Abstract
Patients with melanoma metastatic to regional lymph nodes exhibit a range in tumor progression, survival, and treatment. Current approaches to stratify patients with this stage of disease predominantly involve clinical and histological methods. Molecular classification thus far has focused almost exclusively on genetic mutations. In this study, proteomic data from 69 melanoma lymph node metastases and 17 disease free lymph nodes acquired by histology-directed MALDI imaging mass spectrometry were used to classify tumor from control lymph node and to molecularly sub-classify patients with stage III disease. From these data, 12 survival associated protein signals and 3 recurrence associated signals in the acquired mass spectra were combined to generate a multiplex molecular signature to group patients into either poor or favorable groups for recurrence and survival. Proteins represented in the signature include cytochrome c, s100 A6, histone H4, and cleaved forms of thymosin β-4, thymosin β-10, and ubiquitin. In total over 40 protein signals from the tissue were identified.
Keywords: MALDI, Imaging mass spectrometry, Melanoma, Proteomics, Molecular classification
1. Introduction
Despite continued research and increased public awareness, melanoma accounted for an estimated 62,000 new cases and 8400 deaths in 2008 in the U.S. [1]. Melanoma arises from melanocytes, the melanin (pigment) producing cells found in the skin. Melanocytes occupy the basal layer between the dermis and epidermis, and are normally singly distributed with each melanocyte surrounded by multiple keratinocytes. Melanocyte growth and production of melanin upon exposure to ultraviolet light (UVL) are tightly regulated by these surrounding keratinocytes via highly structured dendritic processes [2,3]. Tumorigenesis of the melanocytes requires independence from keratinocyte regulation and the suppression of pro-apoptotic and anti-proliferative pathways [4].
Staging of melanoma was established by the 2002 American Joint Committee on Cancer (AJCC) Melanoma Staging Committee and is determined by the depth of the primary tumor, ulceration, and presence or absence of regional lymph node involvement (stage III) or distant metastasis (stage IV) [5,6]. Early stage melanoma is easily diagnosed and highly curable, but late stages are resistant to therapy with survival rates often below 15% [7]. The first line of treatment for all stages of melanoma is surgical resection, when feasible. In advanced cases, systemic treatment with α-interferon or dacarbazine, or high-dose interleukin-2 are used, although response rates are low, toxicity is high, and long-term survival benefits are limited [7–9].
The significant variability in survival of patients with stage III melanoma (24–70% 5-year survival) points to an insufficient understanding of the heterogeneity of the disease. Clinical and pathological prognostic factors for survival of patients with stage III disease include tumor thickness, ulceration and mitotic rate of the primary tumor, and the number and size of involved lymph nodes. These factors, however, offer limited biologic or mechanistic understanding of the disease. A need exists to develop molecular markers to categorize the heterogeneity of melanoma tumors for diagnosis, prognosis, and potential treatment efficacy to improve the resources clinicians have for patient evaluation and treatment. So far, molecular classification of melanoma has largely focused on identifying genomic mutations and gene expression profiling [10–12]. Several mutations among key cell signaling pathways have been shown in metastatic melanoma, including BRAF [13] and NRAS of the MAPK pathway, CDKN2A, and APAF-1 [14,15]. Gene expression profiling has been applied to classify and sub-classify primary cutaneous melanoma and advanced stage IV disease with an emphasis on correlation to clinical outcome [16,17]. Gene expression technology provides specific, quantitative information of mRNA production; however, cells use a variety of mechanisms to regulate proteins, from varying translation rates, addition of functional groups, peptide cleavage, and protein recycling.
Proteomic based molecular classification, which provides information about relative protein abundances and post-translational modifications (PTM’s), remains underdeveloped in providing rich, high-throughput analysis for large patient cohorts. Due in part to this, 2D-gel electrophoresis experiments have focused primarily on comparisons of melanocyte and melanoma cell lines [18–21], though these may differ substantially from native tissue environments and fail to account for the population diversity. Alternatively, serum proteomics, which is amenable to higher throughput screening analyses, has been applied to molecular classification of melanoma [22–24]. However, serum remains a significant analytical challenge due to the minute amounts of tumor secreted proteins, high background levels of serum proteins, and fluctuating protein levels [25].
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) is a molecular technology that acquires information from intact proteins directly from thin sections of tissue [26]. The spatial integrity of the proteins within the biopsy section is maintained, allowing specific cellular regions to be analyzed. For the analysis, matrix is applied uniformly or in a discrete grid array across the tissue. A laser irradiates the matrix and ionizes the proteins, generating data containing both the proteins detected and the tissue location. Each acquisition location can then be used as a pixel to generate a molecular image of any analyte detected. In addition to being able to analyze many proteins in parallel, MALDI IMS technology has the benefit of 1) measuring intact proteins with high mass accuracy (typically better than 1 part in 10,000), 2) measurement of intact proteins in their native, biologic state including PTM’s, 3) in-situ analysis directly from tissue with location and cellular specific information and, 4) high sensitivity of intact proteins below 20 kDa (an area often overlooked in other proteomic analyses). MALDI IMS has been employed to generate ion density maps (images) of numerous detected analytes, including pharmaceuticals [27–30], lipids [31], enzymatically cleaved proteins [32], and intact proteins [33–36].
This technology has also been used to identify biomarkers and classifiers for clinical diagnosis and potential treatment of cancer [37], including lung cancer [38], gliomas [39], and breast cancer [40]. MALDI IMS has recently been adapted to accurately target small cellular regions within tissue biopsies, termed histology directed IMS where a pathologist examines a histological stained tissue section to guide and confidently target the cellular regions of interest in an adjacent serial section of the biopsy [41]. In this way, the proteins from tumor foci or regions of adjacent “normal” tissue can be analyzed from each section, and necrosis, autolysis, connective tissue, and blood vessels can be avoided, so as not to unduly complicate the analysis.
In this present study, histology-directed MALDI IMS has been used to identify proteins found in 69 lymph nodes involved by metastatic melanoma and 17 control lymph nodes free of tumor. From the proteomic signatures obtained directly from the tissue, a molecular classification of tumor and control lymph tissue was generated that could differentiate normal and malignant tissue. Further, 62 of the patients with stage III melanoma lymph node metastases were analyzed to identify proteins associated with survival and recurrence. The protein signatures that correlate with these outcomes were identified with high confidence. Beyond diagnostic and prognostic value in assessing melanoma progression, these protein signatures may provide valuable insights in the choice of optimal treatment strategies for individual patients.
2. Methods
2.1. Sample collection
Lymph node metastases were collected from 69 melanoma patients at the time of regional lymph node dissection; 17 control, cancer-free lymph nodes were obtained from patients undergoing partial colectomy for benign disease. All samples were frozen in liquid nitrogen and stored at −80 C until analysis. Seven patients who were provided only palliative care were excluded from survival and recurrence analyses. Melanoma biopsies were acquired in two batches, the first comprising the training set and the second the test set, Table 1.
Table 1.
Clinical characteristics of the training and test cohorts comprised of patients with stage III melanoma. All times are from the date of surgery from which the tissue biopsy was collected.
| Melanoma training and test data sets
| ||
|---|---|---|
| Training | Test | |
| Patients | 43 | 19 |
| Deaths | 18 | 5 |
| Median survival | 19.9 months | N/A |
| Recurrence | 28 | 11 |
| Median rec. time | 7.7 months | 9.1 months |
| Male/female | 30/13 | 14/5 |
| Age | 28–84 years | 36–86 years |
| Median age | 52.6 years | 56.1 years |
2.2. Histology-directed MALDI IMS
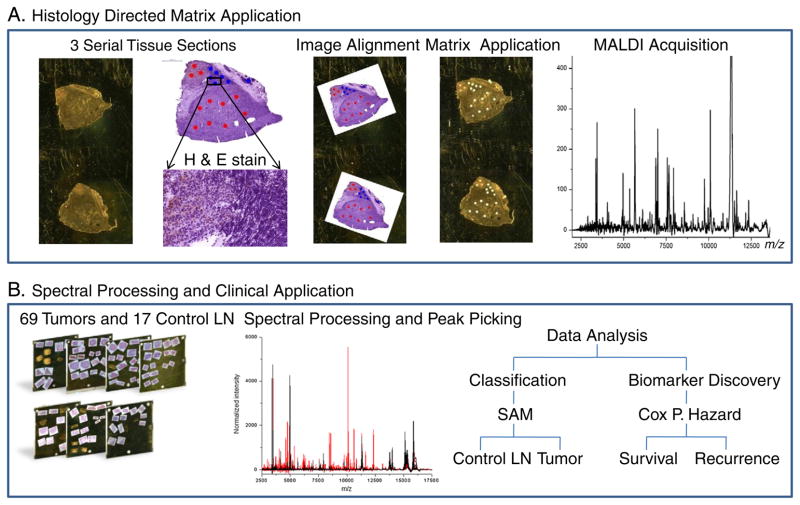
Tissue preparation for MALDI and the histology directed MALDI workflow have been previously described [41,42], and are slightly modified here (Fig. 1A). For each tissue specimen, three 12 μm serial sections were cut on a cryo-microtome. The first and last tissue sections were thaw-mounted onto a gold coated target plate for MALDI analysis. The center tissue section was thaw-mounted onto a glass microscope slide, stained with hematoxylin and eosin (H & E), and photomicrographed at high resolution (Mirax, 3D Histech, Hungary). The total sample set spanned 15 MALDI plates containing over 150 tissue sections. The MALDI plates were immersed in 70% and 90% ethanol (EtOH) for 20 s each, allowed to air-dry, marked with 4 fiducials, a scanned image acquired, and then stored at −80 C under nitrogen until matrix application. A pathologist examined each H & E photomicrograph and selected regions of interest of 200 μm in diameter, corresponding to the size of a matrix spot (Fig. 2). Each marked H & E image was aligned to the two serial sections on the MALDI target plate image. The MALDI target plate and the pixel coordinates of each 200 μm mark were loaded into a Portrait 630 (Labcyte, Sunnyvale, CA), and using 4 fiducials, translated into matrix printing locations.
Fig. 1.
A) Histology-directed MALDI IMS. Cellular regions are selected by a pathologist to ensure melanoma foci are targeted. B) The MALDI spectra from these regions were averaged and compared by SAM (control LN versus tumor-positive LN) and by CPH to determine proteins associated with survival and recurrence.
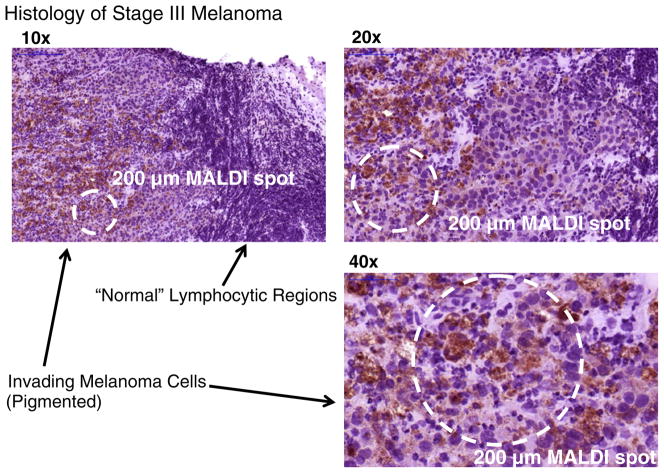
Fig. 2.
H & E stain of stage III melanoma infiltrating lymph node. Brown pigmentation and enlarged nuclei characterize melanoma cells, while lymphocytes appear with much smaller nuclei and with little cytoplasm. Dashed circles indicate the area of a typical 200 μm diameter dried MALDI sample spot.
For each imported printing coordinate, ~180 pL droplets of 20 mg/mL sinapinic acid (SA) in 60/40/0.2 v/v/v acetonitrile/water/trifluoroacetic acid (ACN/H2O/TFA) were printed in 6 successive iterations of 10 drops, ensuring adequate drying time between each iteration. The resultant dried matrix spots were approximately 180 μm in diameter. The coordinates of the positions of each dried matrix spot were transferred to an Autoflex II TOF (Bruker Daltonics, Billerica, MA) equipped with a Smart Beam laser. MALDI spectra were auto-acquired from each matrix spot in linear mode using the auto-execute software in Flexcontrol (Bruker Daltonics). The relevant instrument settings were: ion source 1 (20 kV), ion source 2 (18.65 kV), lens (6.85 kV), real time smooth (High), and detection (m/z 2000–70,000). A total of 500 spectra were summed for each spot using the Random Walk raster pattern, with no evaluation criteria.
2.3. Spectral processing
The overall workflow for processing of the mass spectral data is shown in Fig. 1B. The summed spectra from each matrix spot were pre-processed individually using ProTSMarker (Biodesix, Steamboat Springs, CO). Briefly, the program is an updated version of the software previously described which performs baseline subtraction, spectral smoothing, peak alignment, and peak detection using measured local values across the entire m/z range and total-ion current (TIC) normalization [43]. Outlying spectra of low signal-to-noise, spectra with less than 30% of peaks present, or spectra dominated by hemoglobin (m/z 15,127 and 15,865) were removed. The remaining processed spectra were averaged within each patient to generate a single representative spectrum composed of >15 discrete matrix spots (melanoma specific regions) from each patient resulting in intra-patient coefficients of variation below 30%. The averaged spectra were used in all of the statistical analyses.
2.4. Statistical analysis
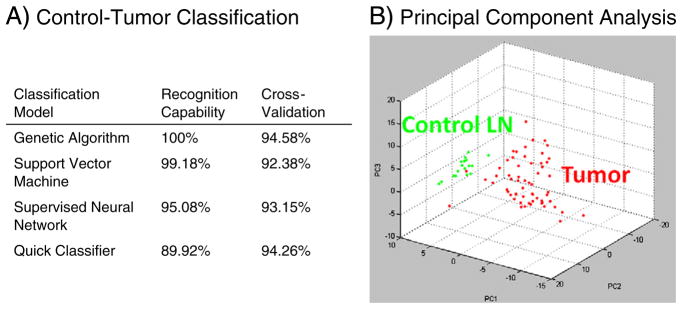
Expression differences and classification models were generated between the control lymph node and tumor. Protein intensity differences between 17 control lymph nodes and 69 metastatic melanoma regions of infiltrated lymph nodes were evaluated by a significance-analysis-of-microarray (SAM) test [44]. Protein peaks that differed by more than 2-fold area under the curve (AUC) with a false-discovery rate (FDR) were considered significant. Data was collected for analytes up to 70 kDa, with visible peaks corresponding to 42, 53, and 66 kDa (tentatively assigned as actin, vimentin, and albumin). However, generally above m/z 25,000 the number and intensity of peaks observed were not sufficient to reliably judge a significant change in expression and features above this m/z were excluded. Classification by protein peaks between control lymph node and tumor was performed using 4 models (genetic algorithm, support vector machine, supervised neural network, and quick classifier) in the ClinProTools software (Bruker Daltonics), Fig. 4. Each model was evaluated by 20% leave out cross-validation over 10 iterations.
Fig. 4.
A) Model algorithms to classify control lymph node and melanoma lymph node metastases by their proteomic signature. B) 3D PCA plot of control lymph node (green) and melanoma lymph node metastases (red).
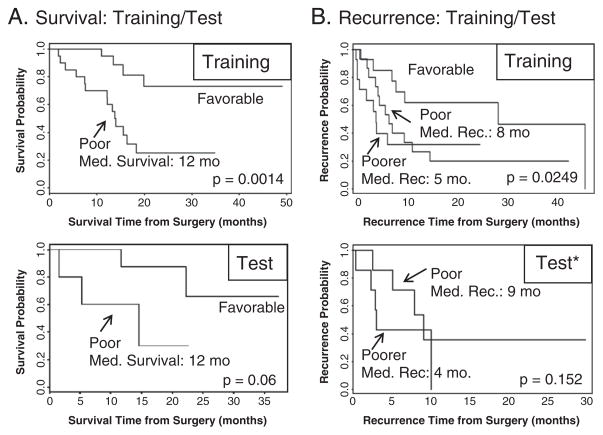
Melanoma biopsies were acquired in two batches, the first comprising the training set and the second the test set, Table 1. Individual proteins associated with survival and time-to-recurrence in stage III melanoma lymph node metastases were first determined in the training set by using a univariate Cox-proportional hazard (CPH) model [45], with an individual p-value of less than 0.05 considered significantly associated. The significant proteins were weighted by their univariate CPH score and combined into a single compound predictor (CP), for both survival and recurrence. Patients in the training set were ranked by their CP score and a division by CP scores of “poor” (Med. Survival/Recurrence <12 months) and “favorable” (Med. Survival/Recurrence >12 months) was made. The set of proteins and their weighted predictive CP score were used to group the test set into both a “poor” and “favorable” group using the same CP score range.
The two cohorts were combined into a single data set and evaluated using the same univariate CPH model and workflow to generate individual proteins and their weights. Additionally, a bootstrap method was applied in which the combined data was resampled with replacement 1000 times and the results ranked across the sampled data (p-value <0.05). Significant proteins from both models were weighted and combined into a single CP score and the patients ranked and plotted by their CP score.
2.5. Protein identification
Proteins were identified using an adaptation of the procedure previously outlined [33,46] in which intact proteins are fractionated, analyzed by MALDI MS and matched to on-tissue m/z values, further purified by gel-electrophoresis, in-gel digested, and the resulting peptides identified by LC–MS/MS. A sample of tissue (~50 mg) was mechanically homogenized in 1 mL of TPER (Pierce) solution on ice and the soluble protein concentration determined using a Bradford analysis and diluted to 200 mg/mL in 98/2/0.1 H2O/ACN/formic acid (FA). The mixture was separated on a Vydac C8 RP HPLC column, using a 96 minute gradient flowing at 0.5 mL/min. Fractions were collected every minute on a 96-well plate and dried. The 96-well fractions were reconstituted in 5 μL of 60/40/0.2 v/v/v ACN/H2O/TFA and 1 μL from each well was spotted onto a MALDI target plate. SA matrix (20 mg/mL 60/40/0.2 v/v/v ACN/H2O/TFA) was mixed on the MALDI target plate and allowed to dry. The protein/matrix spots were analyzed on an Autoflex II linear TOF in an autoexecute mode. The fractions that contained protein peaks of interest were further purified on a tricine gel (Pierce, Rockford, IL), the bands excised, in-gel digested with trypsin (Promega, Madison, WI), and the peptides analyzed by nano-HPLC MS/MS using a C18 capillary column attached to an HCT ion trap (Bruker Daltonics, Billerica, MA). Results were searched using the ipi-human database on SEQUEST and proteins containing at least 2 unique peptides or more than 15% sequence coverage were considered valid. Modifications, including removal of initiator methionine, N-terminal and interior acetylations, disulfide bridges, and signal peptide removal, were accounted for to derive the final mass of each. Several common proteins have been previously identified, including several histones and macrophage inhibitory factor [34,47,48].
Several low-molecular weight proteins were identified by tandem MS from singly charged intact MALDI ions as follows. To improve sensitivity, a small section of tissue was homogenized in 60/40/0.2 v/v/v ACN/H2O/TFA and immediately spotted onto a MALDI target plate. DHB matrix (40 mg/mL in 50/50/0.2 MeOH/H2O/TFA) was mixed with the solution on plate and allowed to air dry. SA and α-cyano matrix were tested, but were not as effective in isolating and fragmenting intact parent ions (data not shown). Linear MS spectra from the tissue homogenate were compared to the spectra obtained by direct tissue MALDI analysis to ensure homology. Proteins of interest were isolated and fragmented directly from the complex protein mixture using an Ultraflex II TOF/TOF (Bruker Daltonics) in LIFT mode. Resultant MS/MS spectra were searched using MASCOT from the BioTools software (Bruker Daltonics) with variable modifications of N-terminal acetylations and identifications with “extensive homology” considered significant.
3. Results
3.1. Analysis and classification of control LN and stage III melanoma
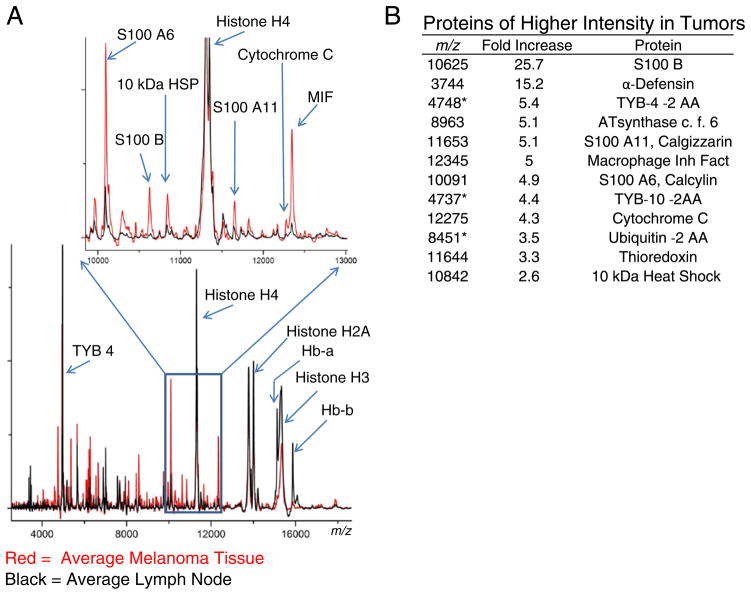
The representative protein profile of control lymph nodes and melanoma metastatic to the lymph nodes were characterized using the histology-directed MALDI MS approach. The peak detection algorithm of ProTSMarker detected a total of 155 protein features in the m/z range 2000–40,000. The SAM results showed 57 protein peaks that exhibited at least a 2-fold intensity difference between control LN and melanoma lymph node metastases (Supplemental Table 1).
Both tissue types consistently display intense peaks at m/z 11,307 (histone H4), m/z 14,007 (histone H2A), and broad peaks at m/z 13,780 (histone H2B) and m/z 15,345 (histone H3) (Fig. 3). The 4 histone species form the chromatin complex that organizes DNA in a highly ordered, compact form. Modifications to the histone species, specifically acetylations and methylations, are highly regulated processes that control the accessibility of chromosomal regions to the transcription machinery [49,50]. Multiply acetylated forms of histone H4 (m/z 11,307, 11,349, and 11,391) and H2A (m/z 14,007, 14,049, and 14,091) are easily distinguishable, but the H2B and H3 histones are observed as broad, unresolved peaks, no doubt due to multiple isoforms and methylations. The histone H3 peak displayed significantly higher intensity in the control LN and the triply acetylated histone H2A at m/z 14,091 was significantly different between the groups with a 2.4-fold increase in the tumor spectra.
Fig. 3.
Comparison of protein signals between control lymph nodes and metastatic melanoma. A) Comparison of averaged spectra of control lymph nodes (black) and melanoma lymph node metastases (red). B) SAM analysis revealing select differentially expressed proteins between LN and melanoma. *Indicates truncated molecular form with 2 amino acids absent from the carboxy-terminus.
Protein peaks observed at higher intensity in the tumor regions included several S100 calcium binding proteins (S100 A6, S100 A11, S100 B), macrophage migration inhibitory factor (MIF) involved in immune response and inhibition of p53 mediated apoptosis [51], heat shock protein 10 kDa 1 (HSP10) that is involved in regulating protein folding with HSP60 and in increasing the anti-apoptotic Bcl-xl and Bcl-2 proteins [52], and the antioxidant thioredoxin [53]. Measurements of high levels of MIF and HSP10 suggest activation of cell survival pathways.
Beyond describing individual protein variations between the groups, the protein signatures were used to generate classification models to accurately distinguish each tissue type (Fig. 4). Each of the classification models showed a high recognition capability, the lowest still recognizing and correctly classifying nearly 90% of the patients. The robustness of each model was additionally evaluated by cross-validation (20% leave out, 10 iterations), with results very near the original recognition. Fig. 4B shows the 3D PCA plot between the cancer (red) and control LN (green), visually characterizing the variation across the tumor samples compared to the compact distribution of the lymph nodes.
3.2. Proteins related to survival and recurrence
The Cox proportional hazard model was applied to examine the hazard potential associated with the proteomic features in the training set. In applying this model to proteomic data, each protein feature can be examined as an independent covariant, evaluating its association with patient survival or time-to-recurrence. Of the 155 proteins detected in the training set, 7 were significantly associated with patient survival and 2 with recurrence (p<0.05, Table 2). Proteins with a ‘poor’ effect increased intensity with worse patient prognosis, and conversely, those with a ‘favorable’ effect increased with better patient prognosis. These were weighted and combined into a single compound predictor (CP) score for survival and recurrence, with m/z 16,789 (p=0.056) included to allow at least 3 proteins for a recurrence compound predictor.
Table 2.
Results of CPH model to determine individual proteins in the training set associated with survival (A) and recurrence (B). “Poor” effect denotes a worse patient prognosis as the protein intensity increases, and likewise, “favorable” denotes a better prognosis as the protein intensity increases.
| m/z | Protein | Effect | P value |
|---|---|---|---|
| A) Test set: survival markers | |||
| 4049 | Poor | 0.021 | |
| 4737 * | TYB-10 * | Poor | 0.023 |
| 4748 * | TYB-4 * | Poor | 0.015 |
| 7045 | Poor | 0.013 | |
| 11,307 | Histone H4 | Favorable | 0.03 |
| 13,778 | Histone H2B | Favorable | 0.018 |
| B) Test set: recurrence markers | |||
| 4283 | Favorable | 0.006 | |
| 12,275 | Cytochrome C | Favorable | 0.01 |
| 16,789 | Calmodulin | Favorable | 0.056 |
Indicates protein form observed has a C-terminal truncation.
For survival, a division from the ranked CP scores of “poor” (median survival ≤12 months) and “favorable” (median survival >12 months) was made (p=0.0014), Fig. 5A top. The set of proteins and their weighted predictive CP score was used to group the test set into both a “poor” and “favorable” group using the same CP score range (p=0.06), Fig. 4A bottom, with the “poor” group having a median survival of roughly 12 months, in line with the prediction.
Fig. 5.
Training and test patient groups were used to generate and validate survival and recurrence compound predictor (CP) protein signatures. The CP was generated from the sum of the significant proteins in the training set, weighted by the proteins’ predictive ability, for both survival and recurrence. A) top: the patients in the training set were ranked and divided by their CP score into 2 groups: “poor” (med. survival ≤12 months) and “favorable” (med. survival >12 months). A) bottom: CP range for “poor” and “favorable” survival was evaluated in the test set, showing comparable median 12 month survival in the “poor” group. B) top: recurrence CP scores divided the training set into “favorable” (med. rec. >12 months) and into two poor groups: “poor” (med. rec.=8 months) and “poorer” (med. rec.=5 months). B) bottom: recurrence CP score evaluated in the test set, which successfully showed comparable “poor” (med. rec.=9 months) and “poorer” (med. rec.=4 months) groups, in line with the predicted score. *The “favorable” group in the recurrence test set was omitted for clarity, see text.
The same general procedure was applied to the training and test set using patient time to recurrence. However, in addition to the “favorable” group (median recurrence >12 months), the recurrence CP scores divided the training set into 2 poor groups: “poor” (median recurrence ~8 months) and “poorer” (median recurrence ~5 months), p=0.0249 Fig. 4B top. The recurrence CP values from the training set were evaluated in the test set, resulting in very comparable “poor” (median recurrence ~9 months) and “poorer” (median recurrence ~4 months) groups, p = 0.152 Fig. 4B bottom. The “favorable” group in the test set contained only 4 patients, making it difficult to draw a reliable conclusion from, and was omitted from the graph for clarity.
Although limited in number, the ability of the predictive CP score to group patients in the test set according to their median survival and recurrence was an encouraging validation of the method and provided confidence for combining the training and test sets into a single cohort to achieve greater statistical power, Table 3. Protein markers from the combined data set were obtained using the same univariate CPH model and p value cutoff as the training set. As an additional test, a bootstrap method was employed that randomly sampled the combined patient data with replacement, ranked each bio-marker using the CPH model on the new sampled data, and repeated the procedure 1000 times.
Table 3.
The training and test set cohorts were pooled and reexamined using both a univariate CPH model and a resampling with replacement bootstrap analysis.
| Pooled patients set
| ||||||
|---|---|---|---|---|---|---|
| Clinical information | Survival protein markers | |||||
| Patients | 62 | m/z | Protein | Effect | P value univariate | P value bootstrap |
| Deaths | 23 | 4026 | Poor | 0.035 | 0.017 | |
| Ave. survival | 22.2 months | 4049 | Poor | 0.0047 | .0049 | |
| Recurrence | 39 | 4737 * | TYB-10 * | Poor | 0.009 | 0.0042 |
| Ave. recurrence | 8.4 months | 4748 * | TYB-4 * | Poor | 0.024 | 0.013 |
| Male/female | 44/18 | 7045 | Poor | 0.032 | 0.021 | |
| Median age | 53.6 years | 8451 * | Ubiquitin | Poor | 0.036 | 0.034 |
| 10,091 | S100 A6(calcyclin) | Poor | 0.05 | 0.041 | ||
| 10,133 | S100 A6+Acetyl | Poor | 0.044 | 0.042 | ||
| 11,307 | Histone H4 | Favorable | 0.016 | 0.021 | ||
| 11,349 | Histone H4+Ac | Favorable | 0.042 | 0.038 | ||
| 13,778 | Histone H2B | Favorable | 0.019 | 0.021 | ||
| 15,336 | Histone H3 | Favorable | 0.038 | 0.027 | ||
| Recurrence protein markers | ||||||
| 12,275 | Cytocrome C | Favorable | 0.0035 | 0.0021 | ||
| 16,791 | Calmodulin | Favorable | 0.0029 | 0.0098 | ||
| 17,922 | Favorable | 0.077 | 0.031 | |||
Indicates protein form observed has a C-terminal truncation.
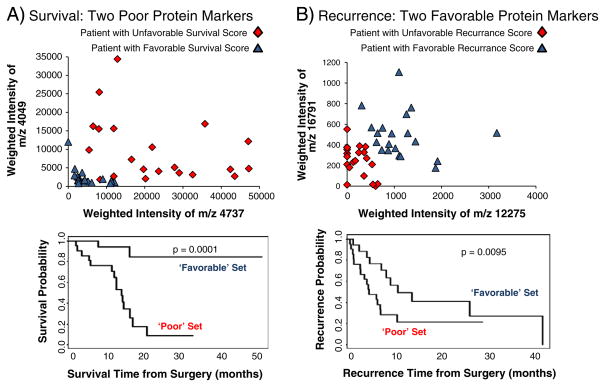
In both the univariate CPH and the bootstrap of the pooled data, all of the original survival and recurrence protein markers from the training set were found, with the exception of m/z 4283. The pooled data analysis was able to identify 6 additional proteins associated with survival. These new markers were weighted and combined into a CP to rank each patient, and the set divided into upper 33% (“unfavorable”, red diamond) and lower 33% (“favorable”, blue triangle) groups for both survival and recurrence. Each patient from the dichotomized groups was plotted using the value of two significant proteins, m/z 4049 and 4737 for survival and m/z 16,791 and 12,275 for recurrence, Fig. 6A–B top. The result shows the spread of patients from two proteins, and the survival and recurrence trends for those groups (p=0.0001 and p=0.0095 respectively), Fig. 6A–B bottom.
Fig. 6.
Graphical representation of proteins related to survival and recurrence. Patients with a high compound predictor (unfavorable outcome, red diamond) were plotted with patients having a low compound predictor (favorable outcome, blue triangle) for both survival and recurrence. A) top: plot of the weighted intensity of two proteins associated with poor survival, m/z 4049 and m/z 4737 (TYB-10, modified). A generally better prognosis is observed as both proteins decrease, as shown in the survival plot (bottom). B) top: plot of the weighted intensity of two proteins associated with favorable recurrence outcome, m/z 16,791 (calmodulin) and m/z 12,275 (CyC), with better prognosis as the intensity of both proteins increase, displaying a longer time to recurrence (bottom).
3.3. Protein identification
MALDI MS produces mass specific data of the intact protein species; however, the m/z alone is often not sufficient to positively identify the protein, as the biologically active protein may have undergone post-translational modifications. Robust protein identification is routinely achieved through protein separation/isolation, proteolytic digestion, sequencing of peptides by LC MS/MS, and matching the peptides to genome-based protein databases. The MALDI peaks confidently identified using this strategy are shown in Supplemental Table 2, with the unique peptides found and % coverage, and the PTM’s that result in the final biologically observed mass. The results show that this workflow was suitable for many of the proteins above ~9 kDa. Proteins of M. W. <6 kDa are difficult to identify from a complex mixture on a 1-D gel or identify by directly digesting the mixture due to the low number of observable peptides generated.
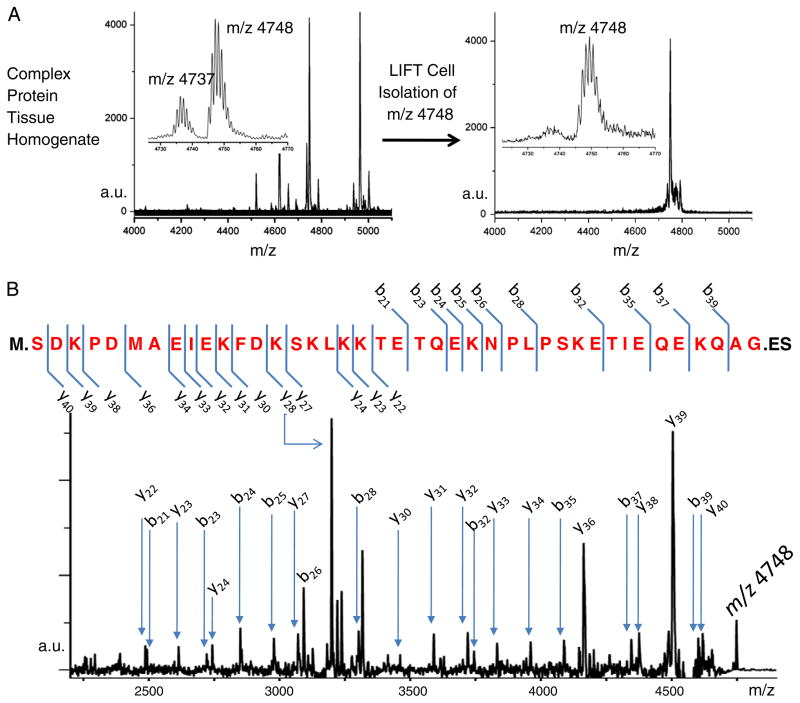
Unknown proteins were identified directly from these clinical samples without LC separation using a MALDI TOF/ TOF top–down approach [54,55]. The ions of interest, corresponding to intact proteins, were isolated and fragmented directly in the mass spectrometer. An example of the top–down MALDI fragmentation and identification from a complex mixture is shown in Fig. 7, where m/z 4748 was isolated, fragmented, and identified as a cleaved form of thymosin β-4 that lacks the 2 C-terminal amino acids. The close proximity signal m/z 4737 was likewise isolated, fragmented, and identified as a thymosin β-10 with the 2 C-terminal amino acids absent. Full length forms of TYB4 (m/z 4964) and TYB10 (m/z 4936) were also isolated, fragmented, and identified. In the case of the ions representing proteins with 2 C-terminal amino acids removed, the protein database did not display this modification.
Fig. 7.
Top–down identification of m/z 4748 (mono-isotopic m/z of 4745) by MALDI TOF–TOF. A) MALDI spectrum of tissue homogenate, with in-source LIFT cell isolation of m/z 4748. B) MALDI MS/MS spectrum of m/z 4748. The protein was identified as thymosin β-4 with the 2 C-terminus amino acids missing. (The final M.W. is given by: 5053–M (131)–ES (277)+acetylation (42)=4748 Da).
4. Discussion
Melanocytes are derived from the neural crest and the predominant cells present in lymph nodes (B and T lymphocytes) are derived from the bone marrow, so it is not surprising that the protein signatures from control lymph nodes and melanoma lymph node metastases exhibit drastically different characteristics. Of the 123 protein features observed, 57 displayed over a 2-fold intensity difference between the groups. Most of the differentially expressed proteins were observed at higher intensity in the tumor specimens, with a smaller number of proteins expressed at higher intensity in control lymph nodes (Supplemental Table 1). This is a common feature of the tumor cellular regions and may be the result of a culmination of rapidly dividing cells, increased cellular density in the tumor regions, and proteins that are specifically over-expressed in the tumor cells. The goal of establishing protein classifiers that distinguish melanoma-involved lymph nodes from control lymph nodes is to improve the targeting, diagnosis and stratification of patients with stage III disease.
Several proteins observed in tumor regions are calcium binding proteins, specifically the S100 family of m/z 10,091 (S100A6, Calcyclin), m/z 10,625 (S100B), and m/z 11,653 (S100A11, Calgizzarin) as well as calmodulin (m/z 16,791). Beyond Ca2+ homeostasis, the S100 proteins are involved in numerous intracellular processes, including regulation of protein phosphorylation, cytoskeleton dynamics, cell growth and differentiation, and inflammatory response [56]. Calcyclin interacts with numerous proteins, including S100B, and its expression is observed in many cancers [57]. The presence of these S100 proteins has been previously observed by MALDI IMS as increased in aggressive human gliomas [39] and wound healing mouse tissue [58]. From the survival analysis, we identified m/z 10,091 (S100A6) as an unfavorable marker, a protein previously identified as a potential marker for aggressive melanoma by mRNA analysis of human xenografts in mice [59]. The bootstrap analysis found calmodulin as a favorable prognostic marker for patient recurrence. Calmodulin is ubiquitously expressed in cells, where it binds to and regulates numerous proteins involved in regulator pathways, including protein kinases and phosphatases as well as in cytokinesis [60]. The protein has been implicated as a prognostic marker in lung cancer [61] and shown to be important in cell adhesion of uveal melanoma cells [62], but its specific association in prolonged recurrence is unknown.
The IHC stain for S100 is routinely used as a histologic marker to confirm the diagnosis of melanoma, but the antibody is strongly associated with S100B and less-specific to other various S100 proteins [63]. In analyzing the tissue via MALDI IMS, each of the detected S100 proteins can be monitored individually from a single experiment. This spatial and protein selectivity allows a combination of the observed S100 features to generate a multiplex signature to improve diagnostics by reducing the influence of less melanoma specific members of the S100 family. For example, although S100B, S100 A6, and S100 A11 display consistent, near 5-fold higher intensity in the melanoma regions, the intensity of S100 A8 (m/z 10,745) is identical between groups and the putative S100 A1 peak (m/z 10,458) is absent in control LN and 1/3 of the tumor samples but is clearly seen in the remaining 2/3 of tumors. Each of the S100 proteins is involved in a wide variety of cellular functions [56], and the ability to monitor these individually by MALDI IMS will be advantageous in understanding their biological roles.
The recognition of cytochrome C (CyC, m/z 12,275) as a protein that may correlate with the behavior of melanoma is intriguing. Cytochrome C expression was significantly more intense in melanoma versus control lymph nodes, presumably due to its role in energy production functions in the inner mitochondrial membrane as part of the electron-transport chain [64]. However, increase in m/z 12,275 correlated favorably to longer time to recurrence, consistent with a role in the intrinsic apoptotic pathway. When released into the cytosol, cytochrome C binds with APAF-1 and initiates the caspase cascade that ultimately leads to apoptosis [65]. The association between cytochrome C expression and increased time to recurrence suggests that melanomas with high expression may have an intact apoptotic pathway and thereby be more responsive to therapy or progress more slowly. Cytochrome C apoprotein is nuclear and does not gain a heme group until reaching the mitochondria. Interestingly, the holocytochrome C (containing the heme group, +615 Da) and not the apocytochrome C (lacking heme group) is required to initiate apoptosis. The form identified in the direct tissue analysis is m/z 12,275, which contains the heme group (the apocytochrome C species presumably at m/z 11,660 was not observed, possibly obscured by the S100 A11 peak m/z 11,651). The increase in m/z 12,275 initially suggested a potential functioning apoptotic pathway in patients with increased time to recurrence; however, with IHC staining for activated caspase-3, only a weak correlation was observed between tissue areas high in m/z 12,275 and activated caspase-3 (data not shown). An increased expression of other anti-apoptotic proteins, such as MIF and HSP10, may be suggestive of a more vigorous anti-apoptotic activity.
One of the more unexpected results is the association of cleaved proteins in patients with low survival. Of the 12 proteins related to survival, 3 are modified forms with two C-terminus peptides missing/removed. Each di-peptide set is unique and the cleaved forms were observed at non-uniform ratios to the intact protein. Ubiquitin (Ub, m/z 8565) appears with its cleaved form missing the C-terminus GG (Ub-GG, m/z 8451) and has been observed in several biological processes [66]. Both thymosin β-4 (TYB4, m/z 4964) and thymosin β-10 (TYB10, m/z 4936) were found to have similarly cleaved forms (TYB4-ES, m/z 4748) and (TYB10-IS, m/z 4737). The thymosin β proteins sequester actin monomers within the cell, inactivating polymerization until needed [67]. The full length thymosin β proteins have been routinely observed across species in MALDI tissue analysis [26,35,39,68] and both thymosin β-4 and β-10 have been implicated in the metastatic progression of melanoma [69,70]. The full length thymosin proteins are often over expressed in metastatic cells, where their over expression causes additional actin sequestration, a less rigid cytoskeleton, and allows more motility in the cell [71,72]. To our knowledge, cleaved forms of TYB4 and TYB10 have not previously been associated with metastatic melanoma or other cancer. The modified forms of the thymosin proteins have been observed (TYB10 natively and TYB4 artificially) and in both cases, though the N-terminus is predominantly responsible for actin binding, the truncated C-terminal form causes a small, but significant loss in binding [73,74]. While an increase in full length thymosin proteins corresponds to metastatic cell potential by means of increased actin sequestration, our analysis suggests that a slight reduction in actin binding among metastatic tumors correlates with a more aggressive tumor. This may give the cells increased rigidity thereby promoting their survival in the foreign lymph environment. Intact forms of thymosin β revealed no significant difference between control and tumor groups or in relation to survival/recurrence, and interestingly, neither did the actin associated proteins cofilin-1 (m/z 18,415) or profilin-1 (m/z 14,968).
Histone H4, H2B, and H3 were found to positively correlate with patient survival. Histone H4 is observed as two clearly resolved isoforms of m/z 11,307 and 11,349. Pesavento et al. [61] showed that these are comprised from the removal of the initiator methionine, an N-terminal acetylation, and a dimethyl modification on residue K20 for m/z 11,307 and an additional acetylation on residue K16 for m/z 11,349. The respective isoforms were shown to comprise 69% and 17% of the total H4 forms during the G0/S0 phase. As the cell cycle passed into G2/M phase for cell division, the relative values fell to 59% and 11%, respectively. In relation to our findings, the positive correlation of these two peaks with survival would clearly be in line with a less rapidly dividing tumor. Decreased intensity of the two H4 peaks indicates significant cell growth in the tumor and would thus correlate unfavorably with survival. Overlapping isoforms of various methylation and acetylation PTMs in histone H3 and H2B register as broad unresolved MALDI peaks, with respective widths of 150 Da and 75 Da, significantly wider than the 30 Da widths of proximite hemoglobin peaks. Improved mass resolution to deconvolute these multiple isoforms will be required in understanding the specific role in survival.
Patients with stage III melanoma exhibit a range of treatment response, survival, and disease progression. Virtually all patients diagnosed with stage III disease have a biopsy, but clinicians have few tools to evaluate the tumor to stratify patients for prognosis or determine the optimal treatment. Here we have acquired protein data directly from human tissue to generate molecular classifiers that distinguish melanoma lymph node metastases from control lymph nodes and identified a set of proteins that correlate with patient prognosis. These protein markers present clinicians with information to aid diagnosis and prognosis for stage III melanoma. The initial findings presented here are promising and the study of larger cohorts would be of immense value in understanding this disease process.
Supplementary Material
Acknowledgments
This research was supported by NIH grant (GM58008-09) and DOD grant (W81XWH-05-1-0179). We wish to thank Dr. Frank Harrell and Pengcheng Lu for their contribution of the bootstrap analysis and helpful discussion.
Definitions
- CPH
Cox-proportional hazard model
- CyC
cytochrome C
- H & E
hematoxylin and eosin
- LN
lymph node
- MALDI IMS
matrix-assisted laser desorption/ionization imaging mass spectrometry
- SLN
sentinel lymph node
- TYB
thymosin β
Footnotes
Supplementary materials related to this article can be found online at doi:10.1016/j.jprot.2011.04.013.
References
- 1.Cancer Facts and Figures 2008. American Cancer Society; 2008. [Google Scholar]
- 2.Slominski A, Tobin DJ, Shibahara S, Wortsman J. Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol Rev. 2004;84(4):1155–228. doi: 10.1152/physrev.00044.2003. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi Y, Brenner M, Hearing VJ. The regulation of skin pigmentation. J Biol Chem. 2007;282(38):27557–61. doi: 10.1074/jbc.R700026200. [DOI] [PubMed] [Google Scholar]
- 4.Mendelsohn J. The molecular basis of cancer. 2. Saunders; Philadelphia: 2001. pp. 385–400. [Google Scholar]
- 5.Greene FL American Joint Committee on Cancer, American Cancer Society. AJCC cancer staging handbook : from the AJCC cancer staging manual. 6. New York: Springer; 2002. p. xv.p. 469. [Google Scholar]
- 6.Balch CM, Buzaid AC, Soong SJ, Atkins MB, Cascinelli N, Coit DG, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19(16):3635–48. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 7.Markovic SN, Erickson LA, Rao RD, Weenig RH, Pockaj BA, Bardia A, et al. Malignant melanoma in the 21st century, part 2: staging, prognosis, and treatment. Mayo Clin Proc. 2007;82 (4):490–513. doi: 10.4065/82.4.490. [DOI] [PubMed] [Google Scholar]
- 8.Garbe C, Eigentler TK. Diagnosis and treatment of cutaneous melanoma: state of the art 2006. Melanoma Res. 2007;17(2):117–27. doi: 10.1097/CMR.0b013e328042bb36. [DOI] [PubMed] [Google Scholar]
- 9.Thompson JF, Scolyer RA, Kefford RF. Cutaneous melanoma. Lancet. 2005;365(9460):687–701. doi: 10.1016/S0140-6736(05)17951-3. [DOI] [PubMed] [Google Scholar]
- 10.Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanoma. J Clin Oncol. 2007;25 (12):1606–20. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- 11.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406(6795):536–40. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 12.Davies MA, Samuels Y. Analysis of the genome to personalize therapy for melanoma. Oncogene. 2010;29(41):5545–55. doi: 10.1038/onc.2010.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 14.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445(7130):851–7. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 15.Anichini A, Mortarini R, Sensi M, Zanon M. APAF-1 signaling in human melanoma. Cancer Lett. 2006;238(2):168–79. doi: 10.1016/j.canlet.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 16.Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98(7):472–82. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 17.Jonsson G, Busch C, Knappskog S, Geisler J, Miletic H, Ringner M, et al. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin Cancer Res. 2010;(13):3356–67. doi: 10.1158/1078-0432.CCR-09-2509. [DOI] [PubMed] [Google Scholar]
- 18.Craven RA, Stanley AJ, Hanrahan S, Totty N, Jackson DP, Popescu R, et al. Identification of proteins regulated by interferon-alpha in resistant and sensitive malignant melanoma cell lines. Proteomics. 2004;4(12):3998–4009. doi: 10.1002/pmic.200400870. [DOI] [PubMed] [Google Scholar]
- 19.Sosa MS, Girotti MR, Salvatierra E, Prada F, de Olmo JA, Gallango SJ, et al. Proteomic analysis identified N-cadherin, clusterin, and HSP27 as mediators of SPARC (secreted protein, acidic and rich in cysteines) activity in melanoma cells. Proteomics. 2007;7(22):4123–34. doi: 10.1002/pmic.200700255. [DOI] [PubMed] [Google Scholar]
- 20.Baruthio F, Quadroni M, Ruegg C, Mariotti A. Proteomic analysis of membrane rafts of melanoma cells identifies protein patterns characteristic of the tumor progression stage. Proteomics. 2008;8(22):4733–47. doi: 10.1002/pmic.200800169. [DOI] [PubMed] [Google Scholar]
- 21.Han MJ, Wang H, Beer LA, Tang HY, Herlyn M, Speicher DW. A systems biology analysis of metastatic melanoma using in-depth three-dimensional protein profiling. Proteomics. 2010;10(24):4450–62. doi: 10.1002/pmic.200900549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Findeisen P, Zapatka M, Peccerella T, Matzk H, Neumaier M, Schadendorf D, et al. Serum amyloid A as a prognostic marker in melanoma identified by proteomic profiling. J Clin Oncol. 2009;27(13):2199–208. doi: 10.1200/JCO.2008.18.0554. [DOI] [PubMed] [Google Scholar]
- 23.Wilson LL, Tran L, Morton DL, Hoon DS. Detection of differentially expressed proteins in early-stage melanoma patients using SELDI-TOF mass spectrometry. Ann NY Acad Sci. 2004;1022:317–22. doi: 10.1196/annals.1318.047. [DOI] [PubMed] [Google Scholar]
- 24.Paulitschke V, Kunstfeld R, Mohr T, Slany A, Micksche M, Drach J, et al. Entering a new era of rational biomarker discovery for early detection of melanoma metastases: secretome analysis of associated stroma cells. J Proteome Res. 2009;8(5):2501–10. doi: 10.1021/pr8010827. [DOI] [PubMed] [Google Scholar]
- 25.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 26.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med. 2001;7(4):493–6. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- 27.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal Chem. 2006;78(18):6448–56. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 28.Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Int J Mass Spectrom. 2007;260(2–3):195–202. [Google Scholar]
- 29.Reyzer ML, Hsieh YS, Ng K, Korfmacher WA, Caprioli RM. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 2003;38(10):1081–92. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh Y, Casale R, Fukuda E, Chen JW, Knemeyer I, Wingate J, et al. Matrix-assisted laser desorption/ionization imaging mass spectrometry for direct measurement of clozapine in rat brain tissue. Rapid Commun Mass Spectrom. 2006;20(6):965–72. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- 31.Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J Am Soc Mass Spectrom. 2008;19(6):882–6. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J Mass Spectrom. 2007;42(2):254–62. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- 33.Burnum KE, Tranguch S, Mi D, Daikoku T, Dey SK, Caprioli RM. Imaging mass spectrometry reveals unique protein profiles during embryo implantation. Endocrinology. 2008;149(7):3274–8. doi: 10.1210/en.2008-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierson J, Norris JL, Aerni HR, Svenningsson P, Caprioli RM, Andren PE. Molecular profiling of experimental Parkinson’s disease: direct analysis of peptides and proteins on brain tissue sections by MALDI mass spectrometry. J Proteome Res. 2004;3(2):289–95. doi: 10.1021/pr0499747. [DOI] [PubMed] [Google Scholar]
- 35.Reyzer ML, Caldwell RL, Dugger TC, Forbes JT, Ritter CA, Guix M, et al. Early changes in protein expression detected by mass spectrometry predict tumor response to molecular therapeutics. Cancer Res. 2004;64(24):9093–100. doi: 10.1158/0008-5472.CAN-04-2231. [DOI] [PubMed] [Google Scholar]
- 36.Thibault DB, Gillam CJ, Grey AC, Han J, Schey KL. MALDI tissue profiling of integral membrane proteins from ocular tissues. J Am Soc Mass Spectrom. 2008;19(6):814–22. doi: 10.1016/j.jasms.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caprioli RM. Deciphering protein molecular signatures in cancer tissues to aid in diagnosis, prognosis, and therapy. Cancer Res. 2005;65(23):10642–5. doi: 10.1158/0008-5472.CAN-04-3581. [DOI] [PubMed] [Google Scholar]
- 38.Yanagisawa K, Shyr Y, Xu BJ, Massion PP, Larsen PH, White BC, et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362(9382):433–9. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz SA, Weil RJ, Thompson RC, Shyr Y, Moore JH, Toms SA, et al. Proteomic-based prognosis of brain tumor patients using direct-tissue matrix-assisted laser desorption ionization mass spectrometry. Cancer Res. 2005;65(17):7674–81. doi: 10.1158/0008-5472.CAN-04-3016. [DOI] [PubMed] [Google Scholar]
- 40.Sanders ME, Dias EC, Xu BJ, Mobley JA, Billheimer D, Roder H, et al. Differentiating proteomic biomarkers in breast cancer by laser capture microdissection and MALDI MS. J Proteome Res. 2008;7(4):1500–7. doi: 10.1021/pr7008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cornett DS, Mobley JA, Dias EC, Andersson M, Arteaga CL, Sanders ME, et al. A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Mol Cell Proteomics. 2006;5(10):1975–83. doi: 10.1074/mcp.M600119-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38(7):699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 43.Norris JL, Cornett DS, Mobley JA, Andersson M, Seeley EH, Chaurand P, et al. Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int J Mass Spectrom. 2007;260(2–3):212–21. doi: 10.1016/j.ijms.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox DR, Oakes D. Analysis of survival data. London, New York: Chapman and Hall; 1984. p. viii.p. 201. [Google Scholar]
- 46.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J Proteome Res. 2006;5(11):2889–900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 47.Chaurand P, Fouchecourt S, DaGue BB, Xu BJ, Reyzer ML, Orgebin-Crist MC, et al. Profiling and imaging proteins in the mouse epididymis by imaging mass spectrometry. Proteomics. 2003;3(11):2221–39. doi: 10.1002/pmic.200300474. [DOI] [PubMed] [Google Scholar]
- 48.Oppenheimer SR, Mi D, Sanders ME, Caprioli RM, Caprioli Molecular analysis of tumor margins by MALDI mass spectrometry in renal carcinoma. J Proteome Res. 2010;9(5):2182–90. doi: 10.1021/pr900936z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–52. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 50.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 51.Mitchell RA, Liao H, Chesney J, Fingerle-Rowson G, Baugh J, David J, et al. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc Natl Acad Sci USA. 2002;99(1):345–50. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Czarnecka AM, Campanella C, Zummo G, Cappello F. Mitochondrial chaperones in cancer: from molecular biology to clinical diagnostics. Cancer Biol Ther. 2006;5(7):714–20. doi: 10.4161/cbt.5.7.2975. [DOI] [PubMed] [Google Scholar]
- 53.Nordberg J, Arner ES. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic Biol Med. 2001;31(11):1287–312. doi: 10.1016/s0891-5849(01)00724-9. [DOI] [PubMed] [Google Scholar]
- 54.Liu Z, Schey KL. Optimization of a MALDI TOF–TOF mass spectrometer for intact protein analysis. J Am Soc Mass Spectrom. 2005;16(4):482–90. doi: 10.1016/j.jasms.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z, Schey KL. Fragmentation of multiply-charged intact protein ions using MALDI TOF–TOF mass spectrometry. J Am Soc Mass Spectrom. 2008;19(2):231–8. doi: 10.1016/j.jasms.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. 2003;60(6):540–51. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- 57.Lesniak W, Slomnicki LP, Filipek A. S100A6 — new facts and features. Biochem Biophys Res Commun. 2009;390(4):1087–92. doi: 10.1016/j.bbrc.2009.10.150. [DOI] [PubMed] [Google Scholar]
- 58.Caldwell RL, Opalenik SR, Davidson JM, Caprioli RM, Nanney LB. Tissue profiling MALDI mass spectrometry reveals prominent calcium-binding proteins in the proteome of regenerative MRL mouse wounds. Wound Repair Regen. 2008;16(3):442–9. doi: 10.1111/j.1524-475X.2007.00351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weterman MA, van Muijen GN, Bloemers HP, Ruiter DJ. Expression of calcyclin in human melanocytic lesions. Cancer Res. 1993;53(24):6061–6. [PubMed] [Google Scholar]
- 60.O’Day DH. CaMBOT: profiling and characterizing calmodulin-binding proteins. Cell Signal. 2003;15(4):347–54. doi: 10.1016/s0898-6568(02)00116-x. [DOI] [PubMed] [Google Scholar]
- 61.Xu BJ, Gonzalez AL, Kikuchi T, Yanagisawa K, Massion PP, Wu H, et al. MALDI-MS derived prognostic protein markers for resected non-small cell lung cancer. Proteomics Clin Appl. 2008;2(10–11):1508–17. doi: 10.1002/prca.200800094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagner M, Bielby S, Rennie IG, Mac Neil S. Attachment of human uveal melanocytes and melanoma cells to extracellular matrix proteins involves intracellular calcium and calmodulin. Melanoma Res. 1997;7(6):439–48. doi: 10.1097/00008390-199712000-00001. [DOI] [PubMed] [Google Scholar]
- 63.Zubovits J, Buzney E, Yu L, Duncan LM. HMB-45, S-100, NK1/C3, and MART-1 in metastatic melanoma. Hum Pathol. 2004;35(2):217–23. doi: 10.1016/j.humpath.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 64.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8(5):405–13. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 65.Ow YL, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9(7):532–42. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 66.Herring K. Identification of protein markers of drug-induced nephrotoxicity by maldi ms: in vivo discovery of ubiquitin-t. Nashville: Vanderbilt University; 2009. [Google Scholar]
- 67.Sun HQ, Yin HL. The beta-thymosin enigma. Ann NY Acad Sci. 2007;1112:45–55. doi: 10.1196/annals.1415.021. [DOI] [PubMed] [Google Scholar]
- 68.Laurent C, Levinson DF, Schwartz SA, Harrington PB, Markey SP, Caprioli RM, et al. Direct profiling of the cerebellum by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: a methodological study in postnatal and adult mouse. J Neurosci Res. 2005;81(5):613–21. doi: 10.1002/jnr.20590. [DOI] [PubMed] [Google Scholar]
- 69.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406 (6795):532–5. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 70.Weterman MA, van Muijen GN, Ruiter DJ, Bloemers HP. Thymosin beta-10 expression in melanoma cell lines and melanocytic lesions: a new progression marker for human cutaneous melanoma. Int J Cancer. 1993;53(2):278–84. doi: 10.1002/ijc.2910530218. [DOI] [PubMed] [Google Scholar]
- 71.Nummela P, Yin M, Kielosto M, Leaner V, Birrer MJ, Holtta E. Thymosin beta4 is a determinant of the transformed phenotype and invasiveness of S-adenosylmethionine decarboxylase-transfected fibroblasts. Cancer Res. 2006;66(2):701–12. doi: 10.1158/0008-5472.CAN-05-2421. [DOI] [PubMed] [Google Scholar]
- 72.Liu CR, Ma CS, Ning JY, You JF, Liao SL, Zheng J. Differential thymosin beta 10 expression levels and actin filament organization in tumor cell lines with different metastatic potential. Chin Med J Engl. 2004;117(2):213–8. [PubMed] [Google Scholar]
- 73.Eadie JS, Kim SW, Allen PG, Hutchinson LM, Kantor JD, Zetter BR. C-terminal variations in beta-thymosin family members specify functional differences in actin-binding properties. J Cell Biochem. 2000;77(2):277–87. doi: 10.1002/(sici)1097-4644(20000501)77:2<277::aid-jcb10>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 74.Hannappel E. beta-Thymosins. Ann NY Acad Sci. 2007;1112:21–37. doi: 10.1196/annals.1415.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.