Abstract
The linking together of molecular fragments that bind to adjacent sites on an enzyme can lead to high affinity inhibitors. Ideally, this strategy would employ linkers that do not perturb the optimal binding geometries of the fragments and do not have excessive conformational flexibility that would increase the entropic penalty of binding. In reality, these aims are seldom realized due to limitations in linker chemistry. Here we systematically explore the energetic and structural effects of rigid and flexible linkers on the binding of a fragment-based inhibitor of human uracil DNA glycosylase. Analysis of the free energies of binding in combination with co-crystal structures shows that the flexibility and strain of a given linker can have a significant impact on binding affinity even when the binding fragments are optimally positioned. Such effects are not apparent from inspection of structures and underscore the importance of linker optimization in fragment-based drug discovery efforts.
Over the last decade fragment-based drug discovery has become a well-established approach for identifying lead compounds with pharmacologic activity 1. The emerging success of this approach as compared to high-throughput chemistry and screening tactics relies on several factors. One important aspect is the greater likelihood that a simple molecule will find a complementary binding site on a protein target as compared to a more complex entity where the probability of finding an exact match between the ligand and the target is small 2. Although a small molecule with few interactions would be expected to bind weakly to a target, molecular simplicity allows for the distinct possibility of finding two small molecules that bind to adjacent sites on the target. This outcome allows for covalent tethering of the two “fragments” into a larger compound that under optimal circumstances may take advantage of the combined binding affinity of the two weakly binding pieces. The energetics of this situation are well-known: if the binding affinities of the two fragments are not perturbed during the process of linking them, then their combined binding energies will be realized in the linked compound. Adding to this desirable energetic outcome will be the significant rotational and translational entropy benefit arising from binding a single linked compound, rather than two fragments 3, 4. Despite the potential energetic benefits of this approach, often the linked fragments bind differently than the free fragments, negating realization of the full energetic benefits of tethering. These observations indicate that the tether may be as important as the fragments in designing high affinity ligands for a target.
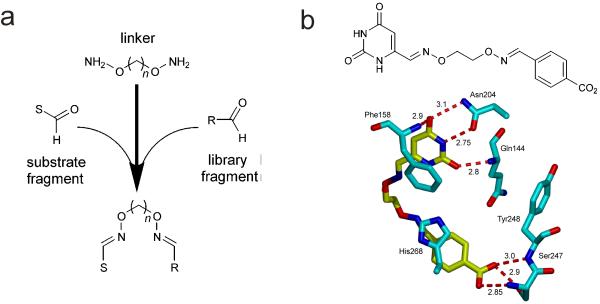
We have been exploring a substrate fragment-based approach for enzyme inhibitor design against several enzymes involved in uracil DNA base excision repair 5-7, which is an important pathway in viral pathogenesis 8, 9, cancer chemotherapy 10, 11 and the development of lymphoid cancers 12-14. The approach relies on using a piece of the full substrate (the substrate fragment) that still binds competitively with the intact substrate to the active site. This substrate fragment can then be modified with a chemical handle to allow its connection via variable length linkers to a library of random molecular fragments. An efficient and economical chemical approach for assembly of substrate-fragment libraries is to use an aldehyde handle on the substrate fragment and bivalent alkyloxyamine linkers to link it to library aldehyde fragments via stable oxime linkages (Fig. 1a) 5, 15. Several small molecule inhibitors of the enzyme human uracil DNA glycosylase (hUNG) with Ki values in the range 0.2 to 10 μM have been discovered by screening linked libraries where 6-formyl uracil was the substrate fragment 5, 7. The structure of one such compound in complex with hUNG has been reported and shows that the uracil fragment binds to the active site with the same aromatic stacking and hydrogen bonding interactions as seen with the isolated uracil fragment (Fig. 1b) 6, and that the 4-carboxybenzaldehyde library fragment π-π stacks with an active site histidine and its carboxylate group mimics a charged hydrogen bonding interaction of a key phosphate in the intact DNA substrate.
Figure 1.
Substrate fragment tethering strategy and application to human uracil DNA glycosylase (hUNG2). (a) The method involves linking a substrate- derived aldehyde fragment to a library of aldehydes using bivalent oxyamine linkers (n = 2 – 6). The tethering reactions are performed in high-throughput and high-yield (>90%) using 96-well plates 5-7. Without the need for purification, the libraries are directly screened against a desired enzyme target to rapidly identify inhibitors. (b) Substrate fragment tethering using 6-formyl uracil (11) as the substrate fragment yielded the first small molecule inhibitor of the DNA repair enzyme hUNG2 (13, KD = 6 μM). The interactions of the uracil and library fragments of dioxime 13 with hUNG2 are shown (PDB ID 2HXM). The tether does not directly interact with the enzyme and has an unusual kinked conformation (see text).
The advantages of substrate fragment linking are nontrivial. First, the substrate fragment targets the linked compound to the active site and serves as a molecular anchor for binding of the library fragment, which in the absence of the uracil anchor typically binds very weakly (Ki ~ 10 mM) 5. Thus, linking to the uracil substrate fragment allows the detection of library fragments whose binding would be difficult or impossible to detect otherwise. Second, it is common that useful binding pockets are found adjacent to active sites in enzymes that bind polymeric substrates, or for those that utilize organic cofactors. Thus, the probability of finding a useful binding pocket for the library fragment is increased because a fertile region is being mined.
What is the energetic impact of the linker shown in Figure 1b, and more generally, for other fragment-based ligands? By inspection of the structure in Figure 1b, the linker shows no apparent interactions with the enzyme target, and the uracil base is presented in the active site in an indistinguishable binding mode as compared to the complex with the free base 16. However, the linker has an unusual conformation suggesting the possibility of linker strain. Another indication of strain is that the binding free energy of the tethered inhibitor is +0.4 kcal/mol less favorable as compared to adding the binding free energies of the component fragments. These observations led us to systematically explore the impact of linker strain and flexibility on the binding energetics in this system. We found that even apparently inert linkers have significant energetic effects on fragment binding that are not predictable from inspection of structures, and that linker optimization should be an integral part of any fragment-based drug discovery project.
RESULTS
Linker design and properties
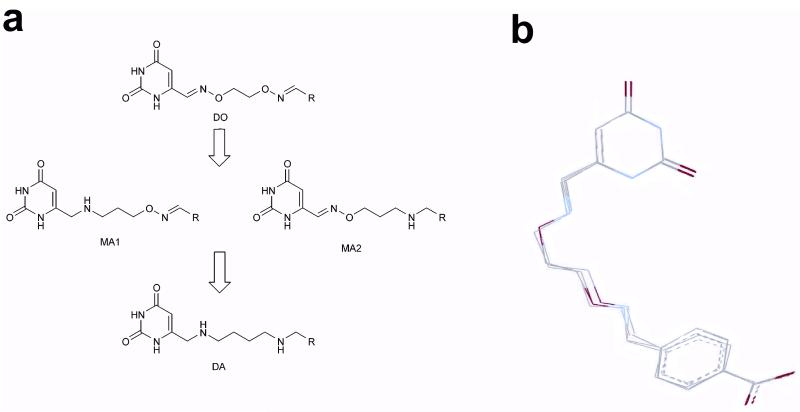
The original substrate fragment tethering methodology employed bis-aminooxyalkane linkers to generate tethered libraries where the uracil substrate fragment (U) and library binding fragments (R) were connected by linkers containing two rigid oxime linkages as shown in Figure 2a (dioximes, DO). As introduced above, structural characterization of the complex between human UNG and one of the more potent inhibitors revealed an unusual conformation of the linker (Fig. 1b), suggesting the presence of strain in the bound ligand. We inferred that a portion of UNG’s binding energy for this inhibitor is used to pay the energetic cost of driving the inhibitor into this unfavorable bound conformation, and that other linkers with different connective properties might avoid this penalty.
Figure 2.
Diversification of rigid bivalent oxime linkers into flexible monoamine and diamine linkers. (a) The rigid and planar sp2 centers of the dioxime (DO) linkers can be systematically converted into more flexible sp3 linkages using amine chemistry. Thus, the original uracil fragment libraries can be transformed into three different amine libraries. The monoamine (MA) libraries have a flexible sp3 amine center at the uracil end of the tether (MA1), or the diversified end of the tether (MA2). The diamine (DA) library has flexible amine centers at both ends of the tether. (b) The oxime and amine linkers can present the uracil and 4-carboxybenzaldehyde (29) binding fragments in the observed productive binding mode shown in Figure 1b. MMF3 molecular mechanics computations were used to superimpose the corresponding MA1, MA2 and DA linker versions with the crystallographically determined bound conformation of 13. In this computation, the uracil and carboxylate atoms of each compound were superimposed and frozen while the linkers were allowed to equilibrate.
To explore connectivity effects in this system, we conceived of a series of linkers where the two rigid oxime linkages of the inhibitor were systematically changed to more flexible amine linkages (Fig. 2a). Thus, the original compound with a DO linker could be transformed into three distinct analogues with an amine linkage on either the uracil side of the linker (monoamine MA1), the library fragment side (monoamine MA2), or both sides (diamine DA). These three linker analogues are superimposable with the bound conformation of the original inhibitor that possessed two oxime linkages, demonstrating that each is capable of forming the same binding interactions (Fig. 2b). Thus, any differences in binding affinities for compounds containing these linkers should reflect the inherent conformational properties of the linker, and how well the linker serves to connect the two interacting fragments.
Synthesis of uracil libraries with flexible amine linkers
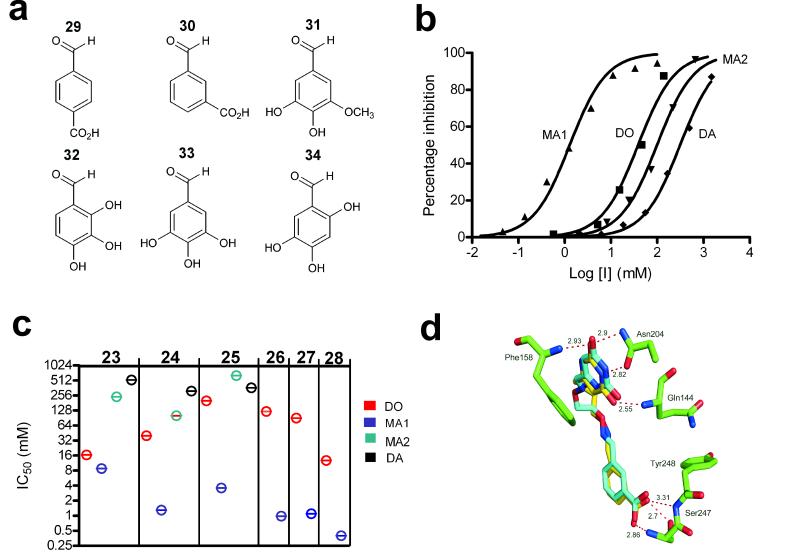
Our previous library screening efforts using 6-formyluracil as the substrate fragment uncovered six aldehyde fragments from a 215 member library that were inhibitory in the context of an oxime linker containing two methylene groups (Fig. 3a) 5, 6. Two of the six library aldehydes were stable aromatic acids, while the remaining four were air sensitive di- or trihydroxybenzaldehydes that were unsuitable for cell culture or structural studies (Fig. 3a). To experimentally examine the impact of linker flexibility in this system, the MA1, DO, MA2 and DA constructs were synthesized as shown in Scheme 1a-d using these six library aldehydes. Briefly, to obtain a linked inhibitor with an amine linkage on the uracil side of the linker, 6-(chloromethyl)uracil (1) was first reacted with the Boc-protected propylamine (2) to give 3, which reacts cleanly with acetyl chloride in cold methanol to give the Boc-deprotected uracil synthon 4 (Scheme 1a). Oxyamine functionality of 4 was then coupled to aldehydes 29 – 34 shown in Figure 3a to give the MA1 series 5 – 10, respectively. The DO derivatives were obtained as previously described from the reaction of O,O’-diaminoethanediol (12) with 6-formyluracil and aldehydes 29 – 34 to give 13 - 18, respectively (Scheme 1b) 5, 6. To obtain MA2 derivatives with an amine linkage on the diversified side of the tether, 6-formyluracil (11) was first reacted with O-(3-aminopropyl)-hydroxylamine (19) to give uracil synthon 20 (Scheme 1c). The three MA2 compounds were synthesized by reaction of 20 with aldehydes 29 – 34 in the presence of NaBH3CN to give monoamines 21, 22 and 23, respectively. Finally, three DA compounds were obtained by first reacting 6-(chloromethyl)uracil (1) with 1,4-diaminobutane (24) to yield 25 (Scheme 1d); the free primary amine of 25 was then coupled to aldehydes 29 – 34 in the presence of NaBH3CN to give diamines 26, 27 and 28, respectively.
Figure 3.
Structures and inhibition profiles of library aldehyde fragments containing DO, MA1, MA2 and DA linkers. (a) Structure of library aldehydes used in the synthesis of DO, MA1, MA2 and DA libraries. (b) Concentration dependence of hUNG2 inhibition by the amine and oxime linker compounds generated from library aldehyde 30. The IC50 values were DO (14) = 40 μM; MA1 (6) = 1.3 μM; MA2 (22) = 100 μM; DA (27) = 315 μM (c) IC50 values for library aldehyde fragments 29 – 34 in the context of DO, MA1, MA2 and DA linkages. For aldehyde fragments 32 – 34, only DO and MA1 linkers were tested. Experiments were repeated in triplicate and errors are standard deviations of the data from the fitted curve. (d) Conformations and interactions of the bound DO (14) and MA1 (6) inhibitors derived from library aldehyde 30 (see Supplemental Figure 1 for electron density map of the complex with 6). The structures of MA2 (22) and DA (27) revealed that, for these compounds, fragment 30 did not interact with its binding site (Supplemental Figure 2 online).
Scheme 1.

Synthesis of rigid and flexible oxime and amine substrate fragment libraries. (a) Monoamine 1 (MA1) library. Compounds 5 - 10 were constructed using the six aldehydes 29 - 34 shown in Figure 3a. (b) Dioxime (DO) library. Compounds 13 - 18 were constructed using the six aldehydes 29 - 34 shown in Figure 3a. (c) Monoamine 2 (MA2) library. Compounds 21 - 23 were constructed using the three aldehydes 29 - 31 shown in Figure 3a. (c) Diamine (DA) library. Compounds 26 – 28 were constructed using the three aldehydes 29 - 31 shown in Figure 3a. DMSO, dimethylsulfoxide; TEA, triethylamine; DMF, dimethylformamide; AcCl, acetyl chloride.
Binding affinities of linked fragments
The various linked fragments in the context of DO, MA1, MA2 and DA linkers were screened for their inhibition of human UNG using a high-throughput fluorescent molecular beacon DNA substrate as previously described 5. Screening was performed using a concentration of the substrate 4-fold less than its Km value, and therefore, the inhibitor IC50 values are only 25 % greater than their true Ki values. Representative inhibition curves for library aldehyde 30 in the context of the four linkers are shown in Figure 3b. IC50 values in the range 1 to 300 μM are observed, with MA1 showing the highest affinity followed by the DO, MA2 and DA forms. For comparison, the uracil fragment binds with a Ki of 700 μM, and fragment 30 binds with an estimated Ki of 10 mM (see below and Supplemental Methods online). Thus, the high KD values for the MA2 and DA constructs, which approximate the uracil fragment alone, suggest that fragment 30 is no longer contributing to binding. In contrast, the best analogue (MA1) has a binding free energy of −8.4 kcal/mol which is 1.3 kcal/mol more negative than expected from the additive binding energies of the uracil and 30 fragments (ΔGadd = −7.1 kcal/mol), but much less than that expected if losses in translational and rotational entropy for binding one bivalent ligand as opposed to two separate fragments are considered 3, 4. The binding of MA1 derived from fragment 30 is largely enthalpy driven as determined by titration calorimetric measurements (ΔG = −7.9 kcal/mol, ΔH = −11.6 kcal/mol, ΔS = −0.012 kcal/mol·K) (Supplementary Figure 1 online; similar measurements with weaker binding compounds were not possible due to solubility limitations). The same trend of binding affinities was observed for library aldehyde fragments 29 and 31 in the context of DO, MA1, MA2 and DA linkers (Fig. 3c), indicating that the linker effects are common to this entire series. To further confirm the generality of these linker trends, we went on to measure the IC50 values for library aldehydes 32 - 34 in the context of the DO and MA1 linkers. Once again, the DO constructs bound 30 to 120 times more weakly than the corresponding MA1 construct (Fig. 3c).
Binding affinity of the individual fragments
To establish that the oxime and amine linker effects reflect how well the linkers present the linked fragments, rather than differential interactions of the two linkers with the enzyme, we synthesized the amine or oxime linker forms that contained only the uracil or benzoate (30) fragment in isolation (see uracil compounds 4 and 20 in Scheme 1, and the corresponding benzoate compounds 35 and 36 in Supplementary Schemes 1 and 2). The uracil amine 4 had a Ki = 700 ± 100 μM, which was similar to that of uracil oxime 20 (Ki = 750 ± 100 μM), making it unlikely that the greater binding affinity of MA1 over MA2 arose from more favorable binding interactions of the enzyme with the amine linkage of MA1 as compared to the oxime functionality of MA2 (Supplementary Figure 2 online). Similarly, the greater binding affinity of MA1 over MA2 cannot be attributed to differential interactions of enzyme with the linkage on the benzoate side because the binding affinity of the benzoate fragment was similar when it was attached via an amine or oxime linkage (3.5 mM concentrations of compounds 35 and 36 inhibited the UNG activity by 18 % and 25 %, respectively) (Supplementary Methods online). Taken together, these findings strongly indicate that the linker plays an important energetic role only when the uracil and benzoate fragments are linked together.
Structures of inhibitor complexes
To gain a better understanding of these linker effects we obtained crystal structures of hUNG bound to the DO, MA1, MA2 and DA analogues 14, 6, 22 and 27, all of which are derived from aldehyde fragment 30. These compounds differ from the previously characterized inhibitor shown in Figure 1B in that 30 has the carboxylate group positioned at the 3-position rather than the 4-position of the benzaldehyde ring. The four complexes were obtained by cocrystallization, and diffraction data were collected to 1.3 Å resolution and refined to Rfactor and Rfree values in the range 0.17 to 0.20 and 0.21 to 0.25, depending on the complex (PDB ID 3FCF, 3FCI, 3FCK, 3FCL). The structures of the tighter binding DO and MA1 complexes are shown in superposition in Figure 3d, and a representative electron density map of MA1 is shown in Supplemental Figure 3 online. Comparison of these two structures with those of the complexes of DA and MA2 (Supplemental Figure 4 online), leads to the conclusion that only the DO and MA1 forms have both the uracil and fragment 30 docked in their respective binding pockets. MA2 shows no electron density for the linker or fragment 30, while DA has its linker directed away from the surface of UNG such that fragment 30 interacts adventitiously with another UNG molecule in the unit cell (Supplemental Figure 4 online). These structural observations are fully consistent with the binding measurements where the MA2 and DA analogues bound with IC50 values approximating the uracil fragment alone and indicate that the linkers in the MA2 and DA constructs have suboptimal connectivity properties that negate binding of the library element.
The discrete binding interactions of UNG with the two halves of the DO and MA1 analogues are essentially identical (Fig. 3d), but the linker of DO assumes the same kinked conformation similar to that previously observed (see Figure 1b). On the uracil side of the linker, stacking interactions with Phe158 are observed, and short hydrogen bonds from the uracil donor and acceptor groups to residues Asn204 and Gln144 are common to both the DO and MA1 forms. On the library side of the linker, the carboxylate groups of DO and MA1 form identical tridentate hydrogen bonding interactions with the backbone amide groups of Ser247 and Tyr248 and the γ hydroxyl of Ser247, all of which bind to an important phosphate group in the DNA substrate 6. Despite nearly identical positioning of the uracil and 30 binding fragments, the linkers of MA1 and DO follow different trajectories. The DO linker assumes a kinked and apparently strained conformation, while the linker conformation of MA1 is unremarkable.
DISCUSSION
The binding studies and the four structures suggest that a flexible amine linkage on the library fragment side of the tether (as in the MA2 and DA analogues) negates binding of the library fragment irregardless of whether an amine or oxime linkage is present on the uracil side of the tether. Conversely, introducing an amine linkage on the uracil side of the tether (i.e. MA1) enhances binding only when an oxime linkage is present on the library side, and cannot rescue the binding deficit brought about by an amine linkage on the library side of the tether. These intriguing positional effects of the oxime and amine linkages provide unambiguous evidence that the tether is not a passive medium for presenting the binding fragments.
The enzyme-ligand system used here and the extensive thermodynamic and structural data provide a favorable opportunity to dissect the energetic contributions of the linker in a fragment-based ligand. First, the linkers do not interact directly with the target, and thus, any observed differences in binding affinities cannot be trivially attributed to such interactions. Importantly, the linkers derived from the various combinations of amine and oxime linkages are each capable of presenting the binding fragments to their respective sites (Fig. 2b), requiring that any observed differences in the binding affinities must be related to the conformational preferences of the given linker, its internal flexibility or linker strain. In addition, these linkers are solvent exposed in the bound state which makes it unlikely that observed differences in binding affinity might be derived from differences in solvation of the various linker forms upon binding. However, despite solvent exposure of the linkers in the free and bound states, it should be noted that the amine linkages are protonated under the conditions of the binding reactions based on the pKa = 9.8 for N-methyl-benzyl amine 17, whereas the oxime linkages are neutral. The electrostatic differences between these linkages might give rise to differential effects on binding, but this possibility is unlikely because (i) the binding affinities of the individual fragments are similar regardless of whether an amine or oxime linkage is employed (see Results), and (ii) there is no correlation between binding affinity of the linked fragments and whether an amine or oxime linkage is present on the uracil side of the linker (Figs. 3 and 4). Taken together, these results indicate that the linker effects arise from how well the linkers present the two fragments to their sites and not electrostatic effects. Finally, the effects of introducing flexible or rigid linkages on each side of the linker can be explored in a combinatorial fashion. This aspect is particularly informative because position dependent effects of the two linkage types, and their energetic communication across the tether, provide an opportunity to deconvolute connectivity effects and understand how linkers with greater internal flexibility influence binding of each individual fragment.
Figure 4.

Free energy changes (ΔΔG) arising from switching between flexible amine and rigid oxime linkages that connect the uracil and benzoic acid (30) binding fragments. Difference free energies are in kcal/mol relative to the DA (27) compound. The individual NH linkages that are changed when switching from DA (27) to MA1 (6), DO (14), or MA2 (22) are numbered as indicated (see text for further details).
The differences in binding free energies between DA, DO, MA2 and MA1 can be most reasonably attributed to two effects: linker strain and favorable freezing of bond rotations when the two amine linkages (1NH and 2NH) are switched to rigid oximes (1ox and 2ox ). Using this framework of linker strain and/or entropy freezing, the free energy diagram shown in Figure 4 can be rationalized. Using DA as the reference state, compound MA2 is generated by the single switch 1NH → 1ox, resulting in a −0.6 kcal/mol enhancement in binding as compared to DA. This 1NH → 1ox switch does not result in binding of fragment 30 to its site, but does freeze rotation of a single bond on the uracil side of tether, negating a potentially unfavorable rotational entropy decrease of the 1NH linkage upon DA binding. The energetic effect of the 1NH → 1ox switch is similar to estimates of −TΔSrot in the range 0.4 to 1 kcal/mol for freezing of single rotatable bonds 18-20. The next compound, DO, is generated by switching the second amine linkage to an oxime (2NH → 2ox), which results in binding of 30 to its site (Fig. 4). Thus, freezing rotations of the linker at a critical position (2NH → 2ox) has allowed realization of the free energy benefit of docking fragment 30 in its binding site (ΔΔG(2NH → 2ox ) = −1.2 kcal/mol). Finally, the highest affinity ligand MA1 is generated from DA by the single switch 2NH → 2ox, which also results in binding of 30 to its site, but with a free energy change that is −2.0 kcal/mol more negative than DO. One potential basis for the enhanced binding of MA1 is that the 1NH linkage relieves the linker strain present in DO (Fig. 3d). However, even though DO and MA1 position the uracil and 30 fragments in an indistinguishable way, it cannot be excluded that part of the enhanced binding affinity of MA1 arises from better positioning of the binding fragments (Fig. 4). In summary, MA1 binds most tightly because the 1NH linkage reduces linker strain leading to optimal positioning of the binding fragments, and the rigid 2ox linkage lowers the single bond rotational entropy at the position where it can have the largest effect on the binding of fragment 30.
These intriguing positional effects of the oxime and amine linkages suggest that binding of a loosely interacting fragment such as 30 will be more highly sensitive to small alterations in linker flexibility than a tighter interacting substrate fragment such as uracil. The substrate fragment has sufficient interactions with its site to remain bound even in the presence of linker strain. In contrast, the binding energy for the library fragment may not be sufficient to overcome a suboptimal linker that has excessive flexibility or strain. In this regard, the uracil fragment serves as a molecular anchor for docking fragment 30 in its site, and the flexibility and strain properties of the linker dictate how much of the binding energy of 30 can be realized.
The ligands explored here are remarkably ordinary in their molecular properties and are also drug like, suggesting that the findings may be general to fragment-based ligand design. The most sobering conclusion is that it is impossible to predict, even with high-resolution structures in hand, how a linker will affect binding of two fragments. Thus, hidden strain and other energetic penalties can only be discovered by iterative optimization and binding measurements. Many fragment based ligands are suboptimal binders because they do not even reach the expectation of additive binding energies of the fragment pieces, let alone the additional expected entropic benefit arising from binding a single tethered molecule as opposed to two fragments see (see examples in references)4, 21. Just one notable example is the widely used immunosuppressant drug FK506 which binds about 3 kcal/mol less tightly than expected from simply summing the binding free energies of its component fragments 22. Although this binding deficit with FK506 cannot necessarily be attributed to the linker without further experimentation, it is clear that even extremely useful fragment based drugs have not reached the theoretical potencies that would be expected, exemplifying the potential for further improvement.
It has been noted that some high-affinity enzyme inhibitors that were not discovered by fragment tethering cannot be parsed into their component fragments that recapitulate the binding modes of the parent linked compound 23. This is strong evidence for nonadditive (cooperative) binding effects in the linked compound that must be in place before one or both fragments can bind to their sites. The significant implication is that such high affinity ligands would be missed in a fragment-based discovery approach because neither fragment has sufficient binding energy for its site to overcome the large rotational and translational entropy losses that occur upon binding of a small molecule to a protein 24, 25. In contrast, substrate fragment tethering has the potential to detect such weak binding fragments because screening is performed while library fragments are already linked to the substrate fragment anchor. Thus, an expanded region of chemical space is explored as compared to screening individual fragments. Nevertheless, the current results also demonstrate how the internal entropy or strain of a tether can also be sufficient to completely negate binding of a tethered fragment that is capable of providing significant binding energy if it were presented properly.
METHODS
General
All chemicals were purchased from commercial sources without further purification. The 1H, 13C-NMR spectra were recorded on a 400 MHz Innova instrument (Varian). The spectra were recorded in deuteriochloroform (CDCl3), hexadeuteriodimethyl sulfoxide (DMSO-d6) or deuterium oxide (D2O). The chemical shifts of protons are given in ppm with TMS as internal standard. The chemical shifts of carbons are obtained in ppm with solvents as internal standards. All compounds assayed were purified by HPLC using aqueous triethylammonium acetate (TEAA) as a running buffer. Therefore, TEAA was not completely removed and it appeared in the NMR spectra. Accordingly, proton and carbon chemical shifts of TEAA were not listed during the characterizations of the compounds. During the purification of catechols, 2-mercaptoethanol (20 mM) was used as an antioxidant. Flash chromatography was carried out with silica (70-230 mesh from Sorbent Technologies) and monitored by thin-layer chromatography (TLC) with silica plates (Merck, Kieselgel 60 F254).
6-[3, N-(tert-Butoxycarbonylaminooxypropylamino)methyl]-uracil (3)
To a solution of N-(3-aminopropoxy)-O-tert-butylcarbamate (0.7 g, 3.7 mmol) in MeOH was added 6-(chloromethyl)uracil (1, 0.3 g, 1.9 mmol). The reaction mixture was refluxed for 48 hours. The precipitate was filtered and the filtrate was then evaporated in vacuo. The residue was purified by column chromatography (CH2Cl2 : MeOH = 20 : 1) to give 3 (0.4 g, 68%) as a white solid. 1H NMR (400 MHz, DMSO-d6): δ 9.91 (br, 1H), 5.45 (s, 1H), 3.70 (t, 2H), 2.48 (t, 2H), 1.62 (qu, 2H); 13C NMR (400 MHz, DMSO-d6): δ 164.9, 156.8, 156.2, 152.3, 98.2, 80.2, 74.3, 49.4, 45.7, 28.7; HRMS (m/z) [M+H]+ calcd for C13H23N4O5 315.1663, found 315.1661.
6-[3-(Aminooxypropylamino)-methyl]-uracil, dihydrochloride (4)
To a solution 3 of (0.4 g, 1.3 mmol) in MeOH was added dropwise acetyl chloride (280 μL, 3.9 mmol) at 0°C. The reaction mixture was stirred at ambient temperature for 4 hrs. The product was precipitated and filtered to give 4 (0.3 g, 80%) as a white solid. 1H NMR (400 MHz, DMSO-d6): δ 11.2 (s, 1H), 11.1 (s, 1H), 10.6 (br, 1H), 5.80 (s, 1H), 4.10 (t, 2H), 3.92 (s, 2H), 3.00 (t, 2H), 2.02 (qu, 2H); 13C NMR (400 MHz, DMSO-d6): δ 164.3, 151.6, 147.3, 102.2, 71.8, 46.3, 44.3, 24.7; HRMS (m/z) [M+H]+ calcd for C8H15N4O3 215.1139, found 215.1139.
3-[(3-Amino-propoxyimino)-methyl]-uracil, hydrochloride (20)
To a solution of 6-formyluracil (11, 96 mg, 0.6 mmol) in DMF were added O-(3-aminopropyl)- hydroxylamine dihydrochloride (19, 100 mg, 0.6 mmol) in H2O. The solution was stirred at 50°C for 4 h. The solvents were evaporated in vacuo and the residue was triturated with MeOH. The resulting precipitate was collected by filtration and washed with MeOH to give 20 (120 mg, 80%) as a brown solid. 1H NMR (400 MHz, DMSO-d6): 11.2 (s, 1H), 10.8 (s, 1H), 8.05 (br, 2H), 7.95 (s, 1H), 5.78 (s, 1H), 4.23 (t, 2H), 2.83 (m, 2H), 1.98 (qu, 2H); 13C NMR (400 MHz, DMSO-d6): δ 164.5, 151.7, 145.1, 143.1, 102.3, 72.4, 36.5, 27.5; HRMS (m/z) [M+H]+ calcd for C8H15N4O3 215.1139, found 215.1139.
6-[4-Aminobutyl)aminomethyl]uracil (25)
To a solution of 1,4-diaminobutane (24, 2 mL, 20 mmol) was added 6-(chloromethyl)uracil (1, 320 mg, 2 mmol) in portions with vigorous stirring at 60°C. After stirring for 30 min, excess 24 was evaporated in vacuo and the residue was triturated and recrystallized twice with 2-propanol to give 25 (150 mg, 35%) as a light brown hygroscopic solid. 1H NMR (400 MHz, DMSO-d6): δ 5.43 (s, 1H), 3.32 (s, 2H), 2.62 (t, 2H), 2.40 (t, 2H), 1.42 (m, 4H); 13C NMR (400 MHz, DMSO-d6): δ 165.0, 156.4, 152.4, 98.2, 49.5, 48.4, 28.7, 27.3, 26.2; HRMS (m/z) [M+H]+ calcd for C9H17N4O2 213.1346, found 213.1346.
Synthesis of MA1 library
To each of six 0.5 mL wells of a microtiter plate was added 15 μL of 4 (20 mM) in DMSO. To each well was added 15 μL of an individual aldehyde 29 – 34 (20 mM) in DMSO. The plate was carefully agitated to make the solution homogenous, and then sealed and incubated at room temperature for 12 h. The yield and purity of the products were >95% based on proton NMR spectra (see Supplemental methods online for spectroscopic characterization of compounds).
Synthesis of MA2 library
To a solution of 20 (0.3 mmol) in MeOH was added aldehyde 29, 30 or 31 (0.6 mmol) in MeOH. The mixture was stood at 50 °C for 5 h. Either NaBH3CN (for the syntheses of 21 and 22) or NaBH4 (for the synthesis of 23) was added to the mixture. After 30 min, the reaction was quenched with 1N HCl. The desired product was purified by direct injection of the reaction mixture onto an Aqua reversed phase C-18 HPLC column (Phenomenex, 250 mm, 10 mm, 5 μm) using gradient elution from 0 to 65% CH3CN in 0.1 M aqueous TEAA buffer over the course of 1 h using UV detection at 320 nm (see Supplemental methods online for spectroscopic characterization of compounds).
Synthesis of DA library
The procedure of synthesis and purification was identical to that described for the MA2 library except that the library was constructed using the uracil synthon 25 and TEA was not added in the reaction mixture (see Supplemental methods online for spectroscopic characterization of compounds).
Synthesis of DO library
The DO compounds were synthesized as described previously with minor modification 5-7. Briefly, to solutions of containing individual aldehydes 29 - 34 (0.20 mmol each) and 6-formyluracil (11, 0.20 mmol) in DMSO were added O,O’-diaminoethanediol (12, 0.20 mmol) in DMSO, and the mixtures were incubated at 37°C for 12 h. The desired compounds were purified by direct injection onto an Aqua reversed phase C-18 HPLC column (Phenomenex, 250 mm, 10 mm, 5 μm) using gradient elution from 0 to 65% CH3CN in 0.1 M aqueous TEAA buffer (pH 7.0) over the course of 1 h using UV detection at 320 nm. The purified compounds were precipitated using ice-cold water, centrifuged, washed twice with ice-cold water and dried in vacuo (see Supplemental methods online for spectroscopic characterization of compounds).
The four new structures have been deposited with PDB codes 3FCF, 3FCI, 3FCK, and 3FCL.
Supplementary Material
Acknowledgement
This work was supported by NIH grant GM56834-12 to J.T.S and GM066895 to L.M.A and a Ruth L. Kirschstein National Research Service Award F31 GM083623 to J.B.P. The content of the publication does not necessarily reflect the views or Policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Note: Supplemental information and chemical compound information is available on the Nature Chemical Biology website.
Author contributions: S.C., performed chemical syntheses, binding measurements and energetic analysis; J.B.P. purified protein samples, crystallized the ligand-protein complexes and performed isothermal titration calorimetry; M.B. collected and analyzed X-ray diffraction data; L.M.A. contributed to data analysis and interpretation; J.T.S, analyzed the data and wrote the paper.
REFERENCES
- 1.Erlanson DA, Wells JA, Braisted AC. Tethering: fragment-based drug discovery. Annu. Rev. Biophys. Biomol. Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 2.Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. J. Chem. Inf. Comput. Sci. 2001;41:856–864. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- 3.Jencks WP. On the attribution and additivity of binding energies. Proc. Natl. Acad. Sci. U. S. A. 1981;78:4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray CW, Verdonk ML. The consequences of translational and rotational entropy lost by small molecules on binding to proteins. J. Comput. Aided Mol. Des. 2002;16:741–753. doi: 10.1023/a:1022446720849. [DOI] [PubMed] [Google Scholar]
- 5.Jiang YL, Krosky DJ, Seiple L, Stivers JT. Uracil-directed ligand tethering: an efficient strategy for uracil DNA glycosylase (UNG) inhibitor development. J. Am. Chem. Soc. 2005;127:17412–17420. doi: 10.1021/ja055846n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krosky DJ, et al. Mimicking damaged DNA with a small molecule inhibitor of human UNG2. Nucleic Acids Res. 2006;34:5872–5879. doi: 10.1093/nar/gkl747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang YL, Chung S, Krosky DJ, Stivers JT. Synthesis and high-throughput evaluation of triskelion uracil libraries for inhibition of human dUTPase and UNG2. Bioorg. Med. Chem. 2006;14:5666–5672. doi: 10.1016/j.bmc.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 8.Priet S, Sire J, Querat G. Uracils as a cellular weapon against viruses and mechanisms of viral escape. Curr HIV Res. 2006;4:31–42. doi: 10.2174/157016206775197673. [DOI] [PubMed] [Google Scholar]
- 9.Fleischmann J, Kremmer E, Greenspan JS, Grasser FA, Niedobitek G. Expression of viral and human dUTPase in Epstein-Barr virus-associated diseases. J. Med. Virol. 2002;68:568–573. doi: 10.1002/jmv.10234. [DOI] [PubMed] [Google Scholar]
- 10.Seiple L, Jaruga P, Dizdaroglu M, Stivers JT. Linking uracil base excision repair and 5-fluorouracil toxicity in yeast. Nucleic Acids Res. 2006;34:140–151. doi: 10.1093/nar/gkj430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischmann J, et al. Expression of deoxyuridine triphosphatase (dUTPase) in colorectal tumours. Int. J. Cancer. 1999;84:614–617. doi: 10.1002/(sici)1097-0215(19991222)84:6<614::aid-ijc13>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 12.Hagen L, et al. Genomic uracil and human disease. Exp. Cell Res. 2006;312:2666–2672. doi: 10.1016/j.yexcr.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Pasqualucci L, et al. Expression of the AID Protein in normal and neoplastic B-cells. Blood. 2004;104:3318–25. doi: 10.1182/blood-2004-04-1558. [DOI] [PubMed] [Google Scholar]
- 14.Ramiro AR, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–9. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maly DJ, Choong IC, Ellman JA. Combinatorial target-guided ligand assembly: identification of potent subtype-selective c-Src inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2419–224. doi: 10.1073/pnas.97.6.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao G, et al. Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited. Proteins. 1999;35:13–24. [PubMed] [Google Scholar]
- 17.Baltas M, et al. Aminolysis of sulfinamoyl-esters, -sulfonamides and -sulfones. Thiooxamate and thiourea formation via a suifine intermediate. Thiophilic or carbophilic reaction? Tetrahedron. 1996;52:14865–14876. [Google Scholar]
- 18.Page MI, Jencks WP. Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc. Natl. Acad. Sci. U. S. A. 1971;68:1678–1683. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohm HJ. The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J. Comput. Aided Mol. Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 20.Bohm HJ. Prediction of binding constants of protein ligands: a fast method for the prioritization of hits obtained from de novo design or 3D database search programs. J. Comput. Aided Mol. Des. 1998;12:309–323. doi: 10.1023/a:1007999920146. [DOI] [PubMed] [Google Scholar]
- 21.Erlanson DA. Fragment-based lead discovery: a chemical update. Curr. Opin. Biotechnol. 2006;17:643–652. doi: 10.1016/j.copbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 23.Babaoglu K, Shoichet BK. Deconstructing fragment-based inhibitor discovery. Nat. Chem. Biol. 2006;2:720–723. doi: 10.1038/nchembio831. [DOI] [PubMed] [Google Scholar]
- 24.Finkelstein AV, Janin J. The price of lost freedom: entropy of bimolecular complex formation. Protein Eng. 1989;3:1–3. doi: 10.1093/protein/3.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Gilson MK, Given JA, Bush BL, McCammon JA. The statistical-thermodynamic basis for computation of binding affinities: a critical review. Biophys. J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.