Abstract
All echinoderms have unique hydraulic structures called tube feet, known for their roles in light sensitivity, respiration, chemoreception and locomotion. In the green sea urchin, the most distal portion of these tube feet contain five ossicles arranged as a light collector with its concave surface facing towards the ambient light. These ossicles are perforated and lined with pigment cells that express a PAX6 protein that is universally involved in the development of eyes and sensory organs in other bilaterians. Polymerase chain reaction (PCR)-based sequencing and real time quantitative PCR (qPCR) also demonstrate the presence and differential expression of a rhabdomeric-like opsin within these tube feet. Morphologically, nerves that could serve to transmit information to the test innervate the tube feet, and the differential expression of opsin transcripts in the tube feet is inversely, and significantly, related to the amount of light that tube feet are exposed to depending on their location on the test. The expression of these genes, the differential expression of opsin based on light exposure and the unique morphological features at the distal portion of the tube foot strongly support the hypothesis that in addition to previously identified functional roles of tube feet they are also photosensory organs that detect and respond to changes in the underwater light field.
Keywords: sea urchin, opsins, photosensory, tube feet, echinoderm
1. Introduction
Green sea urchins (Strongylocentrotus droebachiensis) from the Gulf of Maine, and other echinoderms, undergo an annual reproductive cycle that is regulated by changing autumn (Northern Hemisphere) photoperiod that results in a decrease in the total number of daylight hours. Studies on sea urchins suggest that these changes in photoperiod are the signal that causes the mobilization of nutrients from somatic cells and initiation of gametogenesis in the gonads of both sexes [1–3]. The photoperiod control of gametogenesis requires photosensory capabilities on the surface of the sea urchin to detect and respond to the low irradiances of light in their underwater habitat, which is dominated by the green portion of the spectrum [3]. Sea urchins are clearly capable of sensing changes in the underwater light field of their environment and using this information to modulate both their behaviour [4–6] and physiology [3]. How sea urchins detect these changes has been unclear, because they lack image-forming eyes and light-sensitive eyespots. In the echinoderms, there are several studies that describe the location, distribution and morphology of putative photosensory organs or extra-ocular photoreceptors, while the functional and molecular characteristics are largely unknown or poorly described. Several anatomical features of the sea urchin that might serve as photoreceptors include the spines, pedicellariae and tube feet (i.e. podia), and sea urchins are known to respond to both the visible and ultraviolet components of solar radiation [1,4,6,7], suggesting that their photosensory capability is on the surface of the test because the transmission of visible and ultraviolet radiation through the sea urchin test is less than 1 per cent of the available ambient underwater irradiance [3]. Additionally, recent work on the calcitic microlenses found in light-sensitive brittlestars [8] suggests that the ossicles within their radially arranged arms and underlying tissues may be involved in photoreception, and sea urchins also have ossicles in the distal portion of their tube feet [9].
The study of photoreception in echinoderms, and echinoids in particular, has been of historical interest, and in the past was more commonly referred to as the ‘dermal light sense’ [5,10–13]. Millot & Yoshida [14] described putative photoreceptors in the epithelium of the sea urchin Diadema antillarum and the photosensitivity of its radial nerve. The spectral sensitivity of these photoreceptors peaks between 530 and 550 nm [11], which points towards the involvement of a specific chromophore or pigment. In this study, it was also suggested that the pigments associated with echinoderm photoreceptors might be carotenoids [11], while another study on the sea urchin Centrostephanus longispinus shows peak sensitivity of its isolated chromatophores between 430 and 450 nm based on an action spectrum of chromatophore contraction and expansion [15]. In this latter case, however, the chromatophores were not functionally linked to the nervous system and were not interpreted as being involved in photoreception [15].
Recently, putative opsin proteins have been identified in the optic cushion of asteroids and arm tissue of ophiuroids using both western blots and immunohistochemistry [10]. Opsins have been characterized for many photosensitive prokaryotic and eukaryotic organisms, including sea urchins [16,17]. In vertebrates, multiple opsins are involved both in vision and in extra-ocular photoreception, which regulate circadian clocks and gametogenesis [17,18]. Opsins involved in extra-ocular photoreception share structural features with opsins involved in image-forming eyes, including a lysine retinal attachment site and the Schiff base counterion, but also possess unique features at the gene and protein level. Light appears to be a very important stimulus for a variety of sea urchin functions that may require multiple opsins to confer photosensitivity on time scales from short-term sensitivity (which governs rapid behavioural responses) to long-term circadian rhythms (which control processes such as gametogenesis). Determining which structures in this eyeless organism are capable of photoreception and can express specific opsins should provide useful information on the photobiology of sea urchins generally, and the evolution of photoreceptors and image-forming eyes.
2. Material and methods
(a). Preparation of tube feet for microscopy and immunocytochemistry
Sea urchins (n = 3–5) were immersed in chilled 7 per cent MgCl solution to relax and extend their tube feet. From these sea urchins, 50–75 tube feet were excised for the procedures described below. Several individual tube feet were used to isolate ossicles for scanning electron microscopy (SEM) by fixing in 70 per cent ethanol and macerating in 2.0 per cent KOH for more than 3 days. Additional samples of tube feet for microscopy were placed in primary fixative (2% glutaraldehyde in 0.2 M cacodylate buffer) for 2 h. Fixed tube feet were then dehydrated in an ethanol series, decalcified using 5 per cent ethylenediaminetetra-acetic acid (EDTA) in water and prepared for light (stained with Azure B), SEM or transmission electron microscopy (TEM) or for immunocytochemistry. For SEM and TEM, standard protocols for echinoderm tissues were used as described by Crawford & Burke [19]. For immunocytochemistry, dehydrated tube feet were embedded in Immuno-Bed (Polysciences, Inc., Warrington, PA, USA) overnight under vacuum. Sections (1 µm) were prepared on a JB-4 microtome and mounted on poly-L-lysine coated slides. Immunocytochemical detection and localization of PAX6 protein were performed using a goat PAX6 polyclonal antibody (C-20; Santa Cruz Biotechnology, Santa Cruz, CA, USA) to the C-terminus of human PAX6, and either directly conjugated to QDots (525 green fluorescence; Quantum Dot Corporation, Hayward, CA, USA) or visualized by labelled secondary antibody (Elite Goat IgG ABC kit, Vector Laboratories, Burlingame, CA, USA) and photographed on a Zeiss Axioplan II microscope equipped with epifluorescence and AxioVision v. 4.4 software (Carl Zeiss, Inc., Thornwood, NY, USA). This commercial antibody has been shown to be specific to PAX6 purified protein and cross-reacts with both sea urchin tube feet and sea star optic cushion using western blots and the appropriate negative controls [20].
(b). Green sea urchin opsin primer design and gene amplification
Sequences from the Strongylocentrotus purpuratus Genome Project were searched for matches against the term ‘opsin’. The returned sequences were aligned using Lasergene v. 6 software (DNASTAR, Inc., Madison, WI, USA) and run through BLASTx to match the sequence against the GenBank ‘nr’ database. Two non-degenerate primers were then designed and designated as follows: Opsin Forward (5′-ACCTGCTCGTTGCGACTACG) and Opsin Reverse (5′-GGCCTTCCTGAACTTGGAATT). Approximately 30–50 tube feet were extracted for total RNA using TRIzol Reagent (Sigma-Aldrich, St Louis, MO, USA), and the mRNA population reverse transcribed using a polyT primer and Superscript II (Invitrogen, Carlsbad, CA, USA). The resulting cDNA was used in a polymerase chain reaction (PCR) with the primers described above, and all PCR products were TA cloned (Promega pGEM-T kit) with individual clones grown overnight and miniprepped to produce plasmid DNA for sequencing using an ABI 377 gel sequencer. Rapid amplification of cDNA ends methodology was used in order to obtain full-length amplification of the complete gene. The same cDNA sequence was obtained when the mRNA was pretreated with DNase, suggesting that the extra sequence observed in the cytoplasmic loop is actually present in the mRNA transcript and not the result of contamination from unspliced DNA.
(c). Phylogenetic analysis of green sea urchin opsin
An alignment of 45 opsin amino acid sequences was created using the Mafft alignment program [21]. GenBank accession numbers for the alignment are as follows: Ciona intestinalis opsin (BAB68391); Danio rerio vertebrate ancient long (VAL) opsin (NP_571661) and teleost multiple tissue (TMT) opsin (NP_001112371); Salmo salar vertebrate ancient opsin (O13018); Petromyzon marinus pineal opsin (O42490); bovine rhodopsin (AH001149); Gallus gallus SWS1 (AAA49141), SWS2 (AAA48633), RH2 (AAA48786), LWS (AAA49137), RH1 (BAA00610) and pinopsin (P51475); Geotria australis SWS1 (AAR14684), SWS2 (AAR14681), RhA (AAR14682), RhB (AAR14683) and LWS (AAR14680); Homo sapiens opsin-3 (NP_055137), melanopsin or opsin-4 (NP_150598), opsin-5 (NP_859528) and peropsin (NP_006574); Mus musculus opsin-3 (NP_034228), melanopsin or opsin-4 (NP_038915), opsin-5 (NP_861418) and peropsin (NP_033128); Platynereis dumerilii c-opsin (AAV63834) and r-opsin (CAC86665); Drosophila melanogaster RH1 (NP_524407), RH2 (NP_524398), RH3 (NP_524411), RH4 (NP_476701), RH5 (NP_477096) and RH6 (NP_524368); Limulus polyphemus opsin (P35361); Mizohupecten yessoensis Gq coupled opsin (O15973); Octopus rhodopsin (P09241); Gadus morhua melanopsin (AAO20043); Xenopus laevis melanopsin (AAC41235) and chlamyopsin 4 (XP_001701725). In addition, we included the five opsins identified from the Strongylocentrotus purpuratus genome [16].
The alignment was checked by eye to ensure the characteristic functional regions of the proteins were aligned (e.g. the counterion-113 and lysine-296 residues). The alignment was converted to phylip format using ClustalX [22] and used in ProtTest to identify the best model of protein evolution using the Akaike information criterion [23]. A maximum-likelihood tree was then constructed in genetic algorithm for rapid-likelihood inference [24] by Grid computing [25] through the Lattice Project [26]. The consensus bootstrap tree was then calculated from 1000 bootstrap replicates using PAUP v. 4.0b10 (Sinauer Associates, Sunderland, MA, USA [27]).
A hydrophobicity plot analysis was carried out in order to determine potential transmembrane regions of the green sea urchin opsin protein. The Kyte–Doolittle scale with a window size of 19 was used for delineating the hydrophobic character of the protein [28]. Green sea urchin opsin was analysed along with a subset of opsins including sea urchin, invertebrate and vertebrate opsins with previously characterized transmembrane domains to determine the most likely transmembrane regions of the green sea urchin opsin [29].
(d). Differential gene expression of the green sea urchin opsin gene
qPCR was used to determine the relative levels of mRNA expression of opsin in the tube feet located on different areas of the test of S. droebachiensis (n = 3 urchins). Multiple tube feet (n = 25–50) from the aboral, bottom and oral portions of each individual urchin test (figure 1) were collected and total RNA extracted. The mRNA population was then reverse transcribed as described above and the resulting cDNA was used in a qPCR reaction using the Taqman Fast System (Applied Biosystems, Carlsbad, CA, USA), where triplicate reactions for each set of tune feet from each urchin were run on the 7500 Fast RT-PCR System (Applied Biosystems), at 45 cycles of 95°C with an automatically set critical cycle threshold (Ct). To quantify the opsin transcripts of S. droebachiensis, forward and reverse primers were designed from the sequence data (see above) in combination with a Taqman probe as follows: TB opsin F 5′-AAGCTCCATAGCCCCATCAA, TB opsin R 5′-CGAATAACCACCGACCGTAGAA and TB opsin probe 5′-CGTTAACCTCTCCGCCAGCGACC. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), a housekeeping gene that is constitutively expressed, was used as an internal control to normalize all data. Primer efficiencies, calculated from the raw data, were used to calculate the maximum expression value so that calculations from raw qPCR data used the formula PΔCt, where P = primer efficiency and ΔCt = cycle threshold difference with GAPDH for the sample. Specifically, the ratio of the target gene and the control gene can be determined using the equation
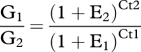 |
and the cycle numbers at which each gene (G) becomes detectable by calculating the Ct and E that is the PCR efficiency for the target and reference genes. The variability between qPCR runs was very low and the biological variability of the relative expression of opsins between tube foot locations is presented.
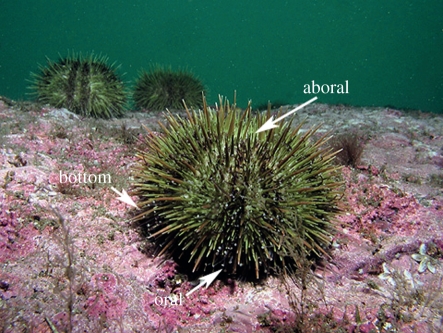
Figure 1.
Underwater photograph of a green sea urchin showing the aboral, bottom (at interface of urchin test and substrate) and oral locations where tube feet were collected. See also fig. 4a,m in [30].
3. Results
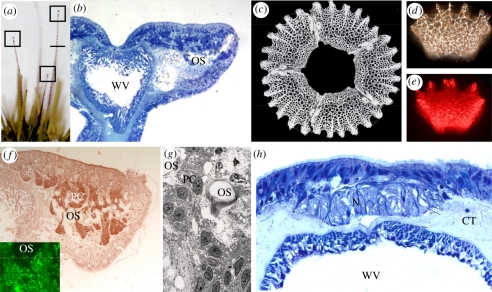
In adult urchins, approximately 1500 tube feet are present and serve as a radially distributed sensory array for these shallow water marine invertebrates (figure 2a). At the distal tip of each tube foot is a group of five independent calcareous ossicles circularly arranged with their concave surfaces facing the ambient underwater light environment (figure 2b,c). When illuminated at any single point on the ossicle using a visible spectrum (400–700 nm) fiberoptic probe, the ossicles scatter light across the entire ossicle (figure 2d). Similarly, when illuminated at any single point on the ossicle using a red (approx. 685 nm) laser the ossicles scatter light across the ossicle (figure 2e). The ossicles of the tube feet are perforated (figure 2c) and contain cells that express PAX6 protein (figure 2f and inset, using PAX6 antibody conjugated to QDots; ossicles removed by the de-calcification process). Note the presence of the pigment cells, probably containing opsin, in and around the ossicles (figure 2g). Nerves are also present along the entire length of the tube foot under the epithelium, forming a nerve plexus (figure 2h) that terminates in a ganglion at the distal portion of the tube foot as previously described for sea urchins [30–32].
Figure 2.
Green sea urchin tube feet. (a) Extended tube feet of a green sea urchin (maximum length of adult tube feet is 7–8 cm). Boxes indicate the distal tips shown in (b) and the line indicates the position of the cross-section shown in (h). (b) Structure of the right-hand side of the distal tip of a tube foot, showing position of ossicles (OS) seen in (c). (c) Array of five single crystalline ossicles removed from the tube foot. (d) Single ossicle under polarized light. (e) Single ossicle showing scattering of laser light directed at lower left corner. (f) Immunocytochemical localization of PAX6 protein in the pigment cells (PC) passing through perforations in the ossicles; inset shows the same PAX6 antibody conjugated to Qdots within pigment cell nuclei. (g) Electron micrograph of pigment cells passing through ossicle. (h) Cross-section at the level shown in figure 1a, indicating the nerve (N) running down one side of the tube foot (CT, connective tissue; WV, water vascular system of tube foot).
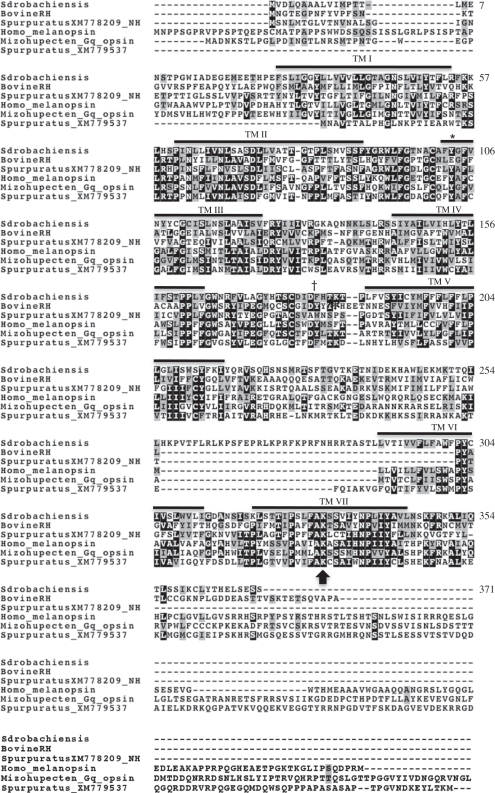
Using primers specific for opsins, a 1.563 Kbp cDNA was amplified from the green sea urchin tube foot. The predicted protein from the corresponding mRNA encodes an opsin protein that has a 1.116 Kbp open reading frame and 371 amino acids (figure 3, GenBank DQ285097). The identity of this protein as an opsin was confirmed by a phylogenetic analysis of the amino acid sequence data (see below). The Kyte–Doolittle hydrophobicity plot analysis of green sea urchin opsin showed seven segments greater than one in the hydropathy score, which represent potential transmembrane domains. These transmembrane domains matched those found in other opsins, including opsins from invertebrates [17,33,34]. The green sea urchin tube foot opsin shares many additional features with known opsins, the most conserved of which is the lysine in the seventh transmembrane domain (TM; K296/K331), which serves as an attachment site for the chromophore retinal [35] and two cysteine residues (C100 and C178) that together form a disulphide bridge essential for stability of opsin tertiary structure [36]. The cytoplasmic carboxy tail of the urchin opsin is only 25 amino acids long and is similar in this respect to the shorter tails of vertebrate visual opsins [37]. Most vertebrate opsins have a negatively charged glutamic acid residue (E113) in TM III that acts as a counterion for the protonated Schiff base [17]. The green sea urchin opsin, however, has a neutrally charged tyrosine residue (E113/Y103) that is common for most non-vertebrate opsins (figure 3) [33] and probably does not act as the counterion. The green sea urchin opsin does have a negatively charged aspartic acid residue (D181) and there is strong evidence that this amino acid acts as the counterion for the protonated Schiff base in the ancestral, non-vertebrate opsins [17,33].
Figure 3.
Amino acid alignment of green urchin opsin and several other opsins. Identical residues between species share a black background and conserved residues share a grey background. Transmembrane domains (TM) are indicated with a solid bar, lysine attachment site for retinal is marked with an arrow, the tyrosine residue (E113/Y103) is marked with an asterisk and the aspartic acid at the proposed counterion position (D181) for the green sea urchin opsin is indicated with a dagger.
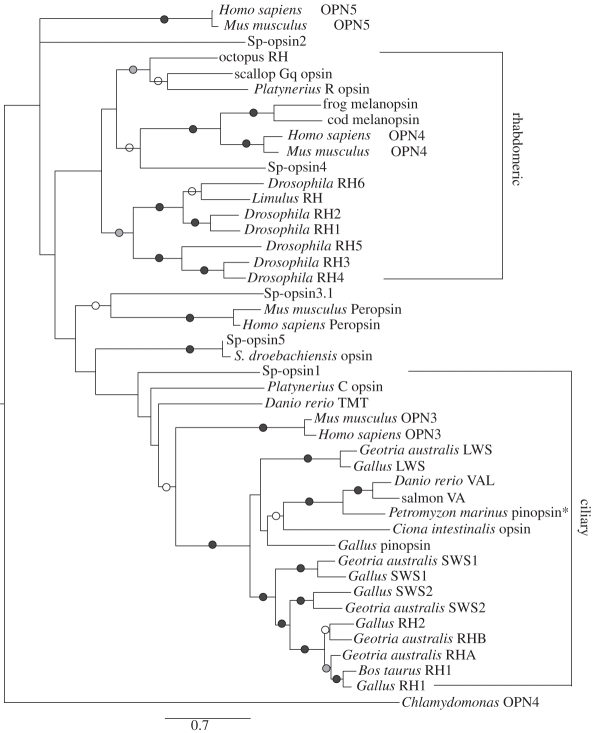
A phylogenetic analysis was performed on 45 opsin sequences, resulting in a bootstrap-supported, maximum-likelihood tree that provides insight into the relationship of this molecule to other opsins (figure 4). ProtTest found the Wheland and Goldman (WAG) [39] protein model to be best, with empirical amino acid frequencies (F option) and four categories of amino acid rate classes (G option). However, including invariant sites (I option) did not improve the model and so was not used. Bootstrap analyses give high support for the majority of the nodes on the tree.
Figure 4.
Phylogenetic analysis: neighbour-joining tree based on amino acid distances that are Poisson-corrected for multiple hits. Scale bar is substitutions per amino acid residue. The range of bootstrap values for significant nodes is indicated. LWS = long-wavelength sensitive, SWS = short-wavelength sensitive, VA = vertebrate ancient, VAL = vertebrate ancient long, RH = rhodopsin, OPN3 = panopsin or encephalopsin, OPN4 = melanopsin, OPN5 = neuropsin, TMT = teleost multiple tissue. An asterisk is located on the pinopsin of the lamprey, Petromyzon marinus, because there is a suggestion that it is actually a VA opsin [38], but the sequence is still listed as a pinopsin. Open circles, 50–75%; filled grey circles, 75–90%; filled black circles, more than 90%.
This analysis included the green sea urchin sequence, as well as sequence data for five urchin genes that encode for opsins from the S. purpuratus purple sea urchin genome project [16]. One of the S. purpuratus opsins (Sp-opsin5) is similar to the green sea urchin opsin, varying only 4 per cent at the nucleotide level and 5.4 per cent in their amino acid sequences. In our analyses and the Raible et al. [16] study, the green sea urchin sequence and Sp-opsin5 are intermediate between the ciliary and rhabdomeric opsins. Interestingly, comparisons of the S. droebachiensis cDNA with the genomic S. purpuratus sequences revealed that the open reading frame of the S. purpuratus homologue occurs without introns. This suggests that this gene is an inserted transcript with the extra sequence in the third cytoplasmic loop, probably resulting from a retrotransposon event.
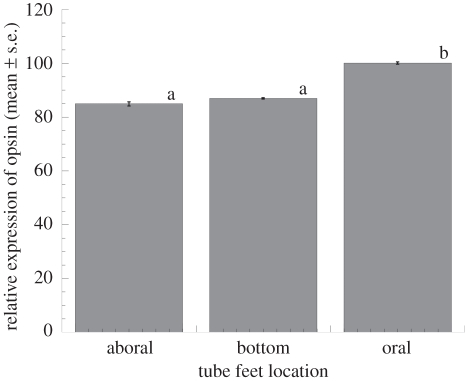
Primer efficiencies for the qPCR of green sea urchin opsin were 99 per cent. The differential expression of the green sea urchin rhabdomeric opsin from tube feet located on different parts of the sea urchin test, relative to GAPDH, showed a significant effect (ANOVA: F = 217.6, p < 0.0001) of tube foot location (figure 5). The tube feet on the oral side of the urchin test, which are exposed to the lowest irradiances of visible light (approx. 5 µmol quanta m−2 s−1; M. Lesser 2006, unpublished data), expressed significantly (Tukey's HSD, p < 0.05) more opsin transcripts than either tube feet from the bottom (approx. 35 µmol quanta m−2 s−1; M. Lesser 2006, unpublished data) or aboral (approx. 65 µmol quanta m−2 s−1; M. Lesser 2006, unpublished data) portions of the test that were exposed to increasing amounts of downwelling irradiance (figure 5) and were not significantly different from each other (Tukey's HSD, p > 0.05).
Figure 5.
Maximum relative expression of the rhabdomeric-like opsin in tube feet from different areas of urchin test (figure 1) compared with the expression of GAPDH in the green sea urchin Strongylocentrotus droebachiensis (mean ± s.e.). As these values are proportions they were a priori transformed (log+1) before analysis by ANOVA and were determined to be normally distributed and homoscedastic. All values were then back-transformed for presentation. Common letters above bars indicate samples that are not significantly different (p < 0.05) from each other using Tukey's HSD multiple comparison testing.
4. Discussion
The tube feet of the green sea urchin include morphological and molecular features consistent with a role in photoreception. Morphologically, the arrangement of the ossicles at the distal portion of the tube feet could function as a light collector in photoreception just as the calcitic microlenses of brittlestars refract and focus light as part of their ability to detect light [8]. As shown here, the presence, structure and morphological arrangement of the ossicles facilitates the scattering of light and the potential for increasing the path length of any individual photon, with a subsequent increase in the ability for light absorption within the ossicle by pigment cell opsins within the ossicle. A nerve plexus is also present along the entire length of the tube foot and extends under the ossicles, providing ultrastructural evidence of nerve transduction to the test and radial nerve. Additionally, the tube feet are used for adhesion and locomotion for which there is evidence of cholinergic-based motor control along the nerve [32,40]. The multiple lines of evidence provided here strongly suggest that tube feet are also photosensory organs [30,41].
The cells within the ossicles also express the transcriptional activator PAX6 protein, a product of the homeotic gene known to control eye development in other organisms [42]. PAX6 is expressed during the development of eyespots in planarians [43] and of structurally advanced image-forming eyes like those in mammals and squid [44,45]. Studies on the fruitfly D. melanogaster suggest that the evolutionarily ancestral role of PAX6 may have been to regulate structural genes involved in photoreception and that it later functioned to regulate the morphogenesis of divergent and complex eye structures, including image-forming eyes, and regulating the expression of rhodopsin [46]. PAX6 has been cloned from the gastrulae of the sea urchin Paracentrotus lividus [47] and is highly conserved with respect to vertebrate genes. A Northern blot analysis of adult P. lividus tissues (including ovary, testis, tube foot, intestine and coelomocytes) demonstrated PAX6 expression only in tube feet, including the tips of the tube feet [47] as we have reported here for the green sea urchin protein. PAX6 sequences from S. purpuratus [30], as well as green sea urchin PAX6 (GenBank DQ230536), are also highly conserved relatives of vertebrate PAX6 genes, including human PAX6.
The most unique feature of green sea urchin tube foot opsin is the extended third cytoplasmic loop between TM V and TM VI, which is 28 amino acid residues longer than the corresponding cytoplasmic loop of all other opsins. Together with amino acids in the third cytoplasmic loop and others in the cytoplasmic carboxy tail, these amino acids are the ones that interact with the α subunit of heterotrimeric G-proteins to initiate the phototransduction cascade [36,48]. All G-protein coupled receptors share a five-residue motif with unknown function in TM VII consisting of the amino acids NPXXY. Green sea urchin opsin has this motif as NPLIY (residues 348–352). The phylogenetic analysis of the green sea urchin opsin suggests that a Gq α is the most likely G-protein involved in the tube foot phototransduction cascade. Alternatively, the unique long third cytoplasmic loop may point to a novel G-protein, as is the case in phototransduction of the scallop, a protostome bivalve mollusc [48]. It is possible that the green sea urchin opsin has no known function or is a pseudogene as we have no protein expression data, but we believe this to be unlikely given the multiple lines of evidence presented here.
The phylogenetic position of the green sea urchin opsin sequence places it between the invertebrate Gq-coupled opsins found in rhabdomeric photoreceptors and the vertebrate Gt α-coupled opsins found in ciliary photoreceptors. However, many features of the green sea urchin opsin—including the presence of a tyrosine at the counterion position (E113/Y103), which is common to non-vertebrate opsins, and the proposed evolutionary history of photoreceptors and associated opsins [33,49]—support the classification of the green sea urchin opsin described here as a rhabdomeric-like opsin similar to Sp-opsin5 from the S. purpuratus urchin genome project, which is homologous to the green sea urchin opsin described here (figure 4). However, in addition to this rhabdomeric-like opsin, S. purpuratus does express a rhabdomeric opsin (Sp-opsin4) that groups with invertebrate Gq-coupled opsins and vertebrate melanopsins, as well as a ciliary opsin (figure 4, Sp-opsin1) that groups with the vertebrate opsins [16]. This expression of rhabdomeric and ciliary opsins is similar to a study by Arendt et al. [33] where they found that the polychaete worm, Platynereis dumerilii, has both vertebrate ciliary and rhabdomeric opsins that are associated with specific photoreceptor cells. Their finding confirms that both types of photoreceptors with distinct opsins are found in the Urbilateria (the last common ancestor of the Bilateria) and that in the evolutionary line leading to vertebrates, which includes sea urchins, both photoreceptor types were selected for, and incorporated into, the evolving vertebrate retina. The rhabdomeric photoreceptor cells may have transformed into ganglion cells expressing melanopsin, acquiring a new role in light detection and signal transduction [49]. First described in Xenopus, melanopsin is now known to confer photosensitivity on non-photosensitive cells types [50,51]. It has been hypothesized that melanopsin sets the circadian clock through non-ocular light detection [52], and is also expressed in sea urchins [30]. Additionally, an encephalopsin homologue has been sequenced from sea urchins [30,41], which is expressed at the tips of the larval arms of plutei and tube feet of adults [16,41], and potentially aids in the detection of light by planktonic larvae vertically migrating in the water column.
The inverse relationship between tube foot position on the test, which equates to differences in light exposure, and expression of the green sea urchin rhabdomeric-like opsin, is phenomenologically similar to the well-known inverse relationship between light and photosynthetic pigments universally expressed from microalgae to higher plants. This inverse relationship between light and opsin expression also occurs in the multi-cellular alga Volvox carteri, where the expression of an opsin (i.e. channelrhodopsin) homologous to animal opsins is inversely related to the amount of light the alga is exposed to, and is also used for phototaxis by the alga [53]. Similarly, the differential expression of the green sea urchin opsin in tube feet may be an important component of a sea urchin's ability to photoacclimatize to different irradiances on multiple timeframes to regulate the phototransduction of this external stimulus for important organismal functions, such as gametogenesis [3]. This may not be the case for all opsin classes in sea urchins, however, as the expression of encephalopsin in the tube feet of the sea urchin Hemicentrotus pulcherrimus showed a more direct relationship with light exposure, where encephalopsin expression decreased under dark conditions [41].
The green sea urchin, like many species of sea urchin, has unique hydraulic structures called tube feet. Within the tube feet there are calcareous ossicles, arranged in such a manner that they could serve as a light collector, and a unique, rhabdomeric-like opsin that is expressed along with PAX6, the master control gene for the development of eyes and photosensory organs in many organisms, including sea urchins [47]. In response to changes in the underwater light field, the light absorbed by the opsins of the pigment cells could potentially stimulate the nerves beneath them and conduct electrical information along the major nerve that passes along one side of each tube foot towards the test of the urchin. We now know that a full array of genes encoding neurotransmitters is present in sea urchins, which could facilitate the transmission of these signals [30]. For temperate invertebrates like the green sea urchin, opsins are well suited to mediate this transduction of an external environmental signal because of their absorption maxima in the blue–green portion of the spectrum that dominates in these underwater habitats [3,54]. The data presented here show that green sea urchins have a rhabdomeric-like opsin. While we have no direct evidence that this opsin is functional, it could be conferring an important capability to detect and transmit differences in light on varying time scales, from seconds and minutes used in the behavioural responses of echonoderms (e.g. [4]) to longer-term changes in irradiance occurring on the scale of months, which are important for processes such as seasonal gametogenesis [3]. Additional work is required to determine the various phototransduction pathways involving these opsins, any additional structures or tissues, where they are expressed, and the temporal scales over which they provide information to modify urchin behaviour and physiology. The simultaneous expression of an opsin gene and the unique morphological features at the distal portion of the tube foot shows that sea urchin tube feet, among their many functions, have also evolved into a unique photosensory organ to sense and respond to changes in the underwater light field, and may also provide useful information related to the origin of eyes in other organisms.
Acknowledgements
This study was supported by New Hampshire Sea Grant Programme grants (R/FMD-14 and R/FMD-166) awarded to C.W.W. and M.P.L., a National Research Initiative Competitive grant (2002-35206-11631) from the USDA and a northeast Regional Aquaculture Center grant (04-15) awarded to C.W.W., a National Science Foundation grant (IBN-0131285) awarded to K.L.C., and grants from the UNH Graduate School, Department of Zoology, Centre for Marine Biology and Sigma Xi Grant-in-aid-of-research awarded to T.M.B. Additionally, thanks to Florian Raible for providing the amino acid sequences for S. purpuratus opsin genes.
References
- 1.Pearse J., Pearse V., Davis K. 1986. Photoperiod regulation of gametogenesis and growth in the sea urchin, Strongylocentrotus purputatus. J. Exp. Zool. 237, 107–118 10.1002/jez.1402370115 (doi:10.1002/jez.1402370115) [DOI] [Google Scholar]
- 2.Walker C. W., Harrington L., Lesser M., Fagerberg W. 2005. Nutritive phagocyte incubation chambers provide a structural and nutritive microenvironment for germ cells of Strongylocentrotus droebachiensis, the green sea urchin. Biol. Bull. 209, 31–48 10.2307/3593140 (doi:10.2307/3593140) [DOI] [PubMed] [Google Scholar]
- 3.Walker C., Unuma T., Lesser M. 2007. Reproduction of sea urchins. In Edible sea urchins: biology and ecology (ed. Lawrence J. M.). Amsterdam, The Netherlands: Elsevier Science [Google Scholar]
- 4.Adams N. 2001. UV radiation evokes negative phototaxis and covering behavior in the sea urchin Strongylocentrotus droebachiensis. Mar. Ecol. Prog. Ser. 213, 87–95 10.3354/meps213087 (doi:10.3354/meps213087) [DOI] [Google Scholar]
- 5.Millot N. 1975. The photosensitivity of echinoids. Adv. Mar. Biol. 13, 1–52 10.1016/S0065-2881(08)60279-5 (doi:10.1016/S0065-2881(08)60279-5) [DOI] [Google Scholar]
- 6.Yerramilli D., Johnsen S. 2010. Spatial vision in the purple sea urchin Strongylocentrotus purpuratus (Echinoidea). J. Exp. Biol. 213, 249–255 10.1242/jeb.033159 (doi:10.1242/jeb.033159) [DOI] [PubMed] [Google Scholar]
- 7.Millot N. 1955. The covering reaction in a tropical sea urchin. Nature 175, 561. 10.1038/175561a0 (doi:10.1038/175561a0) [DOI] [Google Scholar]
- 8.Alzenberg J., Tkachenko A., Weiner S., Addadl L., Hendler G. 2001. Calcitic microlenses as part of the photoreceptor system in brittlestars. Nature 412, 819–822 10.1038/35090573 (doi:10.1038/35090573) [DOI] [PubMed] [Google Scholar]
- 9.Hyman L. H. 1955. Echinodermata. In The invertebrates, vol. 4 (ed. Boell E. J.), pp. 2–13 New York, NY: McGraw-Hill Publications [Google Scholar]
- 10.Johnsen S. 1997. Identification and localization of a possible rhodopsin in the echinoderms Asterias forbesi (Asteroidea) and Ophioderma brevispinum (Ophiuroidea). Biol. Bull. 193, 97–105 10.2307/1542739 (doi:10.2307/1542739) [DOI] [PubMed] [Google Scholar]
- 11.Millott N. 1968. The dermal light sense. Symp. Zool. Soc. Lond. 23, 1–36 [Google Scholar]
- 12.Yoshida M. 1979. Extraocular photoreception. In Handbook of sensory physiology (ed. Autrum H.), pp. 581–640 New York, NY: Springer [Google Scholar]
- 13.Yoshida M., Takasu N., Tamotsu S. 1984. Photoreception in echinoderms. In Photoreception and vision in invertebrates (ed. Ali A.), pp. 743–772 New York, NY: Plenum Press [Google Scholar]
- 14.Millot N., Yoshida M. 1960. The shadow reaction of Diadema antillarum Philippi. I. The spine response and its relation to the stimulus. J. Exp. Biol. 37, 363–375 [Google Scholar]
- 15.Gras H., Weber W. 1983. Spectral light sensitivity of isolated chromatophores of the sea urchin, Centrostephanus longispinus. Comp. Biochem. Physiol. A 76, 279–281 10.1016/0300-9629(83)90327-4 (doi:10.1016/0300-9629(83)90327-4) [DOI] [Google Scholar]
- 16.Raible F., Tessmar-Raible K., Arboleda E., Kaller T., Bork P., Arendt D., Amone M. I. 2006. Opsins and clusters of sensory G-protein coupled receptors in the sea urchin genome. Dev. Biol. 300, 461–475 10.1016/j.ydbio.2006.08.070 (doi:10.1016/j.ydbio.2006.08.070) [DOI] [PubMed] [Google Scholar]
- 17.Shichida Y., Matsuyama T. 2009. Evolution of opsins and phototransduction. Phil. Trans. R. Soc. B 364, 2881–2895 10.1098/rstb.2009.0051 (doi:10.1098/rstb.2009.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster R. G., Soni B. G. 1998. Extraretinal photoreceptors and their regulation of temporal physiology. Rev. Reprod. 3, 145–150 10.1530/ror.0.0030145 (doi:10.1530/ror.0.0030145) [DOI] [PubMed] [Google Scholar]
- 19.Crawford B. J., Burke R. D. 2004. TEM and SEM methods. Method Cell Biol. 74, 411–441 10.1016/S0091-679X(04)74017-0 (doi:10.1016/S0091-679X(04)74017-0) [DOI] [PubMed] [Google Scholar]
- 20.Moody M. L. 2003. The roles of rhodopsin, Pax-6 and c-myc proteins in the light induced gametongenic cycle of the green sea urchin, Strongylocentrotus droebachiensis, 71 pp Masters thesis, University of New Hampshire, Durham, USA [Google Scholar]
- 21.Katoh K., Misawa K., Kuma K., Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 10.1093/nar/gkf436 (doi:10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 10.1093/nar/25.24.4876 (doi:10.1093/nar/25.24.4876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abascal F., Zardoya R., Posada D. 2005. ProtTest: selection of best fit models of protein evolution. Bioinformatics 21, 2104–2105 10.1093/bioinformatics/bti263 (doi:10.1093/bioinformatics/bti263) [DOI] [PubMed] [Google Scholar]
- 24.Zwickl D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum-likelihood criterion. PhD thesis, The University of Texas, Austin, USA [Google Scholar]
- 25.Cummings M. P., Huskamp J. C. 2005. Grid computing. Educause Rev. 40, 116–117 [Google Scholar]
- 26.Bazinet A. L., Cummings M. P. 2008. The Lattice Project: a grid research and production environment combining multiple grid computing models. In Distributed and grid computing: science made transparent for everyone. principles, applications and supporting communities (ed. Weber M. H. W.), pp. 2–13 Marburg, Germany: Rechenkraft.net [Google Scholar]
- 27.Swofford D. L. 2001. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer Associates [Google Scholar]
- 28.Kyte J., Doolittle R. F. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 10.1016/0022-2836(82)90515-0 (doi:10.1016/0022-2836(82)90515-0) [DOI] [PubMed] [Google Scholar]
- 29.Barry T. 2005. Molecular and behavioral investigations of photreception in the green sea urchin, Strongylocentrotus droebachiensis. Masters thesis, University of New Hampshire, Durham, USA [Google Scholar]
- 30.Burke R. D., et al. 2006. A genomic view of the sea urchin nervous system. Dev. Biol. 300, 434–460 10.1016/j.ydbio.2006.08.007 (doi:10.1016/j.ydbio.2006.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Florey E., Cahill M. A. 1977. Ultrastructure of sea urchin tube feet. Cell Tiss. Res. 177, 195–214 10.1007/BF00221081 (doi:10.1007/BF00221081) [DOI] [PubMed] [Google Scholar]
- 32.Florey E., Cahill M. A. 1980. Cholinergic motor control of sea urchin tube feet: evidence for chemical transmission without synapses. J. Exp. Biol. 88, 281–292 [DOI] [PubMed] [Google Scholar]
- 33.Arendt D., Tessmar-Raible K., Snyman H., Dorresteijn A. W., Wittbrodt J. 2004. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science 306, 869–871 10.1126/science.1099955 (doi:10.1126/science.1099955) [DOI] [PubMed] [Google Scholar]
- 34.Nathans J., Hogness D. S. 1983. Isolation, sequence analysis, and intron–exon arrangement of the gene encoding bovine rhodopsin. Cell 34, 807–814 10.1016/0092-8674(83)90537-8 (doi:10.1016/0092-8674(83)90537-8) [DOI] [PubMed] [Google Scholar]
- 35.Wang J. K., McDowell J. H., Hargrave P. A. 1980. Site of attachment of 11-cis-retinal in bovine rhodopsin. Biochem. J. 19, 5111–5117 10.1021/bi00563a027 (doi:10.1021/bi00563a027) [DOI] [PubMed] [Google Scholar]
- 36.Karnik S. S., Sakmar T. P., Chen H. B., Khorana H. G. 1988. Cysteine residues 110 and 187 are essential for the formation of correct structure in bovine rhodopsin. Proc. Natl Acad. Sci. USA 85, 8459–8463 10.1073/pnas.85.22.8459 (doi:10.1073/pnas.85.22.8459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnheiter H. 1998. Evolutionary biology: eyes viewed from the skin. Nature 391, 632–633 10.1038/35487 (doi:10.1038/35487) [DOI] [PubMed] [Google Scholar]
- 38.Moutsaki P., Bellingham J., Soni B. G., David-Gray Z. K., Foster R. G. 2000. Sequence, genomic structure and tissue expression of carp (Cyprinus carpio L.) vertebrate ancient (VA) opsin. FEBS Lett. 473, 316–322 10.1016/S0014-5793(00)01550-7 (doi:10.1016/S0014-5793(00)01550-7) [DOI] [PubMed] [Google Scholar]
- 39.Whelan S., Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699 [DOI] [PubMed] [Google Scholar]
- 40.Santos R., Gorb S., Jamar V., Flammang P. 2005. Adhesion of tube feet to rough surfaces. J. Exp. Biol. 208, 2555–2567 10.1242/jeb.01683 (doi:10.1242/jeb.01683) [DOI] [PubMed] [Google Scholar]
- 41.Ooka S., Katow T., Yaguchi S., Yaguchi J., Katow H. 2010. Spatiotemporal expression pattern of an encephalopsin orthologue of the sea urchin Hemicentrotus pulcherrimus during early development, and its potential role in larval vertical migration. Develop. Growth Differ. 52, 195–207 10.1111/j.1440-169X.2009.01154.x (doi:10.1111/j.1440-169X.2009.01154.x) [DOI] [PubMed] [Google Scholar]
- 42.Halder G., Callaerts P., Gehring W. J. 1995. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267, 1788–1792 10.1126/science.7892602 (doi:10.1126/science.7892602) [DOI] [PubMed] [Google Scholar]
- 43.Callaerts P., Munoz-Marmol A. M., Castillo E., Sun H., Li W.-H., Gehering W. J., Salo E. 1999. Isolation and expression of a Pax-6 gene in the regenerating and intact planarian Dugesia (G) tigrina. Proc. Natl Acad. Sci. USA 96, 558–563 10.1073/pnas.96.2.558 (doi:10.1073/pnas.96.2.558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gehring W. J., Ikeo K. 1999. Pax 6: mastering eye morphogenesis and eye evolution. Trends Gen. 15, 371–377 10.1016/S0168-9525(99)01776-X (doi:10.1016/S0168-9525(99)01776-X) [DOI] [PubMed] [Google Scholar]
- 45.Tomarev S. I., Callaerts P., Kos L., Zinovieva R., Halder G., Gehring W., Piatigorsky J. 1997. Squid Pax-6 and eye development. Proc. Natl Acad. Sci. USA 94, 2421–2426 10.1073/pnas.94.6.2421 (doi:10.1073/pnas.94.6.2421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheng G., Thouvenot E., Schmucker D., Wilson D. S., Desplan C. 1997. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev. 11, 1122–1131 10.1101/gad.11.9.1122 (doi:10.1101/gad.11.9.1122) [DOI] [PubMed] [Google Scholar]
- 47.Czerny T., Busslinger M. 1995. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5). Mol. Cell Biol. 15, 2858–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kojima D., Terakita A., Ishikawa T., Tsukahara Y., Maeda A., Shichida Y. 1997. A novel Go-mediated phototransduction cascade in scallop visual cells. J. Biol. Chem. 272, 22 979–22 982 10.1074/jbc.272.37.22979 (doi:10.1074/jbc.272.37.22979) [DOI] [PubMed] [Google Scholar]
- 49.Arendt D. 2003. Evolution of eyes and photoreceptor cell types. Int. J. Dev. Biol. 47, 563–571 [PubMed] [Google Scholar]
- 50.Panda S., Nayak S. K., Campo B., Walker J. R., Hogenesch J. B., Jegla T. 2005. Illumination of the melanopsin signaling pathway. Science 307, 600–604 10.1126/science.1105121 (doi:10.1126/science.1105121) [DOI] [PubMed] [Google Scholar]
- 51.Qiu X., Kumnalasiri T., Carlson S. M., Wong K. Y., Krishna V., Provencio I., Berson D. M. 2005. Induction of photosensitivity by heterologous expression of melanopsin. Nature 433, 745–749 10.1038/nature03345 (doi:10.1038/nature03345) [DOI] [PubMed] [Google Scholar]
- 52.Foster R. G. 2005. Neurobiology: bright blue times. Nature 433, 698–699 10.1038/433698a (doi:10.1038/433698a) [DOI] [PubMed] [Google Scholar]
- 53.Ebnet E., Fischer M., Deininger W., Hegemann P. 1999. Volvoxrhodopsin, a light-regulated sensory photoreceptor of the spheroidal green alga Volvox carteri. Plant Cell 11, 1473–1484 10.2307/3870976 (doi:10.2307/3870976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cronin T. W. 1986. Photoreception in marine invertebrates. Am. Zool. 26, 403–415 [Google Scholar]