523 [DOI] [PubMed] [Google Scholar]
- McKanna J. A. (1968b). Fine structure of the protonephridial system in Planaria. II. Ductules, collecting ducts, and osmoregulatory cells. Z. Zellforsch. Mikrosk. Anat. 92, 524-535 [DOI] [PubMed] [Google Scholar]
- Mudumana S. P., Hentschel D., Liu Y., Vasilyev A., Drummond I. A. (2008). Odd skipped related1 reveals a novel role for endoderm in regulating kidney versus vascular cell fate. Development 135, 3355-3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford J. W., Sipila P., McMahon J. A., McMahon A. P. (2008). Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev. Biol. 324, 88-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai S., Sugitani Y., Sato H., Ito S., Miura Y., Ogawa M., Nishi M., Jishage K., Minowa O., Noda T. (2003). Crucial roles of Brn1 in distal tubule formation and function in mouse kidney. Development 130, 4751-4759 [DOI] [PubMed] [Google Scholar]
- Nelson F. K., Albert P. S., Riddle D. L. (1983). Fine structure of the Caenorhabditis elegans secretory-excretory system. J. Ultrastruct. Res. 82, 156-171 [DOI] [PubMed] [Google Scholar]
- Newmark P. A., Sánchez Alvarado A. (2000). Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 220, 142-153 [DOI] [PubMed] [Google Scholar]
- Nishinakamura R., Matsumoto Y., Nakao K., Nakamura K., Sato A., Copeland N. G., Gilbert D. J., Jenkins N. A., Scully S., Lacey D. L., et al. (2001). Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development 128, 3105-3115 [DOI] [PubMed] [Google Scholar]
- Osafune K., Takasato M., Kispert A., Asashima M., Nishinakamura R. (2006). Identification of multipotent progenitors in the embryonic mouse kidney by a novel colony-forming assay. Development 133, 151-161 [DOI] [PubMed] [Google Scholar]
- Pearson B. J., Eisenhoffer G. T., Gurley K. A., Rink J. C., Miller D. E., Sánchez Alvarado A. (2009). Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 238, 443-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkerson J. M., Schwartz G. J. (2007). The role of carbonic anhydrases in renal physiology. Kidney Int. 71, 103-115 [DOI] [PubMed] [Google Scholar]
- Quaggin S. E., Kreidberg J. A. (2008). Development of the renal glomerulus: good neighbors and good fences. Development 135, 609-620 [DOI] [PubMed] [Google Scholar]
- Quiring R., Walldorf U., Kloter U., Gehring W. J. (1994). Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science 265, 785-789 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Sánchez Alvarado A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725-757 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Murfitt K. J., Jennings J. R., Sánchez Alvarado A. (2005a). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8, 635-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C., Sánchez Alvarado A. (2005b). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327-1330 [DOI] [PubMed] [Google Scholar]
- Rehorn K. P., Thelen H., Michelson A. M., Reuter R. (1996). A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development 122, 4023-4031 [DOI] [PubMed] [Google Scholar]
- Reisinger E. (1922). Uber die Terminalorgane and das Kanalsystem einiger bekannter Typholoplaniden. Zoologischer Anzeiger 56, 205-224 [Google Scholar]
- Rink J. C., Vu H. T., Sánchez Alvarado A. (2011). The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development 138, 3769-3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert E. E. (1994). Evolutionary origin of the vertebrate nephron. Am. Zool. 34, 542-553 [Google Scholar]
- Ruppert E. E., Smith P. R. (1988). The functional organization of filtration nephridia. Biol. Rev. 63, 231-258 [Google Scholar]
- Sajithlal G., Zou D., Silvius D., Xu P. X. (2005). Eya 1 acts as a critical regulator for specifying the metanephric mesenchyme. Dev. Biol. 284, 323-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A., Newmark P. A., Robb S. M., Juste R. (2002). The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development 129, 5659-5665 [DOI] [PubMed] [Google Scholar]
- Scimone M. L., Meisel J., Reddien P. W. (2010). The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development 137, 1231-1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self M., Lagutin O. V., Bowling B., Hendrix J., Cai Y., Dressler G. R., Oliver G. (2006). Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 25, 5214-5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Delgado-Escueta A. V., Aguan K., Alonso M. E., Shi J., Hara Y., Nishida M., Numata T., Medina M. T., Takeuchi T., et al. (2004). Mutations in EFHC1 cause juvenile myoclonic epilepsy. Nat. Genet. 36, 842-849 [DOI] [PubMed] [Google Scholar]
- Tena J. J., Neto A., de la Calle-Mustienes E., Bras-Pereira C., Casares F., Gomez-Skarmeta J. L. (2007). Odd-skipped genes encode repressors that control kidney development. Dev. Biol. 301, 518-531 [DOI] [PubMed] [Google Scholar]
- van der Flier L. G., Clevers H. (2009). Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71, 241-260 [DOI] [PubMed] [Google Scholar]
- Wagner D. E., Wang I. E., Reddien P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811-816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Lan Y., Cho E. S., Maltby K. M., Jiang R. (2005). Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development. Dev. Biol. 288, 582-594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weavers H., Prieto-Sanchez S., Grawe F., Garcia-Lopez A., Artero R., Wilsch-Brauninger M., Ruiz-Gomez M., Skaer H., Denholm B. (2009). The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature 457, 322-326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T. K., Turner D., Rupp R., Hollenberg S., et al. (1991). The myoD gene family: nodal point during specification of the muscle cell lineage. Science 251, 761-766 [DOI] [PubMed] [Google Scholar]
- Wenemoser D., Reddien P. W. (2010). Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344, 979-991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. A., Webster L. A. (1974). Protonephridia. Biol. Rev. Camb. Philos. Soc. 49, 127-160 [DOI] [PubMed] [Google Scholar]
- Xu P. X., Adams J., Peters H., Brown M. C., Heaney S., Maas R. (1999). Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 23, 113-117 [DOI] [PubMed] [Google Scholar]
- Xu P. X., Zheng W., Huang L., Maire P., Laclef C., Silvius D. (2003). Six1 is required for the early organogenesis of mammalian kidney. Development 130, 3085-3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Liu X., Yue G., Adamian M., Bulgakov O., Li T. (2002). Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J. Cell Biol. 159, 431-440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Hill R. J., Heid P. J., Fukuyama M., Sugimoto A., Priess J. R., Rothman J. H. (1997). end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 11, 2883-2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.