Abstract
The Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF57 protein is an essential multifunctional regulator of gene expression. ORF57 interaction with RNA is necessary for ORF57-mediated posttranscriptional functions, but little is known about the RNA elements that drive ORF57-RNA specificity. Here, we investigate the cis-acting factors on the KSHV PAN RNA that dictate ORF57 binding and activity. We show that ORF57 binds directly to the 5′ end of PAN RNA in KSHV-infected cells. Furthermore, we employ in vitro and cell-based assays to define a 30-nucleotide (nt) core ORF57-responsive element (ORE) that is necessary and sufficient for ORF57 binding and activity. Mutational analysis of the core ORE further suggests that a 9-nt sequence is a specific binding site for ORF57. These studies provide insight into ORF57 specificity determinants and lay a foundation for future analyses of cellular and viral ORF57 targets.
Keywords: ORF57/Mta, Kaposi’s sarcoma-associated herpesvirus, post-transcriptional gene regulation, PAN RNA, RNA-binding, gene expression
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV) is the causative agent of Kaposi’s sarcoma, primary effusion lymphoma (PEL), and some cases of multicentric Castleman’s disease (Cai et al., 2010; Dourmishev et al., 2003; Mesri et al., 2010). KSHV is a member of the gammaherpesvirus family which, like all herpesviruses, is characterized by both latent and lytic phases of infection. During latent infection, the virus expresses only a small subset of its genes that control functions essential for preserving the viral genome and the cells harboring them. Such functions include viral genome maintenance, immune evasion, and control of host cell proliferation. No viral progeny are made during this phase. In contrast, lytic infection involves a regulated cascade of viral gene expression that leads to the production of infectious virions. Both latently and lytically expressed genes have been implicated in KSHV pathogenesis.
The herpesviridae have evolved sophisticated mechanisms to control viral and host gene expression utilizing both viral and cellular factors. Because KSHV has a nuclear dsDNA genome, it is not surprising that KSHV utilizes host factors for transcription and mRNA processing. However, the virus encodes its own proteins that regulate gene expression both transcriptionally and posttranscriptionally (Conrad, 2009; Deng et al., 2007; Glaunsinger and Ganem, 2006; Sinclair, 2003; Staudt and Dittmer, 2007; Swaminathan, 2005). The 51 kDa ORF57 protein (Mta, KS-SM) is a member of a family of proteins conserved throughout the herpesviridae and it is essential for KSHV replication (Boyne and Whitehouse, 2006; Majerciak and Zheng, 2009; Han and Swaminathan, 2006; Majerciak et al., 2007; Ote et al., 2010; Sandri-Goldin, 2008; Toth and Stamminger, 2008). While ORF57 has been implicated in the regulation of KSHV transcription (Kirshner et al., 2000; Malik et al., 2004a; Palmeri et al., 2007), its post-transcriptional activities are more extensively studied. ORF57 has been reported to affect nearly every stage of gene expression including RNA stability, pre-mRNA splicing, RNA export, and translation (Boyne et al., 2008; Boyne et al., 2010; Majerciak et al., 2008; Malik et al., 2004b; Nekorchuk et al., 2007; Palmeri et al., 2007; Sahin et al., 2010).
ORF57 binds to RNA in vitro and in vivo (Boyne et al., 2008; Kang et al., 2011; Majerciak et al., 2006; Nekorchuk et al., 2007; Sahin et al., 2010), but it is currently unknown what dictates the specificity of ORF57 for its targets. Our recent work showed that ORF57 stabilizes PAN RNA (nut-1, T1.1), a nuclear non-coding polyadenylated RNA (Sahin et al., 2010; Sun et al., 1996; Zhong et al., 1996). PAN RNA accumulates to very high levels during lytic infection (Song et al., 2001; Sun et al., 1996) suggesting an important, yet unknown, function in viral replication. ORF57 enhances the abundance of PAN RNA in transfected cells (Kirshner et al., 2000; Nekorchuk et al., 2007; Sahin et al., 2010) and it is essential for PAN RNA accumulation during viral infection (Han and Swaminathan, 2006; Majerciak et al., 2007). Moreover, ORF57 binds directly to PAN RNA in cultured cells and RNA-binding is essential for ORF57 activity (Sahin et al., 2010). A 300-nucleotide (nt) sequence, the ORF57-responsive element (ORE), in the 5′-end of PAN RNA is necessary for binding and for ORF57-responsiveness. However, the minimal ORE has not previously been delineated, nor has it been shown that the ORE is directly and specifically bound by ORF57.
In the present work, we show that ORF57 binds the ORE in infected cells and we identify a 30-nt core ORE consisting of a predicted stem-loop structure. The core ORE was defined based on its activity in three assays. Specifically, ORF57 binds the core ORE in an in vitro label transfer assay, it is sufficient to confer ORF57-responsiveness to an intronless β-globin reporter, and it is necessary for ORF57 responsiveness of PAN RNA. We additionally identified point mutations in the 30-nt core ORE that abrogate binding in vitro and compromise ORF57 response in cells. These analyses implicate a 9-nt sequence in an unstructured loop of the core ORE as a potential ORF57-binding sequence. Recognition of the core ORE by ORF57 is likely sequence-specific rather than structure-based, because alterations in stems adjacent to the 9 nt sequence had little effect on ORF57 binding in vitro. Our analyses also reveal that, while the core ORE is necessary, additional sequences in the full-length ORE are required for robust ORF57-mediated enhancement of PAN RNA levels. These data are the first to clearly delineate an ORF57-interacting RNA sequence of a nuclear target of ORF57. In the long term, these data provide an important foundation to inform future investigations of ORF57 specificity and mechanisms.
RESULTS
ORF57 binds to the 5′ end of PAN RNA in lytically reactivated cells
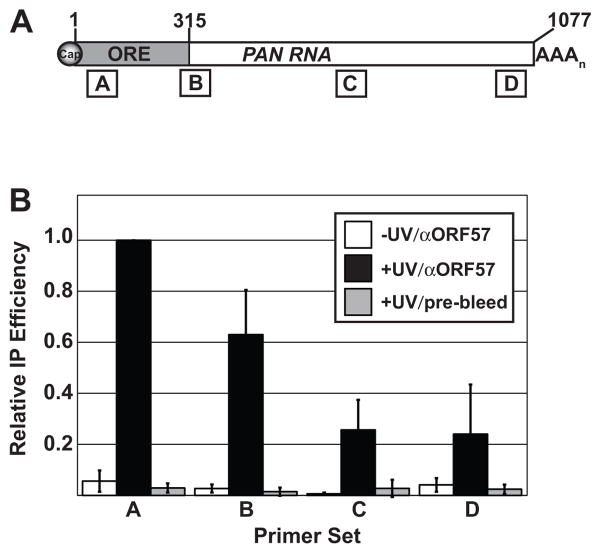
In transiently transfected cells, PAN RNA response to ORF57 is dictated by a 300-nt ORE at the 5′ end of the transcript (nt 13–312), which will be referred to as the full-length ORE throughout this manuscript. Based on these observations, we hypothesized that the 5′ end of PAN RNA contains a high-affinity binding site and that this site is important for ORF57 binding in infected cells (Sahin et al., 2010). We employed an ultraviolet (UV) cross-linking approach to examine whether ORF57 binds to the 5′ end of PAN RNA in KSHV-infected cells (Conrad, 2008; Sahin et al., 2010; Ule et al., 2005). In this assay, lytically reactivated cells are exposed to UV light, lysed, and a partial RNase digestion is performed to cleave the RNA into random fragments prior to immunoprecipitation of the RNA-protein complexes with ORF57 antibodies. The immunoprecipitated RNA fragments are then analyzed by quantitative reverse transcription PCR (qRT-PCR) with primers that span four different regions of PAN RNA (Figure 1A, boxes labeled A–D). Because UV light solely cross-links protein-RNA interactions in very close proximity, the immunoprecipitated RNA fragments represent those that are directly bound by ORF57.
Fig. 1.
ORF57 binds the 5′ end of PAN RNA in lytically reactivated cells. (A) Schematic diagram of PAN RNA with approximate positions of qRT-PCR amplicons shown below. Primer set A, B, C, and D amplify PAN RNA base pairs 50–124, 293–372, 642–728, and 994–1064, respectively. The numbering system for PAN RNA in this manuscript is relative to the start site as defined in Zhong et al. (1996). (B) Results from UV cross-linking immunoprecipitation experiments. The y-axis shows the immunoprecipitation efficiencies relative to that for primer set A (see Materials and Methods). The error bars are standard deviation (n=3).
Examination of the relative immunoprecipitation efficiencies of each segment of PAN RNA shows a clear 5′ bias (Figure 1B, black bars). Because viable cells are exposed to UV light in this procedure, demonstration of a UV-dependent cross-link reveals that the interaction occurs in living cells. Importantly, we see no immunoprecipitation of PAN RNA in the no-UV control (white bars), so we conclude that the interaction with ORF57 exists in lytic phase cells. Additionally, little to no immunoprecipitation was observed when pre-bleed serum was used in place of anti-ORF57 antibodies (gray bars). While we observe a 5′ bias, comparison of the test samples to the no-UV and pre-bleed controls for primer sets B and C show statistically significant immunoprecipitation (p<0.05, Student’s t-test). One interpretation of this observation is that there are multiple binding sites throughout these regions of PAN RNA. Alternatively, it is possible that the RNA was not digested sufficiently for resolution of independent fragments. The values for primer set D at the 3′ end of the transcript do not rise above statistical significance (p>0.1), so our data do not support the existence of an interaction between the 3′ terminus of PAN RNA and ORF57. We conclude that ORF57 binds PAN RNA directly during KSHV lytic infection and that the predominant binding site lies at the 5′ end of the transcript.
ORF57 binds specifically and directly to the PAN ORE
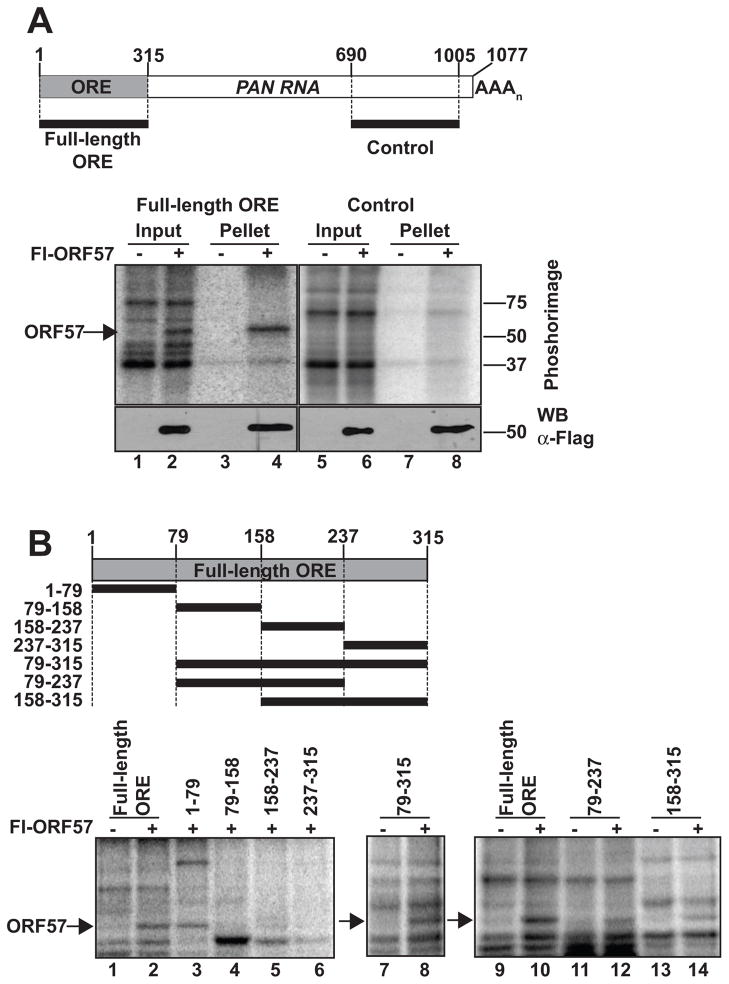
To further explore the interaction between ORF57 and the ORE, we employed an in vitro UV cross-linking, or label transfer, assay (Figure 2). In these assays, a radiolabeled RNA substrate is incubated in cell extract and exposed to UV light to covalently cross-link the proteins bound to the substrate RNA. After cross-linking, the RNAs are digested essentially to completion with RNase. However, small cross-linked RNA fragments (~1–10 nt) are protected from degradation due to the attached protein. The cross-linked protein-RNA complexes can be visualized by Phosphorimager analysis of protein gels by virtue of the radiolabeled RNA. We synthesized uniformly radiolabeled substrates containing the full-length ORE sequence and a control sequence lacking the ORE derived from the 3′ end of PAN RNA (Figure 2A, top). We incubated these RNAs in whole cell lysate from cells expressing Flag-tagged ORF57 (Fl-ORF57) or an empty vector control and performed the label transfer assay. The ORE substrate cross-links to a ~51 kDa protein in extract containing ORF57 but not in those extracts transfected with empty vector (lanes 1,2). The 51 kDa protein is immunoprecipitated with polyclonal antibodies specific for ORF57, confirming the identity of this protein as ORF57 (lanes 3,4). In contrast, the control substrate showed no binding to ORF57 in this assay (lanes 5–8). These experiments demonstrate that ORF57 binds specifically and directly to the ORE in whole cell extract and that those interactions reflect the binding patterns observed in infected cells (Figure 1).
Fig. 2.
ORF57 binds directly to the ORE in vitro. (A) Label transfer and immunoprecipitation assay. Top, Schematic diagram showing the positions of the full-length ORE or control substrate in PAN RNA. Bottom, Results from a representative label transfer assay. Extracts from cells expressing or not expressing flag-tagged ORF57 (Fl-ORF57) were incubated with the indicated substrate as described in the Materials and Methods. The cross-linked, RNase-treated extracts were then immunoprecipitated using anti-flag antibodies; 10% of input is shown. The bottom panels show an anti-flag western blot of the same samples demonstrating expression and immunoprecipitation of Fl-ORF57. (B) Label transfer assays with substrates derived from the ORE region. Top Schematic showing the substrates and their position with respect to the full-length ORE. The bottom panels show the label transfer assay with extract from cells expressing Fl-ORF57 or not as indicated above each lane. The position of ORF57 is indicated by the arrow.
We further exploited the label transfer assay to define the sequences necessary for ORF57 binding. Initially, we divided the full-length ORE into four non-overlapping 79/80-nt substrates (Figure 2B, top and lanes 1–6). The 5′-most of these substrates (nt 1–79) is sufficient for efficient ORF57 cross-linking, while the other three show little to no ORF57 binding. Next, we examined whether PAN RNA nt 1–79 are necessary for ORF57 interaction with the ORE, by deleting this sequence from the full-length ORE (nt 79–315). Surprisingly, this substrate binds to ORF57 even though its three constituent 79-nt fragments do not (lane 8 vs lanes 4–6). One possible explanation for this observation is that a second ORF57 binding site resides near nt 158 or nt 237, the endpoints of the shorter substrates. Therefore, we tested two overlapping fragments including the natural junctions between the individual 79-nt substrates (nt 79–237 and 158–315) and found that both of these fragments bind ORF57 (lanes 11–14). Two models are consistent with these data. It is possible that the RNA sequences surrounding nt 158 and 237 contain two independent ORF57 binding sites. However, based on observations in cultured cells (see below), we favor the model that the first 79 nt of PAN RNA contain a high-affinity ORF57 binding site, while nt 79–315 contain multiple lower-affinity binding sites.
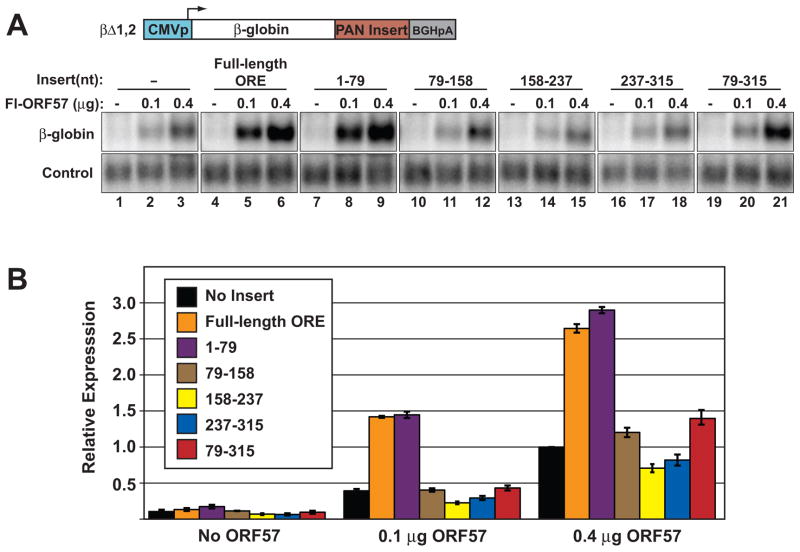
PAN RNA nt 1–79 are sufficient to confer ORF57-responsiveness to a heterologous transcript
We next utilized a β-globin reporter assay (Sahin et al., 2010) to test whether nt 1–79 are sufficient to confer ORF57-responsiveness in cells. We inserted ORE fragments into the 3′ UTR of a β-globin reporter construct from which both β-globin introns had been removed (βΔ1,2; Figure 3A). Our previous studies showed that co-transfection of ORF57 increases βΔ1,2 mRNA levels in a dose-dependent fashion, even in the absence of the ORE (Sahin et al., 2010). However, inclusion of the full-length ORE increases the ORF57-specific response (Figure 3A, lanes 1–6; Figure 3B, black and orange bars). We placed the four 79/80-nt non-overlapping fragments described in Figure 2 into the 3′UTR of βΔ1,2 and determined ORF57-responsiveness by northern blot (lanes 7–18). Quantification of the data shows that the first 79 nt of PAN RNA confer equivalent ORF57-responsiveness to βΔ1,2 as the full-length ORE (Figure 3B, purple bars). Because nt 79–315 maintained binding to ORF57 in label-transfer assays (Figure 2), we also inserted this fragment into βΔ1,2 (lanes 19–21). We observe a minimal ORF57 response with this insert (red bars). The effect is above background only at the highest concentration of ORF57 tested (compare red to black bars for 0.1 μg and 0.4 μg ORF57 samples). Taken with the in vitro results, these data support the model that nt 79–315 have low affinity for ORF57 and thus rely upon greater ORF57 levels to confer ORF57-dependent up-regulation. Most importantly, these data strongly support the conclusion that nt 1–79 contain a high-affinity ORF57 binding site, which is sufficient for ORF57-responsiveness in a heterologous context.
Fig. 3.
The first 79 nt of PAN RNA are sufficient for ORE activity in a heterologous transcript. (A) Top, Schematic diagram of the intronless β-globin reporter. Different portions of the ORE were placed into the 3′ UTR (“PAN Insert”) and tested for ORF57-responsiveness by northern blot. A representative northern blot with β-globin is shown below. The β-globin panels are from the same gel and are shown at the same exposure. The control lanes are probed for a co-transfected loading control. Amounts of co-transfected Fl-ORF57 and the particular insert are given above each lane. (B) Quantification of the northern blot data; error bars are standard deviation (n=3). Each value is relative to the no-insert control with 0.4 μg of ORF57.
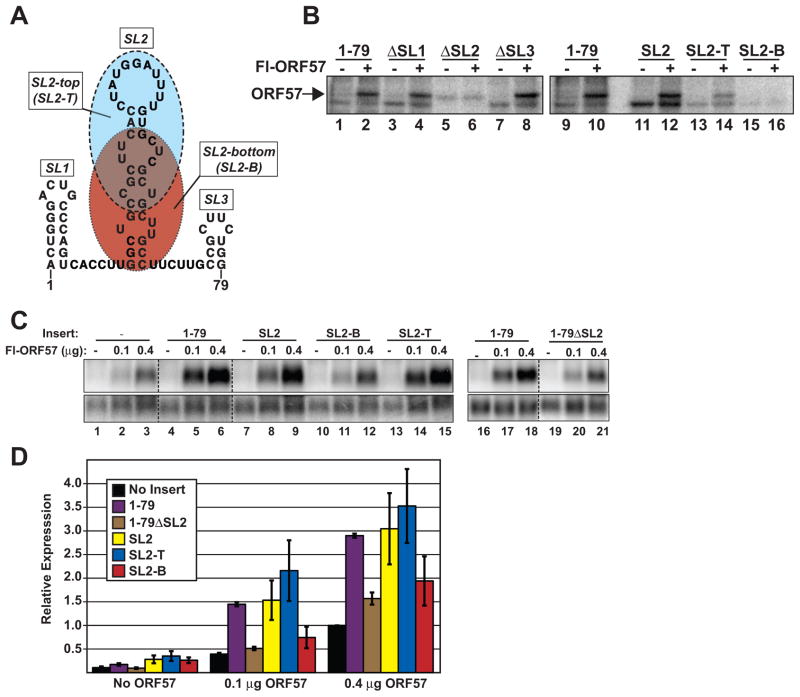
A predicted stem-loop in PAN RNA is necessary for binding in vitro and sufficient to confer ORF57-responsiveness
To determine a core sequence that is sufficient for ORF57 binding in vitro and for its activity in cells, we further dissected PAN RNA nt 1–79. The Mfold RNA secondary structure algorithm (Zuker, 2003) predicted that PAN RNA contains three stem-loop structures within nt 1–79 (Figure 4A; SL1, SL2, SL3). We first determined whether any of these loops are necessary for ORF57 binding in cell extract (Figure 4B). Deletion of SL1 or SL3 from nt 1–79 have no effect on ORF57 binding (Figure 4B; ΔSL1, ΔSL3; lanes 3,4, 7,8), while ORF57 cross-linking is completely lost upon deletion of SL2 (lanes 5,6). The faint protein band that co-migrates with ORF57 in ΔSL2 substrate lanes cannot be ORF57 because it is observed in the no ORF57 extract (lane 5). A substrate that contains the 40-nt SL2 without SL1 or SL3 efficiently cross-links to ORF57 (lanes 11, 12), demonstrating SL2 is sufficient for binding in whole cell extract. Further reduction of SL2 reveals that the cross-linking to ORF57 is driven by sequences in the top of the stem-loop structure. A 30-nt substrate derived from this portion (SL2-T, Figure 4A blue) efficiently cross-links ORF57 in vitro (lanes 13,14), while the bottom portion (Figure 4A; SL2-B) does not bind (lanes 15, 16). These results show that the 30-nt SL2-T sequence is necessary and sufficient to bind ORF57 in cell extract.
Fig. 4.
A 30 nt stem-loop structure is sufficient for ORE activity and binding. (A) Predicted secondary structure of PAN RNA nt 1–79. The three stem loops are labeled SL1, SL2, SL3 and the SL2 “top” (SL2-T) and “bottom” (SL2-B) portions are shown in ovals shaded with blue and red, respectively. (B) Label transfer assay with substrates that delete each of the stem loops (lanes 1–8) in the context of the nt 1–79 fragment. Lanes 11–16 show label transfer with SL2, SL2-T, or SL2-B alone. Cross-linking of each substrate is shown with extract either containing or lacking Fl-ORF57 as indicated. (C) Each of the indicated fragments was inserted into the intronless β-globin construct and examined for ORF57 response by northern blot as described in Figure 3. The dashed lines represent positions where lanes were removed for presentation; the panels displayed are from the same blot at the same exposure. (D) Quantification of the northern blot results was performed as in Figure 3 (n=3).
We next tested whether SL2 is sufficient to confer ORF57-responsiveness in a heterologous context by placing SL2, SL2-T, or SL2-B into the intronless β-globin reporter construct (Figure 4C and 4D). Consistent with the in vitro binding results, SL2 (yellow bars) and SL2-T (blue bars) are both sufficient to confer ORF57-responsiveness comparable to nt 1–79 (purple bars), but SL2-B is not (red bars). Additionally, deletion of SL2 from fragment 1–79 (1-79ΔSL2, brown bars) abrogated ORF57-responsiveness. Both SL2-B and 1-79ΔSL2 show limited, but above background, ORF57-responsiveness at high ORF57 levels (0.4 μg). These data suggest that that the SL2-T region is a core ORE capable of conferring ORF57-binding and response. However, the cis-acting factors responsible for the control of RNA levels by ORF57 are likely significantly more complex than a single short RNA element.
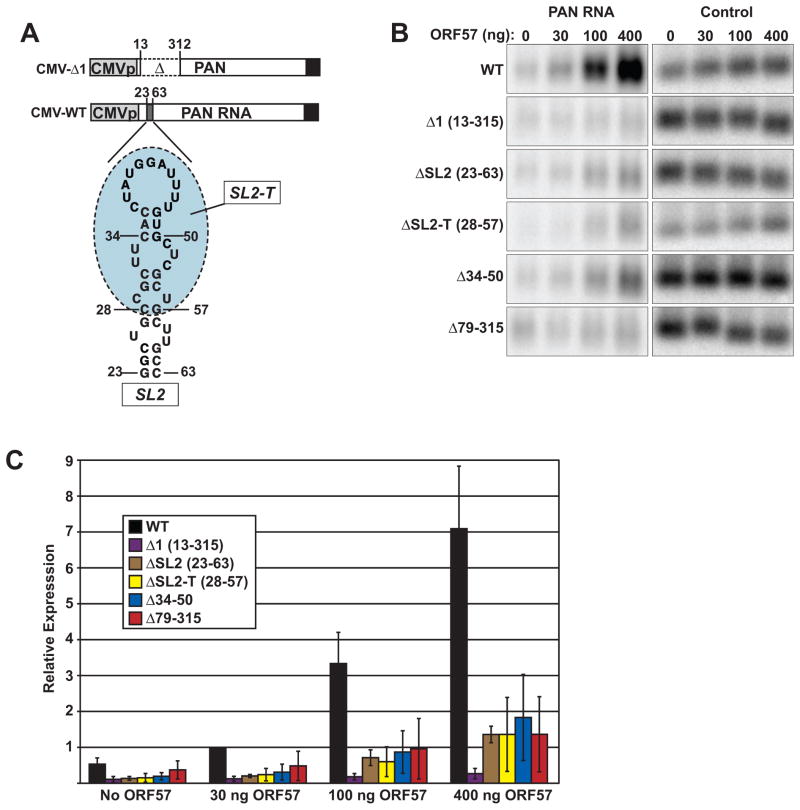
SL2-T is necessary for ORE activity in PAN RNA
The data presented above show that ORF57 binds to the 5′ end of PAN RNA in lytically reactivated PEL cells and that the SL2-T region is sufficient for ORE activity and binding. If this element is indeed a core ORE, removal of SL2-T should decrease PAN RNA levels in the presence of ORF57. To test this, we generated ORE deletions in a previously described ORF57-responsive cytomegalovirus immediate early (CMV) promoter-driven PAN RNA (Sahin et al., 2010) and examined their effects on PAN RNA accumulation in the presence of ORF57 (Figure 5A). We deleted the entire SL2 structure (ΔSL2; nt 23–63), the SL2-T region (ΔSL2-T; nt 28–57), or the predicted upper loop of SL2-T (Δ34-50). The 17 nt deleted in Δ34-50 bind ORF57 in vitro, but are not sufficient to confer ORF57-response in the β-globin reporter assay (data not shown). CMV-Δ1, which lacks the full-length ORE, and CMV-WT are negative and positive controls, respectively. We co-transfected these constructs with increasing amounts of ORF57 (0, 30 ng, 100 ng, and 400 ng) and quantified PAN RNA production by northern blot (Figure 5B and 5C). As previously reported, CMV-Δ1 is completely unresponsive to ORF57 (purple bars). As predicted, deletion of the SL2, SL2-T, or the upper loop of SL2 abrogate PAN RNA accumulation in the presence of ORF57 (brown, yellow, or blue bars, respectively). Thus, SL2, and in particular the SL2-T portion of the predicted stem loop contains a cis-acting activity necessary for the full ORF57 responsiveness in PAN RNA. Therefore, based on the three assays employed in this study, we conclude that the core ORE maps to the 30-nt SL2-T.
Fig. 5.
The core ORE is necessary for ORF57 responsiveness in PAN RNA. (A) Schematic representation of the CMV-driven PAN RNA expression constructs. The wild-type (WT) and Δ1 constructs were previously described (Sahin et al., 2010). Constructs that delete nt 23–63, 28–57, and 34–50 were generated to make the CMV-ΔSL2, CMV-ΔSL2-T, and CMV-Δ34-50 constructs, respectively. (B) Representative northern blot data analyzing the effects of the indicated deletions on ORF57 responsiveness. Panels on the left were probed for PAN RNA, while those on the right were the same blot probed for the endogenous 7SK RNA. The PAN RNA panels are from the same gel and are shown at the same exposure. (C) Quantification of the northern blot data. Samples were normalized to the 30 ng ORF57 samples with CMV-WT; error bars represent standard deviation (n=4).
In vitro binding and β-globin assays suggest that multiple potential binding sites are present in the ORE and that, at least in some contexts, they are sufficient for binding and limited activity. Moreover, deletion of the core ORE does not have as dramatic of an effect on ORE activity as deletion of the full-length ORE. Therefore, we tested a deletion construct (Δ79-315), which leaves SL2 intact, but removes the proposed lower affinity binding sites for ORF57 (Figure 2). In this case, we observe a similar lack of response to ORF57 as we do for ΔSL2 (red bars). One interpretation of this result is that the weak secondary binding sites observed in the in vitro binding experiments are necessary for full ORE activity in the context of PAN RNA. Thus, both the core ORE and secondary sites are necessary for full ORF57-responsiveness in PAN RNA. Alternatively, the core ORE may be sufficient in PAN RNA, however, the ORF57 binding site is masked by alterations in RNA secondary structure introduced by the deletions.
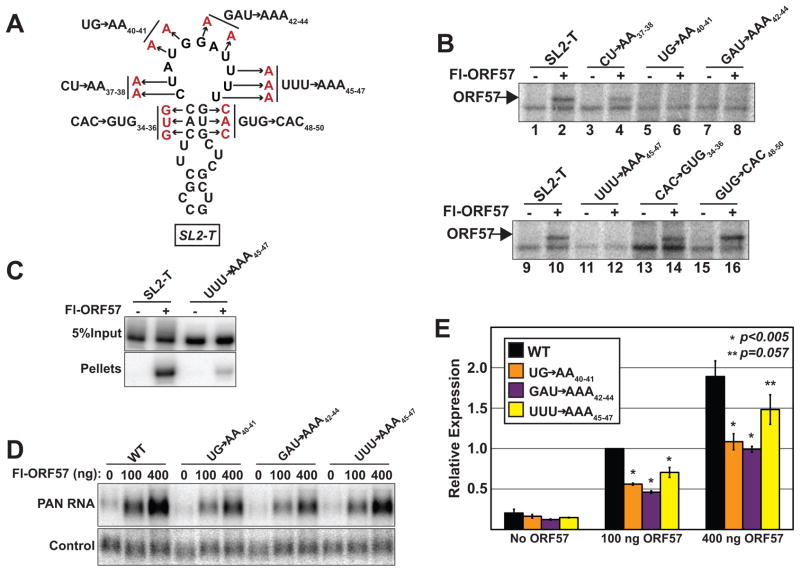
Point mutations in the core ORE abrogate ORF57 binding and activity
To examine activity of the core ORE at the nucleotide level, we generated a series of six point mutants that alter the 11-nt loop or the short stem of SL2-T (Figure 6A). Label transfer assays were employed to test the ability of ORF57 to cross-link each of the mutant substrates (Figure 6B). Mutations in the first two nucleotides of the 11-nt loop have no effect on ORF57 binding (CU→AA37–38 lanes 3,4). In contrast, ORF57 does not bind to any of the other three mutants tested in the SL2-T loop (lanes 5–8, 11,12). We also made mutations that disrupt the base pairing in the three nt stem structure immediately below the SL2-T loop. Neither of these mutations affect ORF57 binding (lanes 13–16), demonstrating that this structure is not necessary for ORF57 binding in vitro. We additionally generated a substrate containing mutations that restore the stem by combining CAC→GUG34–36 with GUG→CAC48–50. As predicted from the lack of effect of the individual triple point mutants, these compensatory mutations had no effect on ORF57 binding in vitro (data not shown). Together, these data suggest that the sequences in nt 40–47 comprise a sequence-specific binding site for ORF57 and that the secondary structure contributes little to binding in vitro.
Fig. 6.
Point mutations in the loop portion of SL2-T abrogate ORF57 binding and response. (A) Schematic diagram of triple and double point mutations generated in SL2-T. The mutations introduced are shown in red adjacent to the name of the mutant. (B) Label transfer assays with mutant substrates. The mutant substrates were generated in the context of the SL2-T sequence, which is used as a positive control (lanes 1, 2, 9, 10). Presence of Fl-ORF57 in the extract is indicated above each lane and the position of ORF57 is given by the arrow. (C) RNA immunoprecipitation of SL2-T and UUU→AAA45–47 substrates. Five percent of Input and 100% of pellets are shown at the same exposure from the same gel of an immunoprecipitation experiment with anti-flag agarose beads. The presence or absence of Fl-ORF57 is indicated above each lane as is the substrate. (D) Representative northern blot data analyzing the effects of the indicated mutations on ORF57 responsiveness probed for PAN RNA or a co-transfected loading control. The PAN RNA panels are from the same gel and are from the same exposure. (E) Quantification of northern blots. Values are relative to the CMV-WT at 100 ng transfection. The error bars are standard deviation (n=3). The p-values are derived from a two-tailed unpaired Student’s t-test comparing each sample to the WT.
One caveat to in vitro UV cross-linking is that, while all bases can be cross-linked, uridine bases cross-link with much greater efficiency (Conrad, 2008). Therefore, it is possible that our adenosine mutations inhibit cross-linking, but not binding to ORF57. This is especially true for the UUU→AAA45–47 mutant which replaces three uridine residues in the putative ORF57 binding site. As a complementary approach, we performed RNA immunoprecipitation from extract in the absence of cross-linking (Figure 6C). Labeled SL2-T or UUU→AAA45–47 substrate was incubated in extract containing or lacking Fl-ORF57 and subsequently immunoprecipitated using anti-flag antibodies. Consistent with the UV cross-linking results, the immunoprecipitation efficiency of SL2-T is significantly more robust (~10-fold) than the UUU→AAA45–47 substrate. Thus, the UUU→AAA45–47 mutation abrogates both cross-linking and binding of ORF57 in vitro, supporting the conclusion that nt 45–47 are part of a sequence-specific binding site for ORF57.
We next introduced the mutations that abrogate binding in vitro into the PAN RNA expression constructs and examined their effects on PAN RNA accumulation in the presence of ORF57 (Figure 6D, 6E). While the mutated constructs retain significant ORF57-responsiveness (Figure 6D), accumulation of PAN RNA is reduced by 2-fold in two of the mutants (UG→AA40–41, GAU→AAA42–44). The two-fold effect is observed both at 100 ng and 400 ng of ORF57 and is highly statistically significant (p<0.005), so we conclude that these specific nucleotides are central to ORF57 activity on PAN RNA. The third mutation tested, UUU→AAA45–47, reduces PAN RNA levels by a small (30%), but statistically significant (p<0.005), quantity when co-transfected with 100 ng ORF57. Upon transfection of four times as much ORF57, the difference still exists, but it is of questionable statistical significance (p=0.057). Taken together, these data suggest that the nucleotide sequence AUGGAUUUU contributes significantly to the binding of ORF57 to PAN RNA, but also demonstrate that other binding sites contribute to the ORF57 response.
DISCUSSION
The ORF57 protein is an essential KSHV factor with multiple roles in viral gene expression (Boyne and Whitehouse, 2006; Conrad, 2009; Majerciak and Zheng, 2009; Swaminathan, 2005). While it likely plays a role in transcription, most of its proposed activities involve posttranscriptional regulation of RNA metabolism and its RNA-binding properties are important for these functions. However, the mechanisms ORF57 uses to recognize its ligand RNAs are unknown. Here, we delineate a core ORF57-responsive element in PAN RNA, a known ORF57 target in both cultured and infected cells (Han and Swaminathan, 2006; Kirshner et al., 2000; Majerciak et al., 2007; Nekorchuk et al., 2007; Sahin et al., 2010). Conservatively, we define the core ORE to be a 30-nt sequence, SL2-T, because it fits all three criteria tested in the present work. That is, SL2-T 1) binds to ORF57 in vitro, 2) enhances intronless β-globin RNA levels in the presence of ORF57, and 3) is necessary for ORF57-responsiveness of PAN RNA. Most likely, the core activity is contained in an even smaller region. Specifically, the nucleotides AUGGAUUUU in the unstructured loop at the top of the core ORE are essential for binding and for full ORF57-responsiveness in PAN RNA. Thus, these nine nucleotides may constitute an ORF57 RNA binding motif.
ORF57 post-transcriptionally enhances the expression of many viral genes as well as various reporter constructs, so it may seem that its binding to RNA is relatively nonspecific. Indeed, in our β-globin assays (Figures 3 and 4) there is a consistent background of ~4–5-fold enhancement by ORF57 in the absence of any ORE. This relatively non-specific effect may be important for the biology of ORF57. In mammalian cells, the stages of nuclear pre-mRNA processing are tightly coupled, and abrogation of one activity often affects other processing steps (Le Hir et al., 2003; Pandit et al., 2008; Reed, 2003; Vinciguerra and Stutz, 2004). Pre-mRNA splicing is central to this coupling, but most KSHV transcripts lack introns (Zheng, 2003). As a result, KSHV genes are missing an important component of efficient expression. One ascribed function of ORF57 is to enhance the expression of intron-lacking viral mRNAs. As such, it seems likely that ORF57 would evolve a relatively promiscuous binding specificity to enable it to bind the large pool of intronless viral mRNAs expressed during lytic phase. Peculiarities of individual viral transcripts may require more robust binding by ORF57 to yield a higher activity. For example, the nuclear accumulation of PAN RNA appears to require a stable complex with ORF57 to protect it from nuclear decay enzymes. Perhaps in these cases, transcripts evolved cis-acting OREs like the one described here.
While this work was in preparation, Zheng and colleagues published a different ORF57-responsive element derived from the viral interleukin-6 (vIL-6) mRNA (Kang et al., 2011). The vIL-6 MRE (Mta-responsive element) overlapped with a miRNA binding site and the authors proposed that binding of ORF57 antagonized the down-regulatory effects of the miRNA. While this mechanism is unlikely to be responsible for ORF57-dependent stabilization of the nuclear PAN RNA, binding of ORF57 to the vIL-6 MRE or to the PAN ORE is essential for its regulatory function. Comparison of the core PAN ORE with the vIL-6 MRE reveals little overall sequence homology. However, there are two interesting similarities. Consistent with the idea that high-affinity ORF57 binding is driven by sequence rather than structure, both elements are predicted to fold into stem-loops, but neither shows particularly strong secondary structures. Even more compelling, the uppermost loops of both the vIL-6 MRE and the PAN ORE share the tetranucleotide GGAU. This element is necessary for ORE activity because mutation of this motif (UG→AA40–41, GAU→AAA42–44) abrogates binding in vitro and ORF57-responsiveness in cells. Thus, these data suggest that the GGAU motif may provide sequence specificity for ORF57 binding that is utilized in multiple transcripts. Interestingly, the herpesvirus saimiri ORF57 protein has been reported to preferentially bind a purine-rich GGAGRG element (Colgan et al., 2009), so there may be at least loose conservation of binding-site specificity with respect to the GGA trinucleotide. Identification and detailed examination of additional ORF57-responsive elements is necessary to test whether this sequence is a general feature of ORF57 regulated RNAs.
In the same study that identified the vIL-6 MRE (Kang et al., 2011), a nonbiased approach was taken to clone viral RNA fragments that interact with ORF57. Consistent with our results, they found that PAN RNA interacted with ORF57; 16 out of 91 PAN RNA clones (18%) overlapped the SL2-T core ORE sequence. Surprisingly, other clones spanned nearly every region of PAN RNA with close to 50% overlapping the ENE, a cis-acting stability element in PAN RNA (Conrad et al., 2006; Conrad et al., 2007; Conrad and Steitz, 2005). No validation of the cloned PAN RNA fragments for their ability to confer ORF57-responsiveness was performed in that study, but our data clearly show that ORF57 binding and response is conferred by sequences residing at the 5′ end of PAN RNA. In fact, deletion of the ENE actually increases ORF57-responsiveness, presumably due to the fact that ORF57’s stabilization effects are greater on an inherently less stable transcript (Sahin et al., 2010). Moreover, our in vivo and in vitro UV cross-linking results do not support a direct interaction between ORF57 and the 3′ end of PAN RNA (Figures 1 and 2).
Our data are consistent, however, with the general model that the core ORE is not the only binding site in PAN RNA for ORF57. Sequences contained in nt 79–315 bind PAN RNA in vitro and deletion or mutation of the core ORE is not sufficient to completely abrogate ORF57-responsiveness (Figures 5 and 6). In contrast, when the full-length ORE is deleted, virtually no ORF57 response is observed. We speculate that the core ORE efficiently recruits ORF57, which has been shown to be in a homomultimer complex of unknown number (Nekorchuk et al., 2007). Once one ORF57 molecule is bound to the ORE, the RNA binding domain(s) on the unbound ORF57 molecule(s) are present in a high local concentration driving binding to weaker sites in adjacent sequences. Similar cooperative models have been proposed for other RNA-binding proteins like hnRNP A1 and the HIV REV protein (Daugherty et al., 2008; Zhu et al., 2001). The presence of high ORF57 levels and/or multiple weak ORF57-binding sites can drive this interaction in the absence of a high-affinity ORE leading to the observed ORF57 promiscuity.
The steady-state analyses of PAN RNA and β-globin used in this study monitored the accumulation of transcript as a measurement of ORF57 activity. These approaches are confounded by the presence of an uncharacterized enhancer of PAN RNA expression that overlaps the full-length ORE (Conrad and Steitz, 2005; Sahin et al., 2010). When driven by the PAN RNA promoter, deletion of the full-length ORE decreases PAN RNA levels by ~5-fold in the absence of ORF57. Use of the CMV promoter decreases the magnitude of the effect of this enhancer, but some up-regulation of RNA levels by this element is still observed. Close examination of the no ORF57 controls in this study suggest a subtle trend of increased RNA accumulation in the presence of the core ORE in the absence of ORF57 (Figures 3–6). To be sure, careful quantification of these data rarely yields a statistically significant effect (data not shown), so it is difficult to interpret the relevance of the trend. One formal possibility is that the core ORE does not drive ORF57-response directly, but rather it is an element that has a function in gene expression that lies upstream of ORF57 activity. However, this idea seems unlikely based on our in vitro binding and β-globin reporter assays. We favor the idea that the element may bind to cellular factors that act in concert with ORF57 to promote gene expression. In the absence of ORF57, the activity of the cellular factor is minimized, but in its presence the effect is enhanced. Consistent with this idea, in vitro binding of baculovirus-expressed ORF57 is enhanced by the presence of cellular extract (Majerciak et al., 2006). Further experimentation is required to unravel the complex molecular mechanisms of these primary and secondary ORF57-responsive elements in PAN RNA.
ORF57 is a multifunctional regulator of gene expression that is essential for KSHV replication. In order to take a meaningful global approach to examine ORF57 targets, we must first employ reductionist techniques to define the requirements for ORF57-RNA interactions. Here we have identified a core element from a natural ORF57 target that is sufficient for ORF57 binding and response. These data provide a foundation to compare ORF57-responsive elements from novel viral and cellular targets as they are uncovered.
MATERIALS AND METHODS
Plasmids
The βΔ1,2, CMV-WT, CMV-Δ1, PANΔ79, PAN-WT and pcFl-ORF57II expression constructs were described previously (Conrad and Steitz, 2005; Conrad et al., 2006; Conrad et al., 2007; Sahin et al., 2010). The PAN RNA inserts for the β-globin reporter assay were generated by PCR amplification of the corresponding sequences using the primers listed in the Supplementary Content with PAN-WT as a template. The PANΔ79-ΔSL2 construct (see below) was used as a PCR template for the ΔSL2 insert. The resulting PCR products were digested with NotI and XhoI and inserted into the βΔ1,2 plasmid cut with the same restriction enzymes. Smaller inserts were created by annealing DNA oligonucleotides with the appropriate NotI and XhoI overhangs. In this case, each oligonucleotide was 5′ phosphorylated with T4 polynucleotide kinase prior to annealing using standard procedures.
Prior to making the CMV-driven PAN RNA expression constructs, the deletions in the ORE were first introduced into PANΔ79 by SOEing PCR (Horton, 1995). The primer sets used are listed in the Supplementary Content. Products were digested with PflMI and NcoI and inserted into the same sites on PANΔ79. NcoI was substituted by AfeI where appropriate. The CMV-driven PAN RNA deletion plasmids were constructed by PCR amplification of PAN RNA sequences from these PANΔ79 derivatives using primers NC872 (5′ agtcctAAGCTTactgggactgcccagtcacc 3′) and NC10 (5′ gggggcccgtcacatttagggcaaagtgg 3′). The PCR products were digested with HindIII and XbaI and ligated into the CMV-WT plasmid cut with the same enzymes. Point mutants in CMV-WT were created using the same SOEing approach and the primers are listed in the supplemental content. The sequence of all PCR-generated inserts was verified.
Cell culture and transfection
TREx BCBL1-Rta cells (Nakamura et al., 2003) were carried in RPMI-1640 media (Sigma) supplemented with 10% tetracycline-free FBS (Clontech), penicillin-streptomycin (Sigma), 2 mM L-glutamate, and 100 μg/ml hygromycin (Sigma). HEK293 cells were grown as previously described in (Sahin et al., 2010).
HEK293 cells were transfected using TransIT-293 (Mirus). β-globin reporter constructs were transfected in 12-well tissue culture plates with a total of 0.8 μg plasmid DNA. A typical transfection contained 0.4 μg of pcFl-ORF57II plus pcDNA3, 0.1 μg of a control plasmid (mgU2-19/30) (Sahin et al., 2010), and 0.3 μg of the β-globin reporter. For the CMV-PAN derivatives, β-globin reporter DNA was substituted by 0.3 μg of a PAN RNA expression construct. Total RNA was harvested 18–24 hrs after transfection using TRI Reagent (Molecular Research Center) and analyzed by northern blotting using standard procedures (Church and Gilbert, 1984; Conrad and Steitz, 2005). Riboprobes against the endogenous 7SK RNA or the co-transfected control were used to normalize RNA recovery and loading.
Antibodies
ORF57 antibodies were made in rabbits (Cocalico Biologicals) using a mixture of two bacterially expressed ORF57 polypeptide sequences, fm9 (amino acids 1–153) and fm7 (amino acids 171–455). Due to propensity of these proteins to precipitate, they were kept under denaturing conditions (0.5% SDS or 8M urea) for the entire preparation. ORF57 antibodies were affinity purified from rabbit serum using the Microlink Peptide Coupling kit (Pierce) as per the manufacturer’s instructions. The affinity purified antibody recognized a single 51-kDa protein on western blots of lysate from infected or transfected ORF57-expressing cells, but not from that of cells that did not express ORF57 (data not shown). Pre-bleed antibodies were purified with Protein A beads (Pierce) using standard procedures.
UV Cross-linking and immunoprecipitation
TREx BCBL1-Rta cells at 5×105 cells/ml were reactivated by treatment with 1 μg/ml doxycycline (Fisher) and 3 mM sodium butyrate (Sigma) for 18–20 hrs. A total of 1×107 cells/condition were washed in 10 mls of phosphate-buffered saline (PBS; Sigma), centrifuged at 700×g for 3 min at 4°C and resuspended in 3 mls of PBS. The cell suspension was transferred to a 10 cm cell culture dish and irradiated on ice at 250 mJ/cm2 ~2–3 cm from the UV source (Spectroline XL-1500, 254nm). After cross-linking, 7 mls of PBS were added to the dish. Cells were transferred to 15 ml conical tubes, and centrifuged at 700×g for 3 min at 4°C. Cell pellets were resuspended in 1ml PBS and centrifuged at 2400×g for 1 min at 4°C. PBS was removed and the pellets were frozen at −80°C (Conrad, 2008). Cell lysis and immunoprecipitation were performed essentially as previously described (Conrad, 2008) except addition of poly(U) RNA was omitted and the QIAshredder step was substituted by digestion with 30 μl of RQ1 DNase (Promega) for 15 min at 25°C. Subsequently, 10 μl of 1μg/ml RNase A (Sigma) was added and incubated at 25°C for 10 min (Ule et al., 2005). For the immunoprecipitation, approximately 8 μg of antibody (pre-bleed or αORF57) were added to the extracts, which were then nutated for 1 hr at 4°C. The extracts were then added to 20 μl of protein-A agarose beads (Pierce) and nutated for an additional hour at 4°C. Washing of bound antibody complexes, elution and RNA recovery were performed exactly as described (Conrad, 2008).
To analyze the RNAs recovered from the UV cross-linking protocol, the input and pellet samples were DNase treated for 1 hr at 37°C in a 20 μl reaction containing 2 μl of RQ1 DNase, 20 units of RNasin Plus (Promega), and DNase Buffer (40 mM Tris pH 8.0, 10 mM MgSO4, 1 mM CaCl2). The reaction volume was brought up to 200 μl in G-50 Buffer (20 mM Tris pH 7.5, 0.25% SDS, 0.3 M sodium acetate, 2 mM EDTA) and then extracted with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1; PCA). An aliquot of the resulting RNAs (~1/7) was ethanol precipitated with 15 μg of glycoblue (Ambion). Reverse transcription reaction was performed using SuperScript™ II RT (Invitrogen) and random hexamers (Sigma) as per the manufacturer’s protocol. No-RT controls were performed in parallel for each sample. Inputs and pellets were diluted 40-fold, 2 μl of which was used as template for real-time PCR as described (Sahin et al., 2010). Real-time primer sets were: A, NC702 (5′ GCTCGCTGCTTGCCTTCTT 3′) and NC703 (5′ CCAAAAGCGACGCAATCAA 3′); B, NC704 (5′ CTTGCGGGTTATTGCATTGG 3′) and NC705 (5′ GACACGTTAAGTATCCTCGCATATCA 3′); C, NC706 (5′ TTTTCCAGTGTAAGCAAGTCGATTT 3′) and NC707 (5′ TGTTCTTACACGACTTTGAAACTTCTG 3′); D, NC708 (5′ TTAACGTGCCTAGAGCTCAAATTAAAC 3′) and NC709 (5′ TTGACCTTTATTTATGTTGTAAGTTGCATTA 3′). Efficiency of the A, B, C and D amplification was determined to be 92%, 91%, 82% and 83%, respectively (Pfaffl, 2001). Relative quantities (RQ) of Inputs and Pellets were determined based on their amplification efficiency and Ct value. The RQ value for the no-RT controls was subtracted from the plus RT samples. Immunoprecipitation efficiency was determined by calculating the pellet/input ratio of the background corrected values. To compare between experiments, these values were then normalized to the”+UV/αORF57” ratio for primer set A.
Label transfer experiments
DNA templates for in vitro transcription of each substrate were generated by PCR amplification using primers containing the T7 promoter sequence (Supplemental Content). In the case of ΔSL2, template was amplified from PANΔ79-ΔSL2, while all others utilized PAN-WT as a template. The smallest T7-DNA templates were made by annealing oligonucleotides. Each oligonucleotide pair was heated at 90°C for 3 min in annealing buffer [10 mM Tris pH 7.5, 150 mM NaCl, 25 mM EDTA, 2.5 μM each oligonucleotide] and then slowly equilibrated to room temperature. Body labeled RNA fragments were generated in a 20 μl reaction containing 50 units of T7 RNA polymerase, 5 μl α-32P-UTP (800 Ci/mmol, 0.010 mCi/μl), 20U RNasin Plus and transcription buffer (40 mM Tris pH7.5, 6 mM MgCl2, 4 mM spermidine, 50 μM UTP, 1 mM ATP, 1 mM CTP, 1 mM GTP, 10 mM DTT). Transcription reaction was run through an illustra MicroSpin™ G-25 column (GE), the RNA was ethanol precipitated with 1M ammonium acetate and glycoblue, and then separated on a urea-PAGE. Full length RNAs were excised from the gel and eluted in G-50 buffer overnight at room temperature. Eluted fractions were extracted with PCA, ethanol precipitated, and resuspended in water. RNA yield was approximated by scintillation (Cerenkov) counting.
Whole cell extracts were made by transfecting a 10 cm dish with 10 μg of either pcDNA3 or pcFl-ORF57II. After 48 hrs, cells were harvested, washed with PBS, and the cell pellets were collected by centrifugation at 2400×g for 1 min at 4°C. The pellets were subsequently resuspended in 300 μl of RSB100T-Plus [10mM Tris pH 7.5, 2.5 mM MgCl2, 100mM NaCl, 0.5% TritonX100, 1mM phenylmethanesulphonylfluoride (PMSF, Sigma), protease inhibitors (cocktail V, Calbiochem)], sonicated three times for 10 sec at 30% amplitude (SONICS Vibra-cell™ VCX130 with a 6 mm probe), and then centrifuged at 16,000×g for 10 min at 4°C. After adding glycerol to 10%, the supernatants were stored at −80°C.
The binding reactions for label transfer assays were performed in a 40 μl reaction volume including 10 μl of whole cell extract, 10 μl RSB100T (10mM Tris pH 7.5, 2.5 mM MgCl2, 100mM NaCl, 0.5% Triton X-100), substrate (~15 nM), 500 μM MgCl2, 80 mM KCl, 1 mM ATP, 20 mM creatine phosphate, and 0.3 mg/ml Torula yeast RNA (Sigma). In some cases 20 μl of extract were used, however, higher amounts of extract did not affect the ORF57 signal strength. Binding was allowed to reach equilibrium by incubating at 30°C for 15 min. The protein-RNA complexes were then cross-linked on ice at 860 mJ/cm2 in a Spectroline XL-1500 (254 nm). Samples were then treated with RNase A at 0.25 mg/ml for 30 min at 25°C. For immunoprecipitation, 80 μl of RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) were added along with ~4.5 μg of affinity purified ORF57 antibody and the extract was nutated for 1 hr at 4°C. This mixture was then transferred to 15 μl of protein-A agarose beads and nutated for an additional hour at 4°C. Beads were washed a total of five times in 1 ml of RIPA buffer. Proteins were eluted by boiling in SDS gel loading buffer. The samples were separated on a 10% SDS-PAGE, dried, and exposed on a Phosphorimager screen.
RNA Immunoprecipitation
Binding reactions for RNA immunoprecipitation experiments were performed as described for the label transfer experiments, except cpm were equilibrated (6×106 cpm), rather than molarity. After binding, the volume was increased to 200 μL with RSB100T and the samples were nutated at 4°C for 1 hr with 15 μl of anti-flag M2 affinity beads (Sigma) pre-blocked with yeast RNA. Beads were washed a total of five times in 750 μl of RSB100T supplemented with 500 μM MgCl2, 80 mM KCl, and 15U of RNasin Plus. Proteins from both inputs and pellets were digested in 200 μl of 4mg/ml proteinase K, 100 mM Tris pH 7.5, 50 mM NaCl, and 10 mM EDTA for 1 hr at 37 °C. Finally, the samples were extracted in an equal volume of PCA, ethanol precipitated, and analyzed by urea-PAGE.
Supplementary Material
Acknowledgments
We thank Olga Hunter for technical assistance with plasmid construction and Dr. Pinghui Feng, Sarah Stubbs, and Stefan Bresson for critical review of this manuscript. We thank Drs. Jae Jung and Pinghui Feng for the TREx BCBL1-Rta cells.
FUNDING
This work was funded by the NIH-NIAID grant AI081710 and by the Welch Foundation research grant I-1732. NKC is a Southwestern Medical Foundation Scholar in Biomedical Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boyne JR, Colgan KJ, Whitehouse A. Recruitment of the complete hTREX complex is required for Kaposi’s sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 2008;4:e1000194. doi: 10.1371/journal.ppat.1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Jackson BR, Taylor A, Macnab SA, Whitehouse A. Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J. 2010;29:1851–1864. doi: 10.1038/emboj.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Whitehouse A. gamma-2 Herpes virus post-transcriptional gene regulation. Clin Microbiol Infect. 2006;12:110–117. doi: 10.1111/j.1469-0691.2005.01317.x. [DOI] [PubMed] [Google Scholar]
- Cai Q, Verma SC, Lu J, Robertson ES. Molecular biology of Kaposi’s sarcoma-associated herpesvirus and related oncogenesis. Advances in Virus Research. 2010;78:87–142. doi: 10.1016/B978-0-12-385032-4.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan KJ, Boyne JR, Whitehouse A. Identification of a response element in a herpesvirus saimiri mRNA recognized by the ORF57 protein. J Gen Virol. 2009;90:596–601. doi: 10.1099/vir.0.007476-0. [DOI] [PubMed] [Google Scholar]
- Conrad NK. Chapter 15. Co-immunoprecipitation techniques for assessing RNA-protein interactions in vivo. Methods Enzymol. 2008;449:317–342. doi: 10.1016/S0076-6879(08)02415-4. [DOI] [PubMed] [Google Scholar]
- Conrad NK. Posttranscriptional gene regulation in Kaposi’s sarcoma-associated herpesvirus. Adv Appl Microbiol. 2009;68:241–261. doi: 10.1016/S0065-2164(09)01206-4. [DOI] [PubMed] [Google Scholar]
- Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell. 2006;24:943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Conrad NK, Shu MD, Uyhazi KE, Steitz JA. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc Natl Acad Sci U S A. 2007;104:10412–10417. doi: 10.1073/pnas.0704187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad NK, Steitz JA. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005;24:1831–1841. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, D’Orso I, Frankel AD. A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol Cell. 2008;31:824–834. doi: 10.1016/j.molcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Liang Y, Sun R. Regulation of KSHV lytic gene expression. Curr Top Microbiol Immunol. 2007;312:157–183. doi: 10.1007/978-3-540-34344-8_6. [DOI] [PubMed] [Google Scholar]
- Dourmishev LA, Dourmishev AL, Palmeri D, Schwartz RA, Lukac DM. Molecular genetics of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol Mol Biol Rev. 2003;67:175–212. doi: 10.1128/MMBR.67.2.175-212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaunsinger BA, Ganem DE. Messenger RNA turnover and its regulation in herpesviral infection. Adv Virus Res. 2006;66:337–394. doi: 10.1016/S0065-3527(06)66007-7. [DOI] [PubMed] [Google Scholar]
- Han Z, Swaminathan S. Kaposi’s sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J Virol. 2006;80:5251–5260. doi: 10.1128/JVI.02570-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- Kang JG, Pripuzova N, Majerciak V, Kruhlak M, Le SY, Zheng ZM. Kaposi’s sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J Virol. 2011;85:2620–2630. doi: 10.1128/JVI.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshner JR, Lukac DM, Chang J, Ganem D. Kaposi’s sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J Virol. 2000;74:3586–3597. doi: 10.1128/jvi.74.8.3586-3597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. Targeted disruption of Kaposi’s sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J Virol. 2007;81:1062–1071. doi: 10.1128/JVI.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerciak V, Yamanegi K, Allemand E, Kruhlak M, Krainer AR, Zheng ZM. Kaposi’s sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J Virol. 2008;82:2792–2801. doi: 10.1128/JVI.01856-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerciak V, Yamanegi K, Nie SH, Zheng ZM. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J Biol Chem. 2006;281:28365–28378. doi: 10.1074/jbc.M603095200. [DOI] [PubMed] [Google Scholar]
- Majerciak V, Zheng ZM. Kaposi’s sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front Biosci. 2009;14:1516–1528. doi: 10.2741/3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik P, Blackbourn DJ, Cheng MF, Hayward GS, Clements JB. Functional cooperation between the Kaposi’s sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J Gen Virol. 2004a;85:2155–2166. doi: 10.1099/vir.0.79784-0. [DOI] [PubMed] [Google Scholar]
- Malik P, Blackbourn DJ, Clements JB. The evolutionarily conserved Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J Biol Chem. 2004b;279:33001–33011. doi: 10.1074/jbc.M313008200. [DOI] [PubMed] [Google Scholar]
- Mesri EA, Cesarman E, Boshoff C. Kaposi’s sarcoma and its associated herpesvirus. Nat Rev Cancer. 2010;10:707–719. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. Global changes in Kaposi’s sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol. 2003;77:4205–4220. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekorchuk M, Han Z, Hsieh TT, Swaminathan S. Kaposi’s sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J Virol. 2007;81:9990–9998. doi: 10.1128/JVI.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ote I, Piette J, Sadzot-Delvaux C. The Varicella-Zoster virus IE4 protein: a conserved member of the herpesviral mRNA export factors family and a potential alternative target in antiherpetic therapies. Biochem Pharmacol. 2010;80:1973–1980. doi: 10.1016/j.bcp.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Palmeri D, Spadavecchia S, Carroll KD, Lukac DM. Promoter- and cell-specific transcriptional transactivation by the Kaposi’s sarcoma-associated herpesvirus ORF57/Mta protein. J Virol. 2007;81:13299–13314. doi: 10.1128/JVI.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- Sahin BB, Patel D, Conrad NK. Kaposi’s sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog. 2010;6:e1000799. doi: 10.1371/journal.ppat.1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri-Goldin RM. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front Biosci. 2008;13:5241–5256. doi: 10.2741/3078. [DOI] [PubMed] [Google Scholar]
- Sinclair AJ. bZIP proteins of human gammaherpesviruses. J Gen Virol. 2003;84:1941–1949. doi: 10.1099/vir.0.19112-0. [DOI] [PubMed] [Google Scholar]
- Song MJ, Brown HJ, Wu TT, Sun R. Transcription activation of polyadenylated nuclear rna by rta in human herpesvirus 8/Kaposi’s sarcoma-associated herpesvirus. J Virol. 2001;75:3129–3140. doi: 10.1128/JVI.75.7.3129-3140.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt MR, Dittmer DP. The Rta/Orf50 transactivator proteins of the gamma-herpesviridae. Curr Top Microbiol Immunol. 2007;312:71–100. doi: 10.1007/978-3-540-34344-8_3. [DOI] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S. Post-transcriptional gene regulation by gamma herpesviruses. J Cell Biochem. 2005;95:698–711. doi: 10.1002/jcb.20465. [DOI] [PubMed] [Google Scholar]
- Toth Z, Stamminger T. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front Biosci. 2008;13:2939–2949. doi: 10.2741/2899. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Vinciguerra P, Stutz F. mRNA export: an assembly line from genes to nuclear pores. Curr Opin Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Zheng ZM. Split genes and their expression in Kaposi’s sarcoma-associated herpesvirus. Rev Med Virol. 2003;13:173–184. doi: 10.1002/rmv.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci U S A. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.