Abstract
Aldehyde dehydrogenases (ALDHs) represent large family members of NAD(P)+-dependent dehydrogenases responsible for the irreversible metabolism of many endogenous and exogenous aldehydes to the corresponding acids. Among 19 ALDH isozymes, mitochondrial ALDH2 is a low Km enzyme responsible for the metabolism of acetaldehyde and lipid peroxides such as malondialdehyde and 4-hydroxynonenal, both of which are highly reactive and toxic. Consequently, inhibition of ALDH2 would lead to elevated levels of acetaldehyde and other reactive lipid peroxides following ethanol intake and/or exposure to toxic chemicals. In addition, many East Asian people with a dominant negative mutation in ALDH2 gene possess a decreased ALDH2 activity with increased risks for various types of cancer, myocardial infarct, alcoholic liver disease, and other pathological conditions. The aim of this review is to briefly describe the multiple post-translational modifications of mitochondrial ALDH2, as an example, after exposure to toxic chemicals or under different disease states and their pathophysiological roles in promoting alcohol/drug-mediated tissue damage. We also briefly mention exciting preclinical translational research opportunities to identify small molecule activators of ALDH2 and its isozymes as potentially therapeutic/preventive agents against various disease states where the expression or activity of ALDH enzymes is altered or inactivated.
Keywords: Aldehyde dehydrogenases, post-translational modifications, cellular defense, drug toxicity, disease states, translational research
1. Introduction
Various reactive aldehydes can be produced from endogenous and exogenous precursors under many different pathophysiological states as well as following exposure to potentially toxic agents including abused substances such as alcohol (ethanol) and cocaine. For instance, in mammals, toxic acetaldehyde can be produced as an intermediate during alcohol metabolism catalyzed by alcohol dehydrogenase (ADH)1 before it is further oxidized to acetic acid by mitochondrial aldehyde dehydrogenase 2 isozyme (ALDH2). Both ADH and ALDH2-mediated reactions require NAD+ as a cofactor. Upon exposure to exogenous toxic chemicals [e.g., carbon tetrachloride (CCl4)] or under physiological conditions [e.g., UV exposure], lipid peroxidation takes place due to increased oxidative stress. As a result, the cellular levels of toxic lipid peroxides such as 4-hydroxynonenal (4-HNE) and malondialdehyde (MDA) are elevated. These aldehydes are highly reactive and thus interact with cellular macromolecules including DNA and proteins [1,2]. Consequently, normal functions of DNA and protein targets are negatively affected, resulting in DNA damage/deletion/mutation and inactivation of proteins, respectively. In severe cases, these toxic aldehydes can directly promote cell/tissue damage (via apoptosis and necrosis) through promoting a loss of mitochondrial potential and activating the mitochondria-dependent cell death pathways [2–5]. However, under normal conditions, these reactive aldehydes can be effectively managed by various antioxidants (e.g., glutathione and ascorbic acid) as well as different metabolic enzymes such as alcohol dehydrogenase, aldehyde dehydrogenase (ALDH), aldo-keto reductase, aldehyde oxidase, glutathione transferase, etc [6–8].
Among the cellular protective enzymes listed above, ALDH isozymes (EC 1.2.1.3) represent NAD(P)+-dependent enzymes involved in the metabolism (oxidation) of various toxic aldehydes of endogenous and exogenous origins into their corresponding acids. In general, the ALDH-mediated reactions are considered irreversible. The ALDH superfamily consists of at least 19 ALDH genes in the human genome and that mutations in some ALDH isozymes are associated with inborn errors with altered aldehyde metabolisms and thus increased susceptibility to certain disease states [9–11]. Although many ALDH isozymes share overlapping substrate specificities and tissue/subcellular distribution, they may have different kinetic parameters such as distinct Km values and catalytic activities toward each substrate compound [9–11]. In addition, the biochemical properties of each homologous ALDH isozyme depend on the species examined. For instance, human cytosolic ALDH1A1 isozyme exhibits a high Km value (a range of 170~190 μM) for acetaldehyde [12]. However, the Km value for acetaldehyde in rodent ALDH1A1 is relatively low (a range of 12 ~ 17 μM measured at pH 7.5), and comparable to that of rodent ALDH2 (0.2 μM) [12]. Therefore, it is reasonable to consider that ALDH2 in humans and ALDH2/1A1 isozymes in rodents likely represent the major enzyme(s) responsible for the metabolism of acetaldehyde produced from ethanol metabolism [12–14].
Because of the important roles of ALDH isozymes in efficiently removing potentially toxic aldehyde compounds, their tissue/subcellular distribution, substrate specificity, regulation of gene expression, and biochemical properties of each ALDH isozyme have been extensively studied [see reviews 9–11]. In general, most ALDH isozymes are expressed in large amounts in the liver while extra-hepatic tissues usually contain lower amounts of ALDH isozymes. However, certain tissues may contain a relatively large amount of a specific ALDH isozyme depending on the unique function of each tissue. For instance, ALDH5A1, NAD+-dependent succinic semialdehyde dehydrogenase involved in the metabolism of a neurotransmitter GABA, is highly expressed in the brain compared to other non-neuronal tissues including the liver [15]. In contrast, ALDH7A1, responsible for cellular protection against salinity, dehydration, and hyper-osmotic stress and betaine aldehyde metabolism, is highly expressed in the kidney, heart, ovary, cochlea, and eye [9,16].
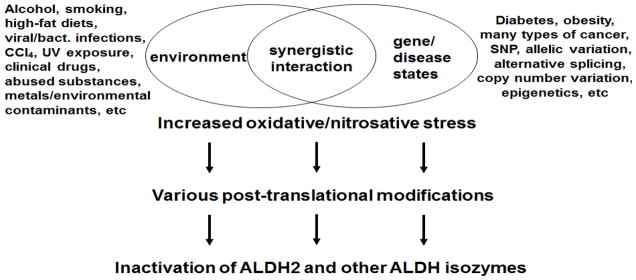
It is now known that the catalytic activities of ALDH2 and ALDH isozymes are usually suppressed under pathophysiological conditions through synergistic interaction between genetic factors (e.g., allelic variation/mutation/single nucleotide polymorphism (SNP)/copy number variation in ALDH genes) [14,17–23] and environmental factors (e.g., alcohol, drugs, smoking, high-fat diet, viral/bacterial infections, toxic chemicals, etc) [24–29] (Figure 1). However, the causal roles of ALDH isozymes in the disease states are incompletely understood. In addition, despite the well-established biochemical properties of each ALDH isozyme, it is poorly understood how these ALDH isozymes are suppressed under some pathological conditions or following exposure to potentially toxic chemicals. We hypothesized that the catalytic activities of ALDH isozymes could be suppressed through post-translational modifications (PTMs) because the amounts of ALDH protein contents seemed unchanged or altered in a small amount (compared with the marked inhibitions of the catalytic activity) after exposure to toxic chemicals including alcohol or following hepatic I/R injury [30–39]. In fact, the mechanisms of PTMs of ALDH isozymes and subsequent changes in their catalytic activities under various pathophysiological conditions have not been studied thoroughly. Therefore, the aim of this review is to briefly describe the multiple PTMs of ALDH isozymes, updating the previous report [40]. As an example, we describe several examples of PTMs of mitochondrial ALDH2 following exposure to various toxic chemicals or under different disease models/states and discuss their functional implications in promoting alcohol/drug-mediated mitochondrial dysfunction and tissue damage. Finally, we briefly mention translational research opportunities in identifying chemicals that can activate ALDH2 and isozymes and thus can be potentially used as preventive/supportive/therapeutic agents against various disease states where ALDH isozymes are inactivated.
Figure 1.
Synergistic interaction between genetic and environmental factors, leading to inactivation of ALDH2 and its isozymes. Various environmental factors (including viral/bacterial infections as listed above) and genetic factors (including SNP and copy number variation) synergistically interact and increase oxidative/nitrosative stress, which promotes various PTMs, leading to inactivation of ALDH2 and other ALDH isozymes.
2. Post-translational modifications of ALDH2 and ALDH isozymes
Under increased oxidative/nitrosative stress, many amino acids such as cysteine (Cys), methionine (Met), tyrosine (Tyr), proline (Pro), histidine (His), lysine (Lys), arginine (Arg), threonine (Thr), phenylalanine (Phe), and tryptophan (Trp) in most proteins can be covalently-modified and their proteins functions are altered (i.e., usually inhibited) [41–43]. It is also true that ALDH isozymes could be modified by various forms of PTMs under pathophysiological conditions, resulting in their inactivation. These PTMs include: oxidation, S-nitrosylation, phosphorylation, nitration, hyper-acetylation, glycosylation, adduct formation, etc. Consequently, the catalytic activities of the ALDH isozymes usually decreased following covalent modifications except for a few cases, as discussed later. We will briefly describe several types of PTMs observed in ALDH2, as an example, under different pathophysiological conditions.
A) Oxidative modifications of critical Cys residues of ALDH2 and isozymes
It is known that Cys residues in most proteins can undergo many different types of oxidative-modifications under oxidative/nitrosative stress [41–43]. The oxidative modifications include: oxidation to sulfenic acid [44], disulfide [45–47], sulfinic/sulfonic acids [48], NO- or peroxynitrite-dependent S-nitrosylation [49,50], NO-independent ADP-ribosylation [51], mixed disulfide formation with glutathione [52], cysteine [53], succinic acid [54], etc. Site-directed mutagenesis of each Cys residue of ALDH2 followed by biochemical characterization demonstrated the active site Cys302 residue [55]. The structural alignment and comparison revealed that the active site Cys residue is 100% conserved among the 17 ALDH isozymes (see Table 1). Because of the suppressed ALDH2 and ALDH1 activities in alcoholic individuals [56,57] or rodents exposed to alcohol [30–35], which increases oxidative/nitrosative stress [58–61], it is expected that the critical Cys residues including the highly-conserved active site Cys could be oxidatively-modified and thus inactivated. In fact, our proteomics analyses showed that cytosolic ALDH1A1, mitochondrial ALDH2, and other isozymes such as ALDH5A1 and ALDH6A1 were oxidatively-modified in rats exposed to alcohol [32–35] or 3,4-methylenedioxymethamphetamine (MDMA, ecstasy) [36] and after hepatic ischemia-reperfusion (I/R) injury [37]. The hepatic ALDH2 activities in these tissue samples were all inhibited but their activities were restored when the activity was measured after pre-incubation with a strong reducing agent dithiothreitol (DTT). To further characterize the nature of Cys modification, we performed immunoblot analysis of the immunoaffinity purified ALDH2 proteins by using the specific antibody against ALDH2 or S-NO-Cys. With the anti-ALDH2 antibody, we detected similar levels of ALDH2 protein (54 kDa) in all 3 mitochondrial samples [i.e., control, alcohol-exposed rats without DTT pre-treatment, and alcohol-exposed rats pretreated with DTT, respectively]. However, we did not detect any immunoreactive band in control rats with the anti-S-NO-Cys antibody. In contrast, one band (54 kDa - same as the immunopurified ALDH2 protein) was recognized in alcohol-exposed rats with the anti-S-NO-Cys antibody. However, the band recognized with the S-NO-Cys antibody disappeared when the mitochondrial proteins were pre-incubated with DTT. The immunoblot data positively correlated with the fluctuations of the ALDH2 catalytic activity in the absence or presence of DTT. All these data strongly suggest that mitochondrial ALDH2 was S-nitrosylated in alcohol-exposed rats [32]. Similar results were also observed with the ALDH2 protein in rat hepatoma H4IIE-C3 cells exposed to NO donors such as S-nitrosoglutathione (GSNO), S-nitroso-N-acetylpenicillamine, and 3-morpholinosydnonimine, respectively [31]. Incubation with DTT or another reducing agent glutathione-ethylester significantly restored the ALDH2 activity suppressed by each NO donor. Immunoblot analysis with anti-S-NO-Cys antibody showed one immunoreactive band (54 kDa, same size as ALDH2) in GSNO-exposed hepatoma cells. The S-nitrosylated ALDH2 band disappeared when GSNO-exposed cells were pretreated with glutathione-ethylester, confirming S-nitrosylation of ALDH2 [31]. Although we have not characterized the mechanism of ALDH2 inhibition in MDMA-exposed rats [36] or mice with hepatic I/R injury [37], it is likely that ALDH2 activities could be inhibited through S-nitrosylation of critical Cys residue(s) because the suppressed ALDH2 activities were recovered in the presence of DTT, as in the case of alcohol-exposed rats [32].
Table 1.
Structural alignment of the region around the active site Cys residue in ALDH isozymes.
| Name | Peptide Sequence | Swiss-Prot |
|---|---|---|
| ALDH1A1 | 275HGVFYHQGQCCIAASRIFVEE295 | P00352 |
| ALDH1A2 | 292QGVFFNQGQCCTAGSRIFVEE312 | O94788 |
| ALDH1A3 | 286QGVFFNQGQCCTAASRVFVEE306 | P47895 |
| ALDH1B1 | 291EALFFNMGQCCCAGSRTFVEE311 | P30837 |
| ALDH1L1 | 679SSVFFNKGENCIAAGRLFVED699 | O75891 |
| ALDH1L2 | 700GAVFFNKGENCIAAGRLFVEE720 | Q3SY69 |
| ALDH2 | 291FALFFNQGQCCCAGSRTFVQE311 | P05091 |
| ALDH3A1 | 216WGKFMNSGQTCVAPDYILCDP236 | P30838 |
| ALDH3A2 | 213WGKYMNCGQTCIAPDYILCEA233 | P51648 |
| ALDH3B1 | 216WFRYFNAGQTCVAPDYVLCSP236 | P43353 |
| ALDH3B2 | 135WFCYFNAGQTCVAPDYVLCSP155 | P48448 |
| ALDH4A1 | 320RSAFEYGGQKCSACSRLYVPH340 | P30038 |
| ALDH5A1 | 312ASKFRNTGQTCVCSNQFLVQR332 | P51649 |
| ALDH6A1 | 289GAAFGAAGQRCMALSTAVLVG309 | Q02252 |
| ALDH7A1 | 302FAAVGTAGQRCTTARRLFIHE322 | P49419 |
| ALDH8A1 | 259RSSFANQGEICLCTSRIFVQK279 | Q9H2A2 |
| ALDH9A1 | 260MANFLTQGQVCCNGTRVFVQK280 | P49189 |
The primary amino acid sequences of the region around the active site cysteine residue of each human ALDH isozyme, as listed in Marchitti et al [10], are aligned except for ALDH16A1 and ALDH18A1, since the active site Cys residues in the latter 2 enzymes are not conserved. The active site Cys (C) and other highly conserved amino acids are presented in bold characters.
We observed that cytosolic ALDH1A1 and ALDH4A1 were oxidatively-modified in alcohol-exposed mice [33] when we performed a redox proteomics approach using biotin-N-maleimide as a sensitive probe for oxidatively-modified Cys residue(s) [30], as recently reviewed [62,63]. Therefore, we investigated whether cytosolic ALDH1 activity was also inhibited in alcohol-exposed rat liver, similar to the suppression of mitochondrial ALDH2. Our subsequent study revealed that cytosolic ALDH1 activity was inhibited in alcohol-exposed rat liver but restored by the presence of DTT, suggesting that cytosolic ALDH1 was reversibly inactivated [34]. Immunoblot analysis with the anti-S-NO-Cys antibody confirmed S-nitrosylation of ALDH1 protein based on the detection of an immunoreactive band (~55 kDa) in the alcohol-exposed rats and its disappearance after pre-incubation with DTT. Furthermore, the pattern of detection and disappearance of the immunoreactive S-NO-Cys band correlated with the ALDH1 activity changes in the absence and presence of DTT, respectively [34].
Recent data suggest that S-nitrosylation of Cys of certain proteins in cardiac tissues may play a protective role against oxidative tissue damage by preventing irreversible hyper-oxidation of Cys residues under increased oxidative stress [64,65], as observed in gram-positive bacteria including Bacillus subtilis, which lacks low molecular antioxidant glutathione [53]. Theoretically, the Cys residues of S-nitrosylated proteins including cardiac ALDH2 should be restored to free sulfhydryl groups when the levels of cellular antioxidants such as glutathione and ascorbate become elevated in the heart. Although this theory may be correct, however, it is unclear whether S-nitrosylated cardiac ALDH2 exhibits a decreased activity than the native protein. It is also unknown whether S-nitrosylated hepatic ALDH1A1 and ALDH2 observed in alcohol-exposed rodents represent a cellular protective mechanism (e.g., prevention from irreversible hyper-oxidation to sulfinic/sulfonic acids), as proposed for the S-nitrosylated proteins in cardiac tissues [64,65] or simply reflect the inactivated proteins per se. Since the liver possesses a high capacity of regenerating or refurbishing the antioxidants through activation of a protective transcription factor Nrf-2 and other genes, the latter case looks more physiological and consistent with the suppressed ALDH activities observed in human alcoholic individuals [56,57]. Thus, based on the conflicting views, the physiological role of S-nitrosylation of hepatic ALDH2 and other ALDH isozymes in alcoholic and nonalcoholic fatty liver or fibrotic/cirrhotic diseases need re-evaluation in the future. In this regard, it would be important to further characterize the covalent modifications of hepatic ALDH2 protein purified from alcoholic individuals compared to control subjects.
B) Phosphorylation of ALDH2 and isozymes
It is well-established that ethanol-inducible cytochrome P450 2E1 (CYP2E1) is a major enzyme responsible for the metabolism of various small molecule toxic compounds including ethanol, acetaminophen, CCl4, dimethylnitrosamine, benzene, etc [58–61,66–68]. The acute toxicity by these agents was prevented or greatly alleviated in Cyp2e1-null mice [69] or pre-treatment with a CYP2E1 inhibitor [70,71]. Acute exposure of ethanol, acetaminophen, or CCl4, increases oxidative/nitrosative stress while each compound markedly inhibits the activity of mitochondrial ALDH2 activity, contributing to mitochondrial dysfunction and severe liver injury (through apoptosis/necrosis) in the pericentral region [69,72]. Because of the ALDH inhibition, the levels of reactive lipid peroxides such as 4-HNE and MDA were increased. Consequently, these elevated toxic peroxides can overwhelm the cellular anti-oxidant defense system, interact with many proteins in different sub-cellular organelles including mitochondria, thus alter their functions, resulting in mitochondria dysfunction and cell death [1–5,42]. In addition, long-term chronic exposure of 4-HNE and MDA can activate hepatic stellate cells, which produce collagen and pro-fibrotic growth factors including transforming growth factor-β, leading to fibrosis and cirrhosis in the liver [73]. Furthermore, these reactive lipid aldehydes can interact with DNA [1,74], causing DNA damage, deletion, and mutations, ultimately contributing to carcinogenesis, as illustrated (Figure 2). Besides these scenarios, other events may also take place cell/tissue damage. For instance, increased oxidative/nitrosative stress can concurrently decrease the levels of anti-oxidants, inhibit anti-oxidant defensive enzymes including mitochondrial superoxide dismutase, activate c-Jun N-terminal protein kinase (JNK)-mediated cell death signaling pathway [5,75], stimulate STAT-1 mediated proinflammatory signaling pathway [76], etc. All these pathways may also contribute to mitochondrial dysfunction and cell/organ damage.
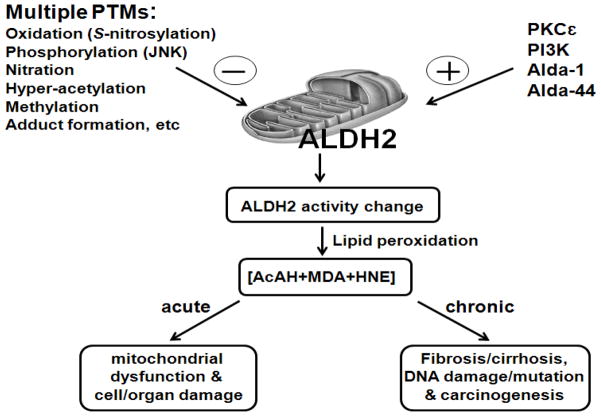
Figure 2.
Biological consequences of PTMs of ALDH2 and isozymes.
Various PTMs listed on the upper left negatively affect the ALDH2 activity, leading to increased levels of toxic aldehyde compounds. These reactive carbonyl compounds can activate the JNK- and p38 kinase-mediated cell death pathway, leading to mitochondrial dysfunction and cell/tissue damage, as recently reported [39]. They can also activate stellate cells to produce collagen and pro-fibrotic growth factors. Furthermore, the reactive carbonyl compounds can interact with mitochondrial DNA, leading to DNA damage, deletion, and mutation, ultimately contributing to carcinogenesis. In contrast, activation of ALDH2 by PKCε, PI3K or small molecule compounds Alda-1 and Alda-44 shown on the upper right can prevent accumulation of lipid peroxides and thus avoid cell/tissue damage or carcinogenesis. The negative and positive signs represent inhibition and activation of ALDH2 activity, respectively.
Despite these acute and chronic effects of various CYP2E1 substrate compounds, the inhibitory mechanism of ALDH2 by each compound is poorly understood. Acute administration of ethanol [75], acetaminophen [69,71], CCl4 [77], 4-HNE [5], or troglitazone [78], a hepatotoxic non-CYP2E1 substrate compound, was shown to activate JNK, contributing to cell death. Based on JNK activation by these compounds, we hypothesized that CCl4 could inhibit ALDH2 through JNK-mediated phosphorylation. However, the direct relationship between the activated JNK and ALDH2 activity has not been studied. Therefore, we recently studied the direct role of JNK in phosphorylating and inactivating ALDH2 in CCl4-exposed rat tissues [39].
Consistent with the earlier data [24,25], our results showed that CCl4 inhibited hepatic ALDH2 activity in a time-dependent manner without altering its protein amount assessed by immunoblot analysis. In contrast, cytosolic ALDH1A1 was not inhibited by CCl4 exposure. Moreover, addition of DTT did not restore the suppressed ALDH2 activity, suggesting that ALDH2 could be inhibited through irreversible covalent modifications such as phosphorylation, based on JNK activation by CCl4 [77]. Since phosphorylated proteins are often detected in more acidic pI regions (i.e., acidic-shifts) than non-phosphorylated native proteins on 2-D gels (with little changes in their mobility on 1-D gels) [79], we performed 2-D gel analysis of ALDH2 protein in control and CCl4-exposed rat liver mitochondria samples. Immunoblot analysis with the anti-ALDH2 antibody revealed that 3 ALDH2 spots were detected in corn oil-treated (vehicle) control samples. However, 2~3 additional immunoreactive ALDH2 spots in acidic pI regions were detected in CCl4-exposed rats. Incubation with alkaline phosphatase significantly restored the suppressed ALDH2 activity to a similar level to that in corn-oil treated control. This activity change was accompanied with concurrent disappearance of acidic ALDH2 spots on 2-D gels. Since both JNK and activated (phosphorylated) JNK were translocated to mitochondria following CCl4 exposure [39], we directly tested whether mitochondrial ALDH2 activity could be inhibited by JNK in in vitro experiments. Incubation of mitochondrial proteins from control rats with catalytically active JNK resulted in significant inhibition of ALDH2 activity, which was accompanied with appearance of acidic ALDH2 spots on 2-D gels. We also performed immunoblot analysis by using the anti-phospho-Ser-Pro or phospho-Thr-Pro antibody, since JNK is a proline-directed protein kinase which phosphorylates Ser or Thr followed by Pro [79]. The anti-phospho-Ser-Pro antibody recognized one band (54 kDa) in the immunoprecipitated ALDH2 protein while no band was detected with the anti-phospho-Thr-Pro antibody. These data demonstrated that Ser residue(s) of ALDH2 could be phosphorylated and thus inactivated. Based on the structural comparison and the differential effects of CCl4 on the activities of ALDH1A1 and ALDH2, Ser463-Pro of rat ALDH2 could be a major site of JNK-mediated phosphorylation [39]. All these results indicate a novel underlying mechanism by which ALDH2 can be inhibited through JNK-mediated phosphorylation, resulting in decreased cellular defense capacity and increased lipid peroxidation. These sequential events likely contribute to the pericentral necrosis in the liver [77]. Furthermore, we expect that ALDH2 could be inactivated by a similar mechanism following exposure to many other hepatotoxic agents such as acetaminophen [71] and troglitazone [78] or under many pathophysiological conditions like UV exposure where JNK is activated and translocated to mitochondria [39,80,81].
Contrary to the JNK-mediated phosphorylation and subsequent inactivation of ALDH2, other protein kinases such as protein kinase Cε (PKCε) can phosphorylate and activate mitochondrial ALDH2. In fact, administration of a small physiological and cardioprotective dose of alcohol stimulated PKCε [82], which was translocated to mitochondria in a time-dependent manner and bound ALDH2 protein [83]. Reciprocal immunoprecipitation experiments with either anti-PKCε antibody or anti-ALDH2 antibody verified that these proteins interact to each other in the mitochondria. In vitro incubation of ALDH2 protein with PKCε promoted phosphorylation of Thr residues of ALDH2. Mass-spectral analysis confirmed that Thr185, Thr412 and possibly Ser279 of ALDH2 could be phosphorylated by PKCε, resulting in ALDH2 activation [84]. Stimulation of ALDH2 by PKCε following exposure to small doses of alcohol [82,83] or small molecule chemical activators such as Alda-1 [84] and Alda-44 [85] correlated with decreased levels of 4-HNE and significant improvement of ischemic cardiac damage in the experimental models. Activation of ALDH2 activity through PKCε-mediated phosphorylation was also supported by the appearance of acidic ALDH2 spots on 2-D gels. By testing the effects of various known activators and inhibitors of ALDH2 including cyanamide, Mochly-Rosen and colleagues concluded that the degree of the myocardial infarct size from the experimental I/R injury inversely correlated to the ALDH2 activities [84,85]. Furthermore, the small molecule activator Alda-1 could support the suppressed activities of heterozygous ALDH2*1/2 or homozygous ALDH2*2/2 variants [84], suggesting that this compound and its structural analogs (including Alda-44) could help many East Asian people (> 0.5 billion people) who carry the dominant negative mutation (Glu487Lys) in the human ALDH2 gene. Recent crystal structural analysis further described that Alda-1 supports ALDH2 activity by working as a chemical chaperone [86]. Taken together, these reports provided convincing evidence that ALDH2 can be activated by PKCε-mediated phosphorylation and that maintaining the ALDH2 activity either direct activation with small activators or indirect activation through PKCε is critically important in cardioprotection. It is of interest whether this type of PKCε-mediated protection exists in other tissues.
In addition, another report recently showed that ALDH2 could be activated through phosphatidyl inositol-3-kinase (PI3K)-dependent phosphorylation, as observed in female rats compared to male counterparts under the experimental models of cardiac I/R injury [87]. The degree of cardiac damage was less significant in females than in males, where Alda-1, a chemical activator of ALDH2 [84], exerted its beneficial effect in males but with little benefits in females. By using the fluorescence 2-D difference (2-D DIGE) proteomics approach to compare the altered levels and PTMs of cardiac mitochondrial proteins, Murphy and colleagues observed that ALDH2 were phosphorylated in greater amounts (appearance of more acidic ALDH2 spots on 2-D gels) in female rats than in males. The acidic ALDH spots, initially observed in female rat hearts, disappeared in ovariectomized female rats, suggesting a role of estrogen in ALDH2 phosphorylation and cardioprotection. Moreover, phosphorylation of ALDH2 in female rats was blocked after pre-incubation with wortmannin, a specific inhibitor of PI3K. In contrast, the effect of wortmannin on the disappearance of acidic ALDH2 spots was not observed in males. These results indicate that the PI3K-mediated phosphorylation (activation) of ALDH2 with decreased levels of 4-HNE in female rats likely contribute to cardioprotection.
Another 2-D DIGE proteomics analysis showed that glycogen-synthase kinase 3 (GSK-3) may control the expression of mitochondrial proteins since the level of an ALDH2 protein spot was slightly but significantly elevated after treatment with a specific GSK-3 inhibitor, which provides cardioprotection [88]. Based on the 2-D DIGE analysis data and no information about the ALDH activity change, it is unlikely that the expression of other ALDH2 spots, usually observed on 2-D gels, is altered under these conditions. Consequently, future studies are needed to determine which amino acids of ALDH2 can be phosphorylated by GSK-3 and how the GSK-3 mediated phosphorylation of ALDH2 affects its overall catalytic activity.
C) Nitration of ALDH2 and isozymes
Recent data showed that ALDH2 plays a central role in the biotransformation (i.e., reduction) of nitroglycerin (glyceryl trinitrate, GTN) used as an anti-ischemic drug to treat angioplasty and cardiovascular diseases [89,90]. However, chronic usage of GTN can lead to inhibition of ALDH2 activity though increased production of reactive oxygen/nitrogen species in the mitochondria and thus causes GTN tolerance (decreased reaction to GTN and decreased vasodilatation) [52,89,91]. Initial studies showed that sulfhydryl groups of ALDH2 were oxidized (to disulfides) or glutathionylated, which could be effectively reduced to free sulfhydryl groups in the presence of DTT or dihydrolipoic acid [52,91]. Another report recently suggested that DTT could not fully restore the suppressed ALDH2 activity, suggesting partially irreversible inactivation of ALDH2 by chronic GTN usage or a bolus dose [92]. The reason for the irreversible inhibition of ALDH2 may reflect hyper-oxidation of critical Cys residues to sulfinic/sulfonic acids, which cannot be reduced by DTT [62,63]. Alternatively, ALDH2 could be inhibited through protein nitration based on the recent data where GTN promotes nitration signaling [93]. In the latter study, ALDH2 activity was mildly inhibited by NO or hydrogen peroxide in the experimental conditions, but was markedly inhibited by incubation with peroxynitrite [93]. Furthermore, in the presence of GTN, ALDH2 could serve as a peroxynitrite synthase, producing increased production of peroxynitrite, which could have nitrated Tyr residue(s) of ALDH2 [49], resulting in its inactivation. Another study provided the direct evidence for nitration of ALDH2 and isozymes. Proteomics identification followed by mass-spectral analysis confirmed that ALDH2 and ALDH7A1 were nitrated in the kidney of spontaneously hypertensive rats [94]. Thus, the potential sites of Tyr nitration(s) of ALDH2 and its implications in GTN bioactivation and cross-tolerance need to be investigated in the future.
D) Acetylation of ALDH2 and isozymes
Protein modifications by acetylation and deacetylation have emerged as an important PTM in regulating the function of many proteins especially in the mitochondria [95]. It is also known that hyper-acetylation of lysine (Lys) residues of many proteins alters their biological functions. For instance, cellular functions of many key regulatory proteins such as p53, a transcription factor, and nuclear histone proteins, involved in controlling the rate of cell growth and normal development, are regulated by acetylation and de-acetylation [96]. Some metabolic enzymes such as long-chain acyl-CoA dehydrogenase, one of the 4 enzymes in the mitochondrial fatty acid β-oxidation pathway, are also regulated by acetylation as demonstrated with the inactivation of this enzyme through hyper-acetylation of its Lys residues, as shown in mice deficient in Sirtuin 3 gene [97]. An earlier proteomics and bioinformatics study showed that ALDH2, ALDH4A1, and ALDH6A1 were acetylated under physiological conditions [98]. Recent proteomics studies revealed that many cytosolic and mitochondrial proteins are hyper-acetylated in alcohol-exposed rats [99] or high-fat exposed mice [100]. Mass-spectral analysis confirmed hyper-acetylation of many cytosolic and mitochondrial proteins including hepatic ALDH1A1 (retinal dehydrogenase), ALDH2, and ALDH3A1 (fatty aldehyde dehydrogenase) in mice exposed to a high-fat diet [100]. Since alcohol metabolism [61] or the high fat diet [100] decreases the levels of NAD+ with diminished ratios of cellular NAD+/NADH, hyper-acetylation of ALDH2 and ALDH isozymes seem to result from decreased activities of sirtuin proteins, which are NAD+-dependent deacetylases [95]. However, the functional roles of acetylation of ALDH2 and other ALDH isozymes have not been studied in recent studies [99,100] and thus warrant further characterizations. Therefore, it would be of interest whether the conserved Lys residues (e.g., such as Lys192 and Lys489 in ALDH2 [101]) are a potential site of acetylation. Furthermore, it would be important to determine the functional roles of acetylation of ALDH2 and ALDH isozymes in various alcohol-related disease states since alcohol exposure decreased the activities and/or protein contents of sirtuin 1 [102,103], sirtuin 3 [104,105], and sirtuin 5 [106].
E) Formation of adducts between ALDH2 and reactive metabolites
The activities of ALDH2 and other ALDH isozymes were inhibited by exposure to many therapeutic drugs such as acetaminophen [28], daunomycin [27], cyanamide [107], daidzin [108], disulfiram [29,109], and pargyline [110]. Many toxic compounds including acrolein, bromobenzene, carbon tetrachloride, 4-HNE, MDMA (ecstasy), reserpine, and thioacetamide can also inhibit ALDH2 activity, as discussed earlier [39,40], although the inhibitory mechanism of ALDH2 activity by each agent is poorly understood. Recent data showed that 4-HNE can inhibit the ALDH2 activity through adduct formation with the active site Cys302 of ALDH2 in alcohol-exposed rodents [111]. Acetaminophen, daunomycin, disulfiram, and MDMA (ecstasy) may inhibit ALDH2 activity by a similar mechanism directly through adduct formation between the critical Cys residue(s) and the reactive metabolite of each of these compounds. For instance, critical Cys residues of ALDH2 could have been conjugated with the reactive metabolite of acetaminophen, N-acetyl-p-benzoquinone imine, leading to its inactivation [28]. Alternatively, all these compounds inhibit ALDH2 activity indirectly through increasing the levels of peroxynitrite, which can S-nitrosylate Cys residues and nitrate Tyr residues [49], elevating lipid peroxides such as 4-HNE which produces an irreversible adduct [42,111], and/or promoting JNK-mediated phosphorylation of ALDH2 [39].
F) Covalent modifications and suppression of ALDH2 in pathological conditions
It is well-established that many East Asian people with facial flushing response upon alcohol consumption possess a dominant negative mutation in human ALDH2 gene [112,113]. Some investigators proposed that ALDH2*2 mutant gene is protective against developing alcoholism and biomedical consequences since individuals with either heterozygous ALDH2*1/2 or homozygous ALDH2*2/2 mutant allele do not usually drink alcohol due to flushing response and uncomfortable feeling with excessive swelling and accelerated heart rates. Biochemically, these individuals have decreased ALDH2 activities which usually result in accumulation of acetaldehyde and other lipid aldehydes [114]. These conditions with elevated levels of acetaldehyde upon alcohol intake have been simulated in experimental animal models such as Aldh2-null mice [115] and UChA rats [116]. In addition to ALDH2 inhibition through genetic mutations [15,18–23], ALDH2 activity or protein amount could be decreased under various pathophysiological conditions such as aged rodents [117], partial hepatectomy [118], and hepatoma [27,119], although the inhibitory mechanisms were not elucidated in each case. Global proteomics analyses followed by mass-spectral analysis identified 3 separate ALDH2 spots on 2-D gels for 10 normal tissues while only 1 most acidic ALDH2 spot was observed in 10 hepatoma tissues [119]. Similar results were also reported by another laboratory, where 11 different hepatoma tissues were analyzed [120]. Based on the 3 ALDH spots (with little change in their molecular sizes) in normal control tissues, we believe that these multiple ALDH2 spots likely represent either hyper-oxidation to sulfinic/sulfonic acids or different states of phosphorylated proteins. Interestingly, at least 3 discrete ALDH3A1 spots, a known marker of hepatic cancer [121], and 4 separate ALDH1A1 spots on 2-D gels were consistently observed in both hepatoma and normal tissues [119]. Regardless of the presence or absence of ALDH isozyme spots in hepatoma tissues, these multiple ALDH1A1, ALDH2, and ALDH3A1 spots with apparently same molecular masses on 2-D gels likely represent PTMs of each ALDH isoform, although we do not know the reason why the 2 basic ALDH2 spots disappeared in hepatoma tissues. The nature and functional role of phosphorylated ALDH2 and isozymes need to be studied.
Other amino acids of ALDH2 and isozymes could be modified by different types of PTMs. In fact, mass spectral analysis confirmed that Met residues of ALDH1A1, ALDH2, and ALDH3A1 were oxidized in hepatoma tissues [119]. Another 2-D DIGE analysis followed by mass-spectral analysis identified 5 acidic ALDH2 spots in non-small cell lung cancer tissues, suggesting multiple PTMs including glycosylation, acetylation, and phosphorylation [122]. Therefore, it is possible that other amino acids of ALDH2 can be covalently modified under different pathological conditions. However, the functional roles of these additional modifications in ALDH2 and isozymes need to be established.
G) ALDH2 as an emerging target for translational research
It is well-established that alcoholic individuals are more susceptible to many disease states [14,20,123]. This is particularly true for many East Asian people with the mutant ALDH2 allele [20–23,113]. Chronic alcohol intake increases the levels of acetaldehyde and lipid peroxides as well as oxidative/nitrosative stress, which lowers the levels of anti-oxidants and inhibits many defensive enzymes such as superoxide dismutases, peroxiredoxin, and ALDH2, most likely through oxidative/nitrosative modifications of critical amino acids in the target proteins [32–34,58–61,124]. Consequently, chronic and excessive alcohol consumption causes alcoholic fatty liver, hepatitis, fibrosis, cirrhosis, and cancer in the liver while alcohol can also damage many other organs including the brain, heart, lung, pancreas, and testis. Furthermore, there are additive or synergistic interactions between alcohol and other potentially genetic factors/toxic substances such as hepatitis viruses, AIDS virus, obesity, diabetes, nicotine (smoking) and drugs including acetaminophen, cocaine, MDMA (ecstasy) (Figure 1). For instance, it is well-established that alcoholic individuals and people who drink alcohol regularly are more susceptible to severe liver damage caused by acetaminophen even at clinically relevant doses [125,126]. The acetamophen-related acute liver damage in alcohol-exposed subjects is believed to be promoted through alcohol-mediated induction of CYP2E1 [123], which can produce more reactive toxic metabolites, eventually contributing to mitochondrial dysfunction and apoptosis/necrosis. Additive or synergistic interactions between hepatitis viruses, diabetes, obesity, or MDMA and alcohol have been also demonstrated or observed in experimental models and human conditions [63,73,123,127–132]. Combination of these comorbidity factors significantly accelerates disease processes or aggravates the existing conditions. Therefore, many chemicals such as cyanamide [107], daidzin [108], disulfiram [109], and specific anti-sense oligonucleotides [133] or siRNAs [134] to ALDH2 have been developed to inhibit ALDH2 activity or suppress ALDH2 gene expression. Some of these ALDH2 inhibitors such as disulfiram and cyanamide have been used in humans as deterrents for alcohol consumption to ultimately prevent alcohol-associated organ damage.
On the other hand, ALDH2 activity and/or expression are suppressed in disease states such as ischemia followed by reperfusion [37,84] and after exposure to many commonly used drugs/chemicals including acetaminophen, carbon tetrachloride, daunomycin, disulfiram, and GTN [24–29,47,52]. Due to the suppression of ALDH2 and possibly other ALDH isozymes, the levels of toxic lipid peroxidation products such as 4-HNE and MDA are likely to be elevated. Consequently, we expect increased rates of mitochondrial dysfunction and cell/organ damage. Based on the pathophysiological implications of the suppressed ALDH2 activity, it is desirable to find small molecule activators of ALDH2 or at least compounds to prevent suppression of ALDH2 to blunt accumulation of toxic aldehydes including acetaldehyde, acrolein, 4-HNE, and MDA. In fact, antioxidants such as DTT and reduced lipoic acid were effective in preventing inactivation of ALDH2, which was suppressed by GTN treatment [52,91].
Numerous reports suggest that inactive or low ALDH2 activity, through either genetic mutations or suppression by toxic compounds, seems to be a major risk factor for various disease states including alcoholic organ damage, cancer, and cardiovasucular diseases [18–28]. Therefore, it is desirable to identify potent activators of ALDH2. After high-throughput screening of a diverse library of small molecules, Mochly-Rosen and colleagues identified chemical activators of ALDH2, Alda-1 [N-(1,3-benzodioxol-5-ylmethyl)-2,6-dichlorobenzamide] and Alda-44, both of which not only directly activate ALDH2 but also significantly protect against ischemic heart damage [84,85]. They also showed an inverse relationship between the ALDH2 activity and the degree of cardiac damage in experimental models of ischemic heart damage [84], providing convincing evidence that ALDH2 may have become a new emerging target of developing medicines for treating cardiovascular diseases. Furthermore, they showed that Alda-1 supports the activities of over-produced recombinant ALDH2*1/2 and ALDH2*2/2 mutant proteins, whose basal activities were relatively low compared with that of ALDH2*1/1 wild-type protein. These results indicate that Alda-1 and its structural analogs including Alda-44 could be used to support many East Asian people who carry the dominant negative mutant ALDH2*1/2 or ALDH2*2/2 gene.
It is well-established that the efficacy and tolerance of GTN therapy in treating cardiovascular diseases depend on mitochondrial ALDH2 activity for its biotransformation. In fact, many East Asian people with a dominant negative mutation in ALDH2*1/2 or ALDH2*2/2 gene exhibit a decreased efficacy of GTN therapy [135]. In addition, since chronic use of GTN develops tolerance (through oxidative inactivation of ALDH2, as discussed before), it is expected that these small molecule activators of ALDH2 could be used to support the sublingual usage of GTN for treating cardiovascular diseases, especially for many East Asian people with a mutant ALDH2 allele since these individuals cannot efficiently produce nitric oxide the active metabolite of GTN and thus have poor therapeutic benefits [135]. Alternatively, another GTN analog such as aminoethyl nitrate (AEN) [136] would be an ideal compound for treating cardiovascular disorders, since AEN does not need to be activated by ALDH2, whose activity can be inhibited through oxidative modification during GTN therapy. Furthermore, although AEN exhibited a similar potency to GTN, it neither seems to increase mitochondrial oxidative stress nor develop in vitro tolerance.
Based on the pathophysiological roles of ALDH2 and recent developments of drug candidates for supporting the ALDH2 activity in ischemic cardiovascular conditions, it is expected that many other small molecule activating compounds would be developed in the future. Some of the drug candidates are likely used to improve our knowledge about the roles of ALDH2 in various pathological conditions and can be used in clinics as therapeutics or supportive agents in many disease states.
Acknowledgments
This research was supported by the Intramural Program Fund at the National Institute on Alcohol Abuse and Alcoholism. Part of this research was also supported by a grant for the Chronic Liver Disease Project (to B.J. Song) from the Center for Biological Modulators in South Korea. We are grateful to Drs. Timothy D. Veenstra, Brian L. Hood, Thomas P. Conrads, Li-Rong Yu, and Xiaoying Ye at the Laboratory of Proteomics and Analytical Technologies, Advanced Technology Program, SAIC-Frederick Inc, Frederick, Maryland for the mass-spectral analysis to determine the identities of the oxidatively-modified proteins in our experimental models. We also thank Dr. Klaus Gawrisch for his support.
Footnotes
The abbreviations used are: AEN, aminoethyl nitrate; ALDH, aldehyde dehydrogenase; ALDH1A1, cytosolic aldehyde dehydrogenase; ALDH2, mitochondrial low-Km aldehyde dehydrogenase 2; CYP2E1, ethanol-inducible cytochrome P450 2E1 isozyme; DIGE, fluorescence 2-D difference gel electrophoresis; DTT, dithiothreitol; GSK-3, glycogen synthase kinase-3; GSNO, nitrosoglutathione; GTN, glyceryl trinitrate; 4-HNE, 4-hydroxynonenal; I/R, ischemia-reperfusion; JNK, c-Jun N-terminal protein kinase; MDA, malondialdehyde; MDMA, 3,4-methylenedioxymethamphetamine; PI3K, phosphatidyl-inositol-3-kinase; PKCε, protein kinase Cε isozyme; PTM, post-translational modification; RNS, reactive nitrogen species; ROS, reactive oxygen species; S-NO-Cys, S-nitrosylated Cys; SNP, single nucleotide polymorphism.
The authors do not have any conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.LoPachin RM, Gavin T, Petersen DR, Barber DS. Molecular mechanisms of 4-hydroxy-2-nonenal and acrolein toxicity: nucleophilic targets and adduct formation. Chem Res Toxicol. 2009;22:1499–508. doi: 10.1021/tx900147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhary P, Sharma R, Sharma A, Vatsyayan R, Yadav S, Singhal SS, Rauniyar N, Prokai L, Awasthi S, Awasthi YC. Mechanisms of 4-hydroxy-2-nonenal induced pro- and anti-apoptotic signaling. Biochemistry. 2010;49:6263–75. doi: 10.1021/bi100517x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raza H, John A. 4-hydroxynonenal induces mitochondrial oxidative stress, apoptosis and expression of glutathione S-transferase A4–4 and cytochrome P450 2E1 in PC12 cells. Toxicol Appl Pharmacol. 2006;216:309–18. doi: 10.1016/j.taap.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Soh Y, Jeong KS, Lee IJ, Bae MA, Kim YC, Song BJ. Selective activation of the c-Jun N-terminal protein kinase pathway during 4-hydroxynonenal induced apoptosis of PC12 cells. Mol Pharmacol. 2000;58:535–41. doi: 10.1124/mol.58.3.535. [DOI] [PubMed] [Google Scholar]
- 6.Siems W, Grune T. Intracellular metabolism of 4-hydroxynonenal. Mol Aspects Med. 2003;24:167–75. doi: 10.1016/s0098-2997(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 7.Alary J, Gueraud F, Cravedi JP. Fate of 4-hydroxynonenal in vivo: disposition and metabolic pathways. Mol Aspects Med. 2003;24:177–87. doi: 10.1016/s0098-2997(03)00012-8. [DOI] [PubMed] [Google Scholar]
- 8.Hartley DP, Ruth JA, Petersen DR. The hepatocellular metabolism of 4-hydroxynonenal by alcohol dehydrogenase, aldehyde dehydrogenase, and glutathione S-transferase. Arch Biochem Biophys. 1995;316:197–205. doi: 10.1006/abbi.1995.1028. [DOI] [PubMed] [Google Scholar]
- 9.Sladek NE. Human aldehyde dehydrogenases: Potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol. 2003;17:7–23. doi: 10.1002/jbt.10057. [DOI] [PubMed] [Google Scholar]
- 10.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Black WJ, Stagos D, Marchitti SA, Nebert DW, Tipton KF, Bairoch A, Vasiliou V. Human aldehyde dehydrogenase genes: alternatively spliced transcriptional variants and their suggested nomenclature. Pharmacogenet Genomics. 2009;19:893–902. doi: 10.1097/FPC.0b013e3283329023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klyosov AA, Rashkovetsky LG, Tahir MK, Keung WM. Possible role of liver cytosolic and mitochondrial aldehyde dehydrogenases in acetaldehyde metabolism. Biochemistry. 1996;35:4445–56. doi: 10.1021/bi9521093. [DOI] [PubMed] [Google Scholar]
- 13.Svanas GW, Weiner H. Aldehyde dehydrogenase activity as the rate-limiting factor for acetaldehyde metabolism in rat liver. Arch Biochem Biophys. 1985;236:36–46. doi: 10.1016/0003-9861(85)90603-4. [DOI] [PubMed] [Google Scholar]
- 14.Day CP, Bashir R, James OF, Bassendine MF, Crabb DW, Thomasson HR, Li TK, Edenberg HJ. Investigation of the role of polymorphisms at the alcohol and aldehyde dehydrogenase loci in genetic predisposition to alcohol-related end-organ damage. Hepatology. 1991;14:798–801. doi: 10.1002/hep.1840140509. [DOI] [PubMed] [Google Scholar]
- 15.Chambliss KL, Caudle DL, Hinson DD, Moomaw CR, Slaughter CA, Jakobs C, Gibson KM. Molecular cloning of the mature NAD(+)-dependent succinic semialdehyde dehydrogenase from rat and human. cDNA isolation, evolutionary homology, and tissue expression. J Biol Chem. 1995;270:461–7. doi: 10.1074/jbc.270.1.461. [DOI] [PubMed] [Google Scholar]
- 16.Brocker C, Lassen N, Estey T, Pappa A, Cantore M, Orlova VV, Chavakis T, Kavanagh KL, Oppermann U, Vasiliou V. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J Biol Chem. 2010;285:18452–63. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal DP, Goedde HW. Pharmacogenetics of alcohol metabolism and alcoholism. Pharmacogenetics. 1992;2:48–62. doi: 10.1097/00008571-199204000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Chao YC, Liou SR, Chung YY, Tang HS, Hsu CT, Li TK, Yin SJ. Polymorphism of alcohol and aldehyde dehydrogenase genes and alcoholic cirrhosis in Chinese patients. Hepatology. 1994;19:360–6. [PubMed] [Google Scholar]
- 19.Higuchi S, Matsushita S, Muramatsu T, Murayama M, Hayashida M. Alcohol and aldehyde dehydrogenase genotypes and drinking behavior in Japanese. Alcohol Clin Exp Res. 1996;20:493–7. doi: 10.1111/j.1530-0277.1996.tb01080.x. [DOI] [PubMed] [Google Scholar]
- 20.Yokoyama A, Muramatsu T, Ohmori T, Yokoyama T, Okuyama K, Takahashi H, Hasegawa Y, Higuchi S, Maruyama K, Shirakura K, Ishii H. Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis. 1998;19:1383–7. doi: 10.1093/carcin/19.8.1383. [DOI] [PubMed] [Google Scholar]
- 21.Muto M, Hitomi Y, Ohtsu A, Ebihara S, Yoshida S, Esumi H. Association of aldehyde dehydrogenase 2 gene polymorphism with multiple oesophageal dysplasia in head and neck cancer patients. Gut. 2000;47:256–61. doi: 10.1136/gut.47.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagi S, Iwai N, Yamauchi R, Kojima S, Yasuno S, Baba T, Terashima M, Tsutsumi Y, Suzuki S, Morii I, Hanai S, Ono K, Baba S, Tomoike H, Kawamura A, Miyazaki S, Nonogi H, Goto Y. Aldehyde dehydrogenase 2 gene is a risk factor for myocardial infarction in Japanese men. Hypertens Res. 2002;25:677–81. doi: 10.1291/hypres.25.677. [DOI] [PubMed] [Google Scholar]
- 23.Jo SA, Kim EK, Park MH, Han C, Park HY, Jang Y, Song BJ, Jo I. A Glu487Lys polymorphism in the gene for mitochondrial aldehyde dehydrogenase 2 is associated with myocardial infarction in elderly Korean men. Clin Chim Acta. 2007;382:43–7. doi: 10.1016/j.cca.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Hjelle JJ, Grubbs JH, Beer DG, Petersen DR. Inhibition of rat liver aldehyde dehydrogenase by carbon tetrachloride. J Pharmacol Exp Ther. 1981;219:821–6. [PubMed] [Google Scholar]
- 25.Hjelle JJ, Grubbs JH, Beer DG, Petersen DR. Time course of the carbon tetrachloride-induced decrease in mitochondrial aldehyde dehydrogenase activity. Toxicol Appl Pharmacol. 1983;67:159–65. doi: 10.1016/0041-008x(83)90220-x. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell DY, Petersen DR. Inhibition of rat liver aldehyde dehydrogenases by acrolein. Drug Metab Dispos. 1988;16:37–42. [PubMed] [Google Scholar]
- 27.Banfi P, Lanzi C, Falvella FS, Gariboldi M, Gambetta RA, Dragani TA. The daunorubicin-binding protein of Mr 54,000 is an aldehyde dehydrogenase and is down-regulated in mouse liver tumors and in tumor cell lines. Mol Pharmacol. 1994;46:896–900. [PubMed] [Google Scholar]
- 28.Landin JS, Cohen SD, Khairallah EA. Identification of a 54-kDa mitochondrial acetaminophen-binding protein as aldehyde dehydrogenase. Toxicol Appl Pharmacol. 1996;141:299–307. doi: 10.1006/taap.1996.0287. [DOI] [PubMed] [Google Scholar]
- 29.Lipsky JJ, Shen ML, Naylor S. Overview-in vitro inhibition of aldehyde dehydrogenase by disulfiram and metabolites. Chem Biol Interact. 2001;130–2:81–91. doi: 10.1016/s0009-2797(00)00224-6. [DOI] [PubMed] [Google Scholar]
- 30.Suh SK, Hood BL, Kim BJ, Conrads TP, Veenstra TD, Song BJ. Identification of oxidized mitochondrial proteins in alcohol-exposed human hepatoma cells and mouse liver. Proteomics. 2004;4:3401–12. doi: 10.1002/pmic.200400971. [DOI] [PubMed] [Google Scholar]
- 31.Moon KH, Kim BJ, Song BJ. Inhibition of mitochondrial aldehyde dehydrogenase by nitric oxide-mediated S-nitrosylation. FEBS Lett. 2005;579:6115–20. doi: 10.1016/j.febslet.2005.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon KH, Hood BL, Kim BJ, Hardwick JP, Conrads TP, Veenstra TD, Song BJ. Inactivation of oxidized and S-nitrosylated mitochondrial proteins in alcoholic fatty liver of rats. Hepatology. 2006;44:1218–30. doi: 10.1002/hep.21372. [DOI] [PubMed] [Google Scholar]
- 33.Kim BJ, Hood BL, Aragon RA, Hardwick JP, Conrads TP, Veenstra TD, Song BJ. Increased oxidation and degradation of cytosolic proteins in alcohol-exposed mouse liver and hepatoma cells. Proteomics. 2006;6:1250–60. doi: 10.1002/pmic.200500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moon KH, Abdelmegeed MA, Song BJ. Inactivation of cytosolic aldehyde dehydrogenase via S-nitrosylation in ethanol-exposed rat liver. FEBS Lett. 2007;581:3967–72. doi: 10.1016/j.febslet.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song BJ, Moon KH, Olsson N, Salem N., Jr Prevention of alcoholic fatty liver and mitochondrial dysfunction in the rat by long-chain polyunsaturated fatty acids. J Hepatol. 2008;49:262–73. doi: 10.1016/j.jhep.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moon KH, Upreti VV, Yu LR, Lee IJ, Ye X, Eddington ND, Veenstra TD, Song BJ. Mechanism of 3,4-methylenedioxymethamphetamine (MDMA, ecstasy)-mediated mitochondrial dysfunction in rat liver. Proteomics. 2008;8:3906–18. doi: 10.1002/pmic.200800215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon KH, Hood BL, Mukhopadhyay P, Rajesh M, Abdelmegeed MA, Kwon YI, Conrads TP, Veenstra TD, Song BJ, Pacher P. Oxidative inactivation of key mitochondrial proteins leads to dysfunction and injury in hepatic ischemia reperfusion. Gastroenterology. 2008;135:1344–57. doi: 10.1053/j.gastro.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Upreti VV, Eddington ND, Moon KH, Song BJ, Lee IJ. Drug interaction between ethanol and 3,4-methylenedioxy-methamphetamine (“ecstasy”) Toxicol Lett. 2009;188:167–72. doi: 10.1016/j.toxlet.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moon KH, Lee YM, Song BJ. Inhibition of hepatic mitochondrial aldehyde dehydrogenase by carbon tetrachloride through JNK-mediated phosphorylation. Free Radic Biol Med. 2010;48:391–8. doi: 10.1016/j.freeradbiomed.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon KH, Kim BJ, Wan J, Lee YM, Abdelmegeed MA, Song BJ. Reversible inactivation of mitochondrial aldehyde dehydrogenase by translational modifications in hepatoma cells and alcohol-fed rat livers. In: Weiner H, Plapp BV, Lindahl R, Maser E, editors. Enzymology and Molecular Biology of Carbonyl Metabolism. Vol. 13. Purdue University Press; West Lafayette, Indiana, USA: 2007. pp. 22–32. [Google Scholar]
- 41.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–6. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 42.Petersen DR, Doorn JA. Reactions of 4-hydroxynonenal with proteins and cellular targets. Free Radic Biol Med. 2004;37:937–45. doi: 10.1016/j.freeradbiomed.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeMaster EG, Quast BJ, Redfern B, Nagasawa HT. Reaction of nitric oxide with the free sulfhydryl group of human serum albumin yields a sulfenic acid and nitrous oxide. Biochemistry. 1995;34:11494–9. doi: 10.1021/bi00036a023. [DOI] [PubMed] [Google Scholar]
- 45.DeMaster EG, Redfern B, Quast BJ, Dahlseid T, Nagasawa HT. Mechanism for the inhibition of aldehyde dehydrogenase by nitric oxide. Alcohol. 1997;14:181–9. doi: 10.1016/s0741-8329(96)00142-5. [DOI] [PubMed] [Google Scholar]
- 46.Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 2004;279:21749–58. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- 47.Daiber A, Oelze M, Coldewey M, Bachschmid M, Wenzel P, Sydow K, Wendt M, Kleschyov AL, Stalleicken D, Ullrich V, Mulsch A, Munzel T. Oxidative stress and mitochondrial aldehyde dehydrogenase activity: a comparison of pentaerythritol tetranitrate with other organic nitrates. Mol Pharmacol. 2004;66:1372–82. doi: 10.1124/mol.104.002600. [DOI] [PubMed] [Google Scholar]
- 48.Woo HA, Jeong W, Chang TS, Park KJ, Park SJ, Yang JS, Rhee SG. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J Biol Chem. 2005;280:3125–8. doi: 10.1074/jbc.C400496200. [DOI] [PubMed] [Google Scholar]
- 49.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls: The cytotoxic potential of superoxide and nitric oxide. J Biol Chem. 1991;266:4244–50. [PubMed] [Google Scholar]
- 50.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–66. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 51.McDonald LJ, Moss J. Nitric oxide-independent, thiol-associated ADP ribosylation inactivates aldehyde dehydrogenase. J Biol Chem. 1993;268:17878–82. [PubMed] [Google Scholar]
- 52.Wenzel P, Hink U, Oelze M, Schuppan S, Schaeuble K, Schildknecht S, Ho KK, Weiner H, Bachschmid M, Münzel T, Daiber A. Role of reduced lipoic acid in the redox regulation of mitochondrial aldehyde dehydrogenase (ALDH-2) activity. Implications for mitochondrial oxidative stress and nitrate tolerance. J Biol Chem. 2007;282:792–9. doi: 10.1074/jbc.M606477200. [DOI] [PubMed] [Google Scholar]
- 53.Hochgräfe F, Mostertz J, Pöther DC, Becher D, Helmann JD, Hecker M. S-cysteinylation is a general mechanism for thiol protection of Bacillus subtilis proteins after oxidative stress. J Biol Chem. 2007;282:25981–5. doi: 10.1074/jbc.C700105200. [DOI] [PubMed] [Google Scholar]
- 54.Frizzell N, Rajesh M, Jepson MJ, Nagai R, Carson JA, Thorpe SR, Baynes JW. Succination of thiol groups in adipose tissue proteins in diabetes: succination inhibits polymerization and secretion of adiponectin. J Biol Chem. 2009;284:25772–81. doi: 10.1074/jbc.M109.019257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farres J, Wang TT, Cunningham SJ, Weiner H. Investigation of the active site cysteine residue of rat liver mitochondrial aldehyde dehydrogenase by site-directed mutagenesis. Biochemistry. 1995;34:2592–8. doi: 10.1021/bi00008a025. [DOI] [PubMed] [Google Scholar]
- 56.Palmer KR, Jenkins WJ. Aldehyde dehydrogenase in alcoholic subjects. Hepatology. 1985;5:260–3. doi: 10.1002/hep.1840050218. [DOI] [PubMed] [Google Scholar]
- 57.Agarwal DP, Harada S, Goedde HW, Schrappe O. Cytosolic aldehyde dehydrogenase and alcoholism. Lancet. 1983;1(8314–5):68. doi: 10.1016/s0140-6736(83)91600-8. [DOI] [PubMed] [Google Scholar]
- 58.Koop DR. Oxidative and reductive metabolism by cytochrome P450 2E1. Faseb J. 1992;6:724–30. doi: 10.1096/fasebj.6.2.1537462. [DOI] [PubMed] [Google Scholar]
- 59.Song BJ, Koop DR, Ingelman-Sundberg M, Nanji A, Cederbaum AI. Ethanol-inducible cytochrome P450 (CYP2E1): biochemistry, molecular biology and clinical relevance: 1996 update. Alcohol Clin Exp Res. 1996;20(Suppl):138A–46A. doi: 10.1111/j.1530-0277.1996.tb01764.x. [DOI] [PubMed] [Google Scholar]
- 60.Caro AA, Cederbaum AI. Oxidative stress, toxicology, and pharmacology of CYP2E1. Annu Rev Pharmacol Toxicol. 2004;44:27–42. doi: 10.1146/annurev.pharmtox.44.101802.121704. [DOI] [PubMed] [Google Scholar]
- 61.Purohit V, Gao B, Song BJ. Molecular mechanisms of alcoholic fatty liver. Alcohol Clin Exp Res. 2009;33:191–205. doi: 10.1111/j.1530-0277.2008.00827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song BJ, Suh SK, Moon KH. A simple method to systematically study oxidatively modified proteins in biological samples and its applications. Methods Enzymol. 2010;473:251–64. doi: 10.1016/S0076-6879(10)73013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song BJ, Moon KH, Upreti VV, Eddington ND, Lee IJ. Mechanisms of MDMA (Ecstasy)-induced oxidative stress, mitochondrial dysfunction, and organ damage. Curr Pharm Biotechnol. 2010;11:434–43. doi: 10.2174/138920110791591436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun J, Steenbergen C, Murphy E. S-nitrosylation: NO-related redox signaling to protect against oxidative stress. Antioxid Redox Signal. 2006;8:1693–705. doi: 10.1089/ars.2006.8.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun J, Murphy E. Protein S-nitrosylation and cardioprotection. Circ Res. 2010;106:285–96. doi: 10.1161/CIRCRESAHA.109.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guengerich FP, Kim DH, Iwasaki M. Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem Res Toxicol. 1991;4:168–79. doi: 10.1021/tx00020a008. [DOI] [PubMed] [Google Scholar]
- 67.Sohn DH, Yun YP, Park KS, Veech RL, Song BJ. Post-translational reduction of cytochrome P450IIE by CCl4, its substrate. Biochem Biophys Res Commun. 1991;179:449–54. doi: 10.1016/0006-291x(91)91391-o. [DOI] [PubMed] [Google Scholar]
- 68.Lieber CS. The discovery of the microsomal ethanol oxidizing system and its physiologic and pathologic role. Drug Metab Rev. 2004;36:511–29. doi: 10.1081/dmr-200033441. [DOI] [PubMed] [Google Scholar]
- 69.Abdelmegeed MA, Moon KH, Chen C, Gonzalez FJ, Song BJ. Role of cytochrome P450 2E1 in protein nitration and ubiquitin-mediated degradation during acetaminophen toxicity. Biochem Pharmacol. 2010;79:57–66. doi: 10.1016/j.bcp.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeong KS, Soh Y, Jeng J, Felder MR, Hardwick JP, Song BJ. Cytochrome P450 2E1 (CYP2E1)-dependent production of a 37-kDa acetaldehyde-protein adduct in the rat liver. Arch Biochem Biophys. 2000;384:81–7. doi: 10.1006/abbi.2000.2119. [DOI] [PubMed] [Google Scholar]
- 71.Bae MA, Pie JE, Song BJ. Acetaminophen induces apoptosis of C6 glioma cells by activating the c-Jun NH2-terminal protein kinase-related cell death pathway. Mol Pharmacol. 2001;60:847–56. [PubMed] [Google Scholar]
- 72.Recknagel RO, Glende EA, Jr, Dolak JA, Waller RL. Mechanisms of carbon tetrachloride toxicity. Pharmacol Ther. 1989;43:139–54. doi: 10.1016/0163-7258(89)90050-8. [DOI] [PubMed] [Google Scholar]
- 73.Lieber CS. Alcoholic fatty liver: its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol. 2004;34:9–19. doi: 10.1016/j.alcohol.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 74.Minko IG, Kozekov ID, Harris TM, Rizzo CJ, Lloyd RS, Stone MP. Chemistry and biology of DNA containing 1, N(2)-deoxyguanosine adducts of the alpha, beta-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem Res Toxicol. 2009;22:759–78. doi: 10.1021/tx9000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee YJ, Aroor AR, Shukla SD. Temporal activation of p42/44 mitogen-activated protein kinase and c-Jun N-terminal kinase by acetaldehyde in rat hepatocytes and its loss after chronic ethanol exposure. J Pharmacol Exp Ther. 2002;301:908–14. doi: 10.1124/jpet.301.3.908. [DOI] [PubMed] [Google Scholar]
- 76.Yoo SH, Park O, Henderson LE, Abdelmegeed MA, Moon KH, Song BJ. Lack of PPARα exacerbates lipopolysaccharide-induced liver toxicity through STAT1 inflammatory signaling and increased oxidative/nitrosative stress. Toxicol Lett. 2011;202:23–9. doi: 10.1016/j.toxlet.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mendelson KG, Contois LR, Tevosian SG, Davis RJ, Paulson KE. Independent regulation of JNK/p38 mitogen-activated protein kinases by metabolic oxidative stress in the liver. Proc Natl Acad Sci USA. 1996;93:12908–13. doi: 10.1073/pnas.93.23.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bae MA, Song BJ. Critical role of c-Jun N-terminal protein kinase activation in troglitazone-induced apoptosis of human HepG2 hepatoma cells. Mol Pharmacol. 2003;63:401–8. doi: 10.1124/mol.63.2.401. [DOI] [PubMed] [Google Scholar]
- 79.Kim BJ, Ryu SW, Song BJ. JNK- and p38 kinase mediated phosphorylation of Bax leads to its activation, mitochondrial translocation and apoptosis of human hepatoma HepG2 cells. J Biol Chem. 2006;281:21256–65. doi: 10.1074/jbc.M510644200. [DOI] [PubMed] [Google Scholar]
- 80.Kharbanda S, Saxena S, Yoshida K, Pandey P, Kaneki M, Wang Q, Cheng K, Chen YN, Campbell A, Sudha T, Yuan ZM, Narula J, Weichselbaum R, Nalin C, Kufe D. Translocation of SAPK/JNK to mitochondria and interaction with Bcl-x(L) in response to DNA damage. J Biol Chem. 2000;275:322–7. doi: 10.1074/jbc.275.1.322. [DOI] [PubMed] [Google Scholar]
- 81.Hanawa N, Shinohara M, Saberi B, Gaarde WA, Han D, Kaplowitz N. Role of JNK translocation to mitochondria leading to inhibition of mitochondria bioenergetics in acetaminophen-induced liver injury. J Biol Chem. 2008;283:13565–77. doi: 10.1074/jbc.M708916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen C-H, Gray MO, Mochly-Rosen D. Cardioprotection from ischemia by a brief exposure to physiological levels of ethanol: role of epsilon protein kinase C. Proc Natl Acad Sci USA. 1999;96:12784–9. doi: 10.1073/pnas.96.22.12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Churchill EN, Disatnik MH, Mochly-Rosen D. Time-dependent and ethanol-induced cardiac protection from ischemia mediated by mitochondrial translocation of varepsilonPKC and activation of aldehyde dehydrogenase 2. J Mol Cell Cardiol. 2009;46:278–84. doi: 10.1016/j.yjmcc.2008.09.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen C-H, Budas GR, Churchill EN, Disatnik M-H, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321:1493–5. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Budas GR, Disatnik MH, Chen CH, Mochly-Rosen D. Activation of aldehyde dehydrogenase 2 (ALDH2) confers cardioprotection in protein kinase C epsilon (PKCepsilon) knockout mice. J Mol Cell Cardiol. 2010;48:757–64. doi: 10.1016/j.yjmcc.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perez-Miller S, Younus H, Vanam R, Chen CH, Mochly-Rosen D, Hurley TD. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat Struct Mol Biol. 2010;17:159–64. doi: 10.1038/nsmb.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lagranha CJ, Deschamps A, Aponte A, Steenbergen C, Murphy E. Sex differences in the phosphorylation of mitochondrial proteins result in reduced production of reactive oxygen species and cardioprotection in females. Circ Res. 2010;106:1681–91. doi: 10.1161/CIRCRESAHA.109.213645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wong R, Aponte AM, Steenbergen C, Murphy E. Cardioprotection leads to novel changes in the mitochondrial proteome. Am J Physiol Heart Circ Physiol. 2010;298:H75–91. doi: 10.1152/ajpheart.00515.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sydow K, Daiber A, Oelze M, Chen Z, August M, Wendt M, Ullrich V, Mulsch A, Schulz E, Keaney JF, Jr, Stamler JS, Munzel T. Central role of mitochondrial aldehyde dehydrogenase and reactive oxygen species in nitroglycerin tolerance and cross-tolerance. J Clin Invest. 2004;113:482–9. doi: 10.1172/JCI19267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Z, Foster MW, Zhang J, Mao L, Rockman HA, Kawamoto T, Kitagawa K, Nakayama KI, Hess DT, Stamler JS. An essential role for mitochondrial aldehyde dehydrogenase in nitroglycerin bioactivation. Proc Natl Acad Sci USA. 2005;102:12159–64. doi: 10.1073/pnas.0503723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Daiber A, Wenzel P, Oelze M, Schuhmacher S, Jansen T, Münzel T. Mitochondrial aldehyde dehydrogenase (ALDH-2)-maker of and marker for nitrate tolerance in response to nitroglycerin treatment. Chem Biol Interact. 2009;178:40–7. doi: 10.1016/j.cbi.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 92.Beretta M, Sottler A, Schmidt K, Mayer B, Gorren AC. Partially irreversible inactivation of mitochondrial aldehyde dehydrogenase by nitroglycerin. J Biol Chem. 2008;283:30735–44. doi: 10.1074/jbc.M804001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oelze M, Knorr M, Schell R, Kamuf J, Pautz A, Art J, Wenzel P, Munzel T, Kleinert H, Daiber A. Regulation of human mitochondrial aldehyde dehydrogenase (ALDH-2) activity by electrophiles in vitro. J Biol Chem. 2011;286:8893–900. doi: 10.1074/jbc.M110.190017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tyther R, Ahmeda A, Johns E, Sheehan D. Proteomic identification of tyrosine nitration targets in kidney of spontaneously hypertensive rats. Proteomics. 2007;7:4555–64. doi: 10.1002/pmic.200700503. [DOI] [PubMed] [Google Scholar]
- 95.Lu Z, Scott I, Webster BR, Sack MN. The emerging characterization of lysine residue deacetylation on the modulation of mitochondrial function and cardiovascular biology. Circ Res. 2009;105:830–41. doi: 10.1161/CIRCRESAHA.109.204974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fu M, Wang C, Zhang X, Pestell RG. Acetylation of nuclear receptors in cellular growth and apoptosis. Biochem Pharmacol. 2004;68:1199–208. doi: 10.1016/j.bcp.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 97.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–5. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 99.Shepard BD, Tuma DJ, Tuma PL. Chronic ethanol consumption induces global hepatic protein hyperacetylation. Alcohol Clin Expt Res. 2010;34:280–91. doi: 10.1111/j.1530-0277.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Vanhove JL, Watson PA, Birdsey N, Bao J, Gius D, Sack MN, Jing E, Kahn CR, Friedman JE, Jonscher KR. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2011;433:505–14. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sheikh S, Ni L, Hurley TD, Weiner H. The potential roles of the conserved amino acids in human liver mitochondrial aldehyde dehydrogenase. J Biol Chem. 1997;272:18817–22. doi: 10.1074/jbc.272.30.18817. [DOI] [PubMed] [Google Scholar]
- 102.You M, Liang X, Ajmo JM, Ness GC. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am J Physiol Gastrointest Liver Physiol. 2008;294:G892–8. doi: 10.1152/ajpgi.00575.2007. [DOI] [PubMed] [Google Scholar]
- 103.Lieber CS, Leo MA, Wang X, Decarli LM. Effect of chronic alcohol consumption on Hepatic SIRT1 and PGC-1alpha in rats. Biochem Biophys Res Commun. 2008;370:44–8. doi: 10.1016/j.bbrc.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 104.Picklo MJ., Sr Ethanol intoxication increases hepatic N-lysyl protein acetylation. Biochem Biophys Res Commun. 2008;376:615–9. doi: 10.1016/j.bbrc.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 105.Shulga N, Pastorino JG. Ethanol sensitizes mitochondria to the permeability transition by inhibiting deacetylation of cyclophilin-D mediated by sirtuin-3. J Cell Sci. 2010;123:4117–27. doi: 10.1242/jcs.073502. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 106.Lieber CS, Leo MA, Wang X, Decarli LM. Alcohol alters hepatic FoxO1, p53, and mitochondrial SIRT5 deacetylation function. Biochem Biophys Res Commun. 2008;373:246–52. doi: 10.1016/j.bbrc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 107.Svanas GW, Weiner H. Enzymative requirement for cyanamide inactivation of rat liver aldehyde dehydrogenase. Biochem Pharmacol. 1985;34:1197–204. doi: 10.1016/0006-2952(85)90495-2. [DOI] [PubMed] [Google Scholar]
- 108.Keung WM, Klyosov AA, Vallee BL. Daidzin inhibits mitochondrial aldehyde dehydrogenase and suppresses ethanol intake of Syrian golden hamsters. Proc Natl Acad Sci USA. 1997;94:1675–9. doi: 10.1073/pnas.94.5.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moncada C, Fuentes N, Lladser A, Encina G, Sapag A, Karahanian E, Israel Y. Use of an “acetaldehyde clamp” in the determination of low-KM aldehyde dehydrogenase activity in H4-II-E-C3 rat hepatoma cells. Alcohol. 2003;31:19–24. doi: 10.1016/j.alcohol.2003.06.004. [DOI] [PubMed] [Google Scholar]
- 110.Lebsack ME, Petersen DR, Collins AC, Anderson AD. Preferential inhibition of the low Km aldehyde dehydrogenase activity by pargyline. Biochem Pharmacol. 1977;26:1151–4. doi: 10.1016/0006-2952(77)90060-0. [DOI] [PubMed] [Google Scholar]
- 111.Doorn JA, Hurley TD, Petersen DR. Inhibition of human mitochondrial aldehyde dehydrogenase by 4-hydroxynon-2-enal and 4-oxonon-2-enal. Chem Res Toxicol. 2006;19:102–10. doi: 10.1021/tx0501839. [DOI] [PubMed] [Google Scholar]
- 112.Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci USA. 1984;81:258–61. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6:e50. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang RS, Nakajima T, Kawamoto T, Honma T. Effects of aldehyde dehydrogenase-2 genetic polymorphisms on metabolism of structurally different aldehydes in human liver. Drug Metab Dispos. 2002;30:69–73. doi: 10.1124/dmd.30.1.69. [DOI] [PubMed] [Google Scholar]
- 115.Isse T, Matsuno K, Oyama T, Kitagawa K, Kawamoto T. Aldehyde dehydrogenase 2 gene targeting mouse lacking enzyme activity shows high acetaldehyde level in blood, brain, and liver after ethanol gavages. Alcohol Clin Exp Res. 2005;29:1959–64. doi: 10.1097/01.alc.0000187161.07820.21. [DOI] [PubMed] [Google Scholar]
- 116.Quintanilla ME, Tampier L, Sapag A, Israel Y. Polymorphisms in the mitochondrial aldehyde dehydrogenase gene (Aldh2) determine peak blood acetaldehyde levels and voluntary ethanol consumption in rats. Pharmacogenet Genomics. 2005;15:427–31. doi: 10.1097/01213011-200506000-00009. [DOI] [PubMed] [Google Scholar]
- 117.Chen JJ, Yu BP. Detoxification of reactive aldehydes in mitochondria: effects of age and dietary restriction. Aging (Milano) 1996;8:334–40. doi: 10.1007/BF03339590. [DOI] [PubMed] [Google Scholar]
- 118.Watanabe A, Hobara N, Nakatsukasa H, Shiota T, Kobayashi M, Nagashima H. Impaired acetaldehyde metabolism in partially hepatectomized rats. Res Exp Med. 1985;185:13–20. doi: 10.1007/BF01851523. [DOI] [PubMed] [Google Scholar]
- 119.Park KS, Cho SY, Kim H, Paik YK. Proteomic alterations of the variants of human aldehyde dehydrogenase isozymes correlate with hepatocellular carcinoma. Int J Cancer. 2002;97:261–5. doi: 10.1002/ijc.1585. [DOI] [PubMed] [Google Scholar]
- 120.Kim J, Kim SH, Lee SU, Ha GH, Kang DG, Ha NY, Ahn JS, Cho HY, Kang SJ, Lee YJ, Hong SC, Ha WS, Bae JM, Lee CW, Kim JW. Proteome analysis of human liver tumor tissue by two-dimensional gel electrophoresis and matrix assisted laser desorption/ionization-mass spectrometry for identification of disease-related proteins. Electrophoresis. 2002;23:4142–56. doi: 10.1002/elps.200290032. [DOI] [PubMed] [Google Scholar]
- 121.Shibuya A, Takeuchi A, Shibata H, Saigenji K, Yoshida A. Immunohistochemical study of hepatocellular carcinoma-specific aldehyde dehydrogenase. Alcohol Alcohol Suppl. 1994;29:119–23. [PubMed] [Google Scholar]
- 122.Oshita F, Morita A, Ito H, Kameda Y, Tsuchiya E, Asai S, Miyagi Y. Proteomic screening of completely resected tumors in relation to survival in patients with stage I non-small cell lung cancer. Oncol Rep. 2010;24:637–45. doi: 10.3892/or_00000902. [DOI] [PubMed] [Google Scholar]
- 123.Zakhari S, Li TK. Determinants of alcohol use and abuse: Impact of quantity and frequency patterns on liver disease. Hepatology. 2007;46:2032–9. doi: 10.1002/hep.22010. [DOI] [PubMed] [Google Scholar]
- 124.Venkatraman A, Landar A, Davis AJ, Chamlee L, Sanderson T, Kim H, Page G, Pompilius M, Ballinger S, Darley-Usmar V, Bailey SM. Modification of the mitochondrial proteome in response to the stress of ethanol-dependent hepatotoxicity. J Biol Chem. 2004;279:22092–101. doi: 10.1074/jbc.M402245200. [DOI] [PubMed] [Google Scholar]
- 125.McClain CJ, Kromhout JP, Peterson FJ, Holtzman JL. Potentiation of acetaminophen hepatotoxicity by alcohol. JAMA. 1980;244:251–3. [PubMed] [Google Scholar]
- 126.Seeff LB, Cuccherini BA, Zimmerman HJ, Adler E, Benjamin SB. Acetaminophen hepatotoxicity in alcoholics. A therapeutic misadventure. Ann Intern Med. 1986;104:399–404. doi: 10.7326/0003-4819-104-3-399. [DOI] [PubMed] [Google Scholar]
- 127.Szabo G, Wands JR, Eken A, Osna NA, Weinman SA, Machida K, Wang JH. Alcohol and hepatitis C virus--interactions in immune dysfunctions and liver damage. Alcohol Clin Exp Res. 2010;34:1675–86. doi: 10.1111/j.1530-0277.2010.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hassan MM, Hwang LY, Hatten CJ, Swaim M, Li D, Abbruzzese JL, Beasley P, Patt YZ. Risk factors for hepatocellular carcinoma: synergism of alcohol with viral hepatitis and diabetes mellitus. Hepatology. 2002;36:1206–13. doi: 10.1053/jhep.2002.36780. [DOI] [PubMed] [Google Scholar]
- 129.Diehl AM. Obesity and alcoholic liver disease. Alcohol. 2004;34:81–7. doi: 10.1016/j.alcohol.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 130.Carmiel-Haggai M, Cederbaum AI, Nieto N. Binge ethanol exposure increases liver injury in obese rats. Gastroenterology. 2003;125:1818–33. doi: 10.1053/j.gastro.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 131.Reid LW, Elifson KW, Sterk CE. Ecstasy and gateway drugs: initiating the use of ecstasy and other drugs. Ann Epidemiol. 2007;17:74–80. doi: 10.1016/j.annepidem.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pontes H, Duarte JA, de Pinho PG, Soares ME, Fernandes E, Dinis-Oliveira RJ, Sousa C, Silva R, Carmo H, Casal S, Remião F, Carvalho F, Bastos ML. Chronic exposure to ethanol exacerbates MDMA-induced hyperthermia and exposes liver to severe MDMA-induced toxicity in CD1 mice. Toxicology. 2008;252:64–71. doi: 10.1016/j.tox.2008.07.064. [DOI] [PubMed] [Google Scholar]
- 133.Karahanian E, Ocaranza P, Israel Y. Aldehyde dehydrogenase (ALDH2) activity in hepatoma cells is reduced by an adenoviral vector coding for an ALDH2 antisense mRNA. Alcohol Clin Exp Res. 2005;29:1384–9. doi: 10.1097/01.alc.0000174909.91034.7c. [DOI] [PubMed] [Google Scholar]
- 134.Cortínez G, Sapag A, Israel Y. RNA interference against aldehyde dehydrogenase-2: development of tools for alcohol research. Alcohol. 2009;43:97–104. doi: 10.1016/j.alcohol.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 135.Li Y, Zhang D, Jin W, Shao C, Yan P, Xu C, Sheng H, Liu Y, Yu J, Xie Y, Zhao Y, Lu D, Nebert DW, Harrison DC, Huang W, Jin L. Mitochondrial aldehyde dehydrogenase-2 (ALDH2) Glu504Lys polymorphism contributes to the variation in efficacy of sublingual nitroglycerin. J Clin Invest. 2006;116:506–11. doi: 10.1172/JCI26564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schuhmacher S, Schulz E, Oelze M, König A, Roegler C, Lange K, Sydow L, Kawamoto T, Wenzel P, Münzel T, Lehmann J, Daiber A. A new class of organic nitrates: investigations on bioactivation, tolerance and cross-tolerance phenomena. Br J Pharmacol. 2009;158:510–20. doi: 10.1111/j.1476-5381.2009.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]