Abstract
OBJECTIVE
Obesity is characterized by chronic oxidative stress. Fibroblast growth factor 21 (FGF21) has recently been identified as a novel hormone that regulates metabolism. NFE2-related factor 2 (Nrf2) is a transcription factor that orchestrates the expression of a battery of antioxidant and detoxification genes under both basal and stress conditions. The current study investigated the role of Nrf2 in a mouse model of long-term high-fat diet (HFD)-induced obesity and characterized its crosstalk to FGF21 in this process.
RESEARCH DESIGN AND METHODS
Wild-type (WT) and Nrf2 knockout (Nrf2-KO) mice were fed an HFD for 180 days. During this period, food consumption and body weights were measured. Glucose metabolism was assessed by an intraperitoneal glucose tolerance test and intraperitoneal insulin tolerance test. Total RNA was prepared from liver and adipose tissue and was used for quantitative real-time RT-PCR. Fasting plasma was collected and analyzed for blood chemistries. The ST-2 cell line was used for transfection studies.
RESULTS
Nrf2-KO mice were partially protected from HFD-induced obesity and developed a less insulin-resistant phenotype. Importantly, Nrf2-KO mice had higher plasma FGF21 levels and higher FGF21 mRNA levels in liver and white adipose tissue than WT mice. Thus, the altered metabolic phenotype of Nrf2-KO mice under HFD was associated with higher expression and abundance of FGF21. Consistently, the overexpression of Nrf2 in ST-2 cells resulted in decreased FGF21 mRNA levels as well as in suppressed activity of a FGF21 promoter luciferase reporter.
CONCLUSIONS
The identification of Nrf2 as a novel regulator of FGF21 expands our understanding of the crosstalk between metabolism and stress defense.
Fibroblast growth factor 21 (FGF21) belongs to the family of atypical FGFs that lack the conventional heparin-binding domain (1,2) and can thus diffuse away from their tissues of origin to function as hormones. FGF21 is abundantly expressed in liver, pancreas, and white adipose tissue (WAT) (3,4). It signals through cell-surface complexes of FGF receptors with the transmembrane protein β-Klotho (5–8). In contrast to the broadly expressed FGF receptors, β-Klotho is expressed in a limited set of tissues that include liver, WAT, pancreas, and testes, thus probably directing FGF21 activity to these target organs (8,9).
Pharmacological studies have shown that FGF21 has broad metabolic actions in obese rodents and primates (rev. in 10). Transgenic mice overexpressing FGF21 in liver had improved insulin sensitivity and glucose clearance and reduced plasma triglyceride concentrations and were resistant to weight gain when fed a high-fat diet (HFD) (11). Similar effects were observed in obese insulin-resistant ob/ob or db/db mice or in Zucker diabetic fatty rats after administration of recombinant FGF21 (11,12). Administration of FGF21 to HFD-induced obese mice increased fat utilization and energy expenditure and reduced plasma glucose, insulin, and lipid concentrations, as well as hepatic triglyceride concentrations (11–13). FGF21 also improved hepatic and peripheral insulin sensitivity in both lean and HFD-induced obese mice (12). In diabetic rhesus monkeys, FGF21 caused significant decreases in fasting plasma glucose, insulin, and triglycerides, while also lowering LDL cholesterol, increasing HDL cholesterol, and causing modest weight loss (14).
Because of its beneficial effects on carbohydrate and lipid metabolism, FGF21 is a candidate for the treatment of type 2 diabetes and the metabolic syndrome (10,15). However, not enough is known about the regulation of endogenous FGF21 in metabolic disease. Elucidating the factors that control FGF21 expression and activity in lean and obese states could lead to new therapeutic strategies.
The vertebrate transcription factor NFE2-related factor 2 (Nrf2) is a member of the cap’n’collar family that has emerged as a master regulator of the cell’s redox status and detoxification responses (16–19). The Nrf2 system mediates both the regulation of baseline antioxidant capacity and the stress response during acute oxidative challenges. In mice and human cultured cells, dozens of protective genes are induced in response to oxidative and electrophilic chemical challenges in an Nrf2-dependent manner (rev. in 20). Stress-activated Nrf2 upregulates the transcription of its target genes by binding to antioxidant response element sequences in their cognate promoters (21). In addition to its ubiquitous antioxidant and detoxification target genes, Nrf2 also has groups of tissue-specific targets. In the liver, the majority of these genes encode proteins specific to the synthesis and metabolism of fatty acids and other lipids (22,23). Interestingly, these metabolic genes are primarily repressed—rather than induced—by Nrf2 via mechanisms that are unknown.
An abundance of studies have documented broad protective functions of Nrf2 against numerous disorders caused or aggravated by oxidative stress. This result is evident in Nrf2 knockout (Nrf2-KO) mice as increased incidence of cancer, pulmonary disease, and inflammatory conditions and markedly increased sensitivity to environmental toxicants, inhaled irritants, and chemical carcinogens (rev. in 18–21). Beyond acute transient stress responses to oxidative insults, Nrf2 was recently shown to have important functions in long-term adaptations to metabolic conditions, such as the response to calorie restriction, as well as the control of lipid metabolism under both normal diets and HFDs (rev. in 24). The genetic or pharmacological activation of Nrf2 in the liver resulted in reduction of liver lipid levels (25). Nrf2-KO mice fed a short-term (4 weeks) HFD had higher expression levels of lipogenic and cholesterologenic transcription factors and their target genes in the liver, as well as higher liver concentrations of free fatty acids; this high-fat regimen repressed Nrf2 expression and activity in wild-type (WT) mice (26). In addition, activation of Nrf2 by the synthetic triterpenoid CDDO-Im (2-Cyano-3,12-dioxooleana-1,9-dien-28-imidazolide) prevented HFD-induced obesity in WT mice (23). On the other hand, Nrf2-KO mice had impaired adipogenesis caused by a role of Nrf2 as a cell-autonomous regulator of adipocyte differentiation; thus, Nrf2-KO mice had decreased adipose tissue mass with smaller adipocytes and were found to be protected against HFD-induced obesity (27).
Given the role of Nrf2 as a regulator of lipid homeostasis, its pharmacological manipulation might be beneficial in treating metabolic disorders such as dyslipidemia and obesity. Indeed, the activation of Nrf2 by the synthetic triterpenoid CDDO-Im in WT mice prevented the increase of adipose tissue mass, body weight, and serum triglyceride levels, as well as the fatty liver caused by an HFD; this protection against obesity was abolished in Nrf2-KO animals (23). Moreover, statins have been shown to activate Nrf2 in cell culture as well as in vivo (28,29), which may account for some of their antioxidant effects (30). It is important to elucidate further the mechanisms by which Nrf2 regulates metabolism and to identify the metabolic control systems with which it interacts. The current study investigated the role of Nrf2 during a long-term (180 days) HFD and identified Nrf2 as a novel regulator of FGF21 during HFD-induced obesity.
RESEARCH DESIGN AND METHODS
Mice.
C57BL6 J Nrf2+/− mice, originally developed by Prof. M. Yamamoto (31), were obtained from RIKEN BRC (Tsukuba, Japan). WT and Nrf2-KO mice were generated by mating Nrf2+/− male and female mice; the offspring were genotyped as previously described (31). Male WT and Nrf2-KO mice (9–10 weeks old) were fed a standard diet (SD) (10 kcal % fat) or an HFD (60 kcal % fat; Research Diets, New Brunswick, NJ) for 180 days. Mice were housed in the animal facility of the University of Patras Medical School in temperature-, light- and humidity-controlled rooms with a 12-h light/dark cycle. All animal procedures were approved by the institutional review board of the University of Patras Medical School and were in accordance with European Commission Directive 86/609/EEC.
Evaluation of insulin sensitivity.
Blood glucose was measured using a Precision Xceed glucose meter (Abbott, Chicago, IL) in blood collected from the tail of the mouse. To assess glucose tolerance, a single dose of d-glucose (100 mg/mL in water, 10 mL/kg) was injected intraperitoneally after 18 h fasting with free access to water. To assess insulin tolerance, a single dose of Actrapid regular insulin (Novo Nordisk, Copenhagen, Denmark) (0.75 units/kg) was injected intraperitoneally after 4 h fasting with free access to water.
Measurements of hormones and metabolites.
Plasma was collected using heparin as an anticoagulant and was centrifuged at 2,000g for 20 min at 4°C. Plasma measurements were conducted following the manufacturer’s instruction for each assay. Enzyme-linked immunosorbent assays were used for plasma leptin (ALPCO, Salem, NH), insulin (ALPCO), and FGF21 (R&D Systems, Minneapolis, MN). Nonesterified fatty acids (NEFAs) were measured using the NEFA C kit (Wako Chemicals, Osaka, Japan), and β-hydroxybutyrate was measured using the β-hydroxybutyrate (ketone body) assay kit (Cayman Chemical, Ann Arbor, MI). Glucose, cholesterol, HDL cholesterol, and triglycerides were measured using an Olympus AU640 analyzer (Hamburg, Germany).
Cell culture studies.
The (–1497+5) promoter FGF21-Luc plasmid was a gift from Prof. S. Kliewer (UT Southwestern Medical Center) (32). The mouse Nrf2 cDNA was amplified by PCR with the primers 5′-CCG-CTC-GAG-ATG-ATG-GAC-TTG-GAG-TTG-CC-3′ and 5′-CGG-GGT-ACC-CTA-GTT-TTT-CTT-TGT-ATC-TGG-CTT-CTT-3′ using mouse liver cDNA as the template and was cloned into the XhoI/KpnI sites of pcDNA 3.1 Hygro(–) (Invitrogen, Carlsbad, CA). A dominant-negative mutant of Nrf2 (DN-Nrf2) lacking the transcription activation domain was generated by PCR as previously described (33) and cloned into the pcDNA 3.1 Hygro(–). ST-2 cells, a mouse cell line derived from bone marrow stroma that expresses both functional Nrf2 (28,30) and FGF21 (present work) (DSMZ, German Collection of Microorganisms and Cell Cultures), were grown in RPMI supplemented with 10% FBS and antibiotics (streptomycin/penicillin) in a humidified 5% CO2 atmosphere. Using lipofectamine (Invitrogen) according to the manufacturer’s instructions, ST-2 cells were transiently transfected with pcDNA 3.1 Hygro(–)-mNrf2 or empty pcDNA 3.1 Hygro(–), (−1497+5) promoter FGF21-Luc, and pCMV-β-galactosidase (Clontech, Mountain View, CA) for monitoring transfection efficiency. After 48 h, the cells were analyzed for luciferase activity using the Promega (Madison, WI) luciferase assay system with reporter lysis buffer according to the manufacturer’s instructions; luciferase activities were normalized to respective β-galactosidase activities.
Quantification of gene expression levels.
Liver and WAT were submerged immediately after collection in RNA later solution (Ambion, Foster City, CA). Total RNA was isolated using the TRIzol reagent (Invitrogen) and further purified using the RNeasy mini-kit (Qiagen, Hilden, Germany). A DNAse (TurboDNAse; Ambion) digestion step was included to prevent genomic DNA contamination. cDNA was synthesized using the Superscript first-strand synthesis system (Invitrogen), and real-time PCRs were performed in triplicate on a Step One Plus instrument (Applied Biosystems, Foster City, CA) using TaqMan Gene Expression assays on demand (Applied Biosystems): FGF21, Mm00840165_g1; PGC-1α, Mm00447183_m1; PEPCK, Mm00440636; PPARα, Mm00440939_m1; GAPDH, 4352339E. Relative mRNA levels were calculated by the comparative threshold cycle method using GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as the housekeeping gene. Relative Nrf2 mRNA expression levels were determined by SYBR Green (Applied Biosystems) RT-PCR and calculated by the relative standard curve method using TATA-binding protein (TBP) as the housekeeping gene with previously described primers (34).
Statistical analysis.
In cell culture studies, three different experiments were performed on three different days. In each experiment, at least three wells were used and averaged together. In animal studies, eight mice were used in each study group. Data were expressed as the mean ± SEM. Student t test or one-way ANOVA followed by Tukey test were performed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA). P < 0.05 was considered significant.
RESULTS
Nrf2 deficiency has a long-term protective effect against HFD-induced obesity.
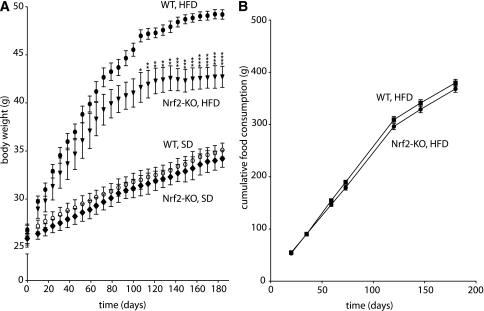
Nrf2-KO and WT mice were fed an SD (10 kcal % fat) or an HFD (60 kcal % fat) for 180 days. The initial weight of Nrf2-KO mice was not significantly different from WT mice, and no difference in weight was observed between Nrf2-KO and WT mice over time on the SD (Fig. 1A). On the HFD, both genotypes gained weight initially at about the same rate. However, the Nrf2-KO mice plateaued at a lower weight; the difference in weight between the two genotypes became statistically significant at 90 days (Fig. 1A). The difference in weight gained was not due to a difference in food consumption, which was similar between Nrf2-KO and WT mice (Fig. 1B).
FIG. 1.
A: Nrf2-KO mice fed an HFD for 180 days gained less weight over time than WT mice. B: Cumulative food consumption is similar between Nrf2-KO and WT mice fed an HFD for 180 days. Data show means ± SEM. n = 8 for each genotype. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (WT HFD vs. Nrf2-KO HFD).
Nrf2 deficiency preserves insulin sensitivity during long-term HFD.
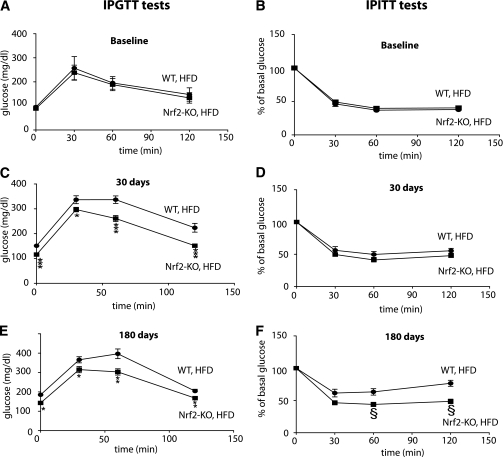
Insulin sensitivity can be indicated by increased glucose tolerance and/or reduced insulin tolerance, as assessed by an intraperitoneal glucose tolerance test and intraperitoneal insulin tolerance test, respectively. Baseline insulin sensitivity was similar between Nrf2-KO and WT mice (Fig. 2A and B). After 30 days on an HFD, even before the divergence of their body weights, Nrf2-KO mice had lower basal glucose levels and were more glucose tolerant (Fig. 2C), yet not more insulin sensitive (Fig. 2D) than WT mice. After 180 days on the high-fat regimen, Nrf2-KO mice were more glucose tolerant (Fig. 2E) and more insulin sensitive (Fig. 2F) than WT mice.
FIG. 2.
Nrf2-KO mice on HFD are less insulin resistant than WT mice. Results from glucose and insulin tolerance tests in WT and Nrf2-KO mice fed an HFD are shown. Tests were performed at baseline (A, B) and after 30 (C and D) and 180 (E and F) days on an HFD. Data are means ± SEM. n = 8 for each genotype. *P < 0.05, **P < 0.01, ***P < 0.001, §P < 0.01. IPGTT, intraperitoneal glucose tolerance test; IPITT, intraperitoneal insulin tolerance test.
Nrf2 alters the metabolic profile after long-term HFD.
Blood was collected from WT and Nrf2-KO mice after 180 days on an HFD after an overnight fast. Nrf2-KO mice had lower basal glucose levels (P < 0.05), lower HDL cholesterol levels (P < 0.005), and higher triglycerides levels (P < 0.005) than their WT counterparts. Furthermore, Nrf2-KO mice had lower leptin levels (P < 0.05), consistent with their lower body weights. No difference in these plasma parameters was found between Nrf2-KO and WT mice on an SD (Table 1).
TABLE 1.
Plasma metabolic parameters of WT and Nrf2-KO mice fed an SD or HFD for 180 days
| SD | HFD | |||
|---|---|---|---|---|
| WT | Nrf2-KO | WT | Nrf2-KO | |
| Glucose (mg/dL) | 90 ± 7.2 | 88 ± 7.7 | 185.2 ± 12.3 | 143 ± 13.5* |
| Cholesterol (mg/dL) | 91.2 ± 8.4 | 89 ± 8.8 | 206.6 ± 17.2 | 162.9 ± 22.82 |
| HDL cholesterol (mg/dL) | 71.3 ± 5.7 | 68.7 ± 5.9 | 154.2 ± 7.9 | 107.1 ± 15.8† |
| Triglycerides (mg/dL) | 59.3 ± 6.8 | 71.3 ± 7.9 | 81.4 ± 10.05 | 119.3 ± 6.2† |
| Insulin (ng/mL) | 0.9 ± 0.12 | 0.7 ± 0.18 | 1.5 ± 0.16 | 0.9 ± 0.3 |
| Leptin (pg/mL) | 4,128 ± 418 | 3,896 ± 402 | 24,830 ± 2,310.4 | 17,770 ± 1,620.1* |
| Free fatty acid (mmol/L) | NA | NA | 0.331 ± 0.02 | 0.336 ± 0.04 |
| β-Hydroxybutyrate (mmol/L) | NA | NA | 1.439 ± 0.14 | 1.483 ± 0.14 |
| FGF21 (pg/mL) | 492 ± 50 | 488 ± 61 | 4,472 ± 626.4 | 9,518 ± 817.8‡ |
Data are means ± SEM. n = 8. Mice were fast overnight before plasma collection. NA, not available.
*P < 0.05.
†P < 0.005.
‡P < 0.0001 (WT vs. Nrf2-KO).
Elevated FGF21 plasma levels in Nrf2-KO mice fed a long-term HFD.
No difference in FGF21 levels between Nrf2-KO and WT mice were found when the mice were maintained on an SD (Table 1). Long-term HFD caused a significant increase in FGF21 plasma levels in WT mice, as expected from previous work (35); a significant increase was also observed in Nrf2-KO mice. Importantly, however, the HFD-induced plasma levels of FGF21 were significantly greater (more than twofold) in Nrf2-KO mice than in WT mice (P < 0.0001) (Table 1).
Nrf2 deficiency increases expression of FGF21 in liver and WAT during long-term HFD.
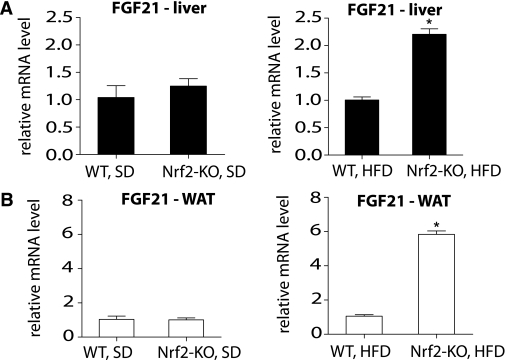
To identify the source of the increased FGF21 plasma levels in Nrf2-KO mice on an HFD, FGF21 mRNA expression levels were quantified in two major sources of circulating FGF21: the liver and WAT. No difference in the FGF21 mRNA expression levels was observed in either tissue in mice on an SD (Fig. 3A and B). However, the mRNA expression level of FGF21 was significantly elevated in both the liver and WAT of Nrf2-KO mice on an HFD (Fig. 3A and B).
FIG. 3.
FGF21 mRNA levels are elevated in the liver (A) and WAT (B) of Nrf2-KO mice fed an HFD for 180 days. FGF21 mRNA levels were measured by quantitative RT-PCR in liver and WAT from WT and Nrf2-KO mice (n = 8 for each genotype) fed an SD or HFD. The RT-PCR was performed in triplicate wells for each sample. Bars show means ± SEM. *P < 0.001.
Nrf2 deficiency increases the expression of FGF21-regulated genes after long-term HFD.
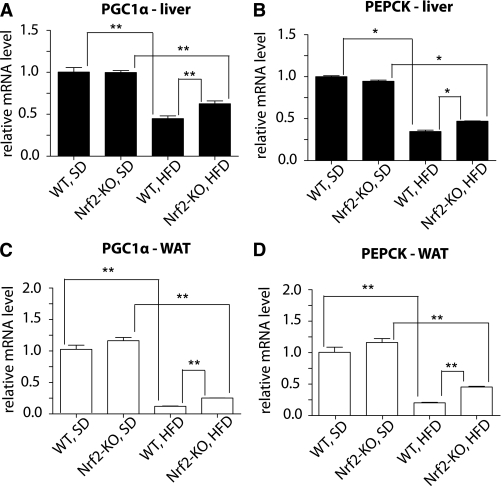
Previous work has shown that FGF21 injection (12) or transgenic expression (36) induces the expression of the metabolic genes PGC-1α (peroxisome proliferator–activated receptor-γ coactivator 1-α) and PEPCK (phosphoenolpyruvate carboxykinase). PGC-1α is a transcriptional coactivator essential for coordinating gluconeogenesis and fatty acid oxidation, and PEPCK is a key gluconeogenic enzyme that is downstream of PGC-1α (37). Therefore, it was examined whether the increased mRNA expression and elevated plasma levels of FGF21 in Nrf2-KO mice were accompanied by increased PGC-1α and/or PEPCK expression. No difference was observed in either gene’s mRNA levels between the two genotypes under SD (Fig. 4). Under HFD, levels of PGC-1α and PEPCK were significantly lower compared with SD (Fig. 4), consistent with previous work (38–42). Notably, Nrf2-KO mice on an HFD had significantly higher mRNA levels of PGC-1α and PEPCK than WT mice in both the liver (P < 0.001 and P < 0.05, respectively) (Fig. 4A and B) and WAT (P < 0.001 for both genes) (Fig. 4C and D).
FIG. 4.
mRNA levels of FGF21-regulated genes are elevated in liver and WAT of Nrf2-KO mice fed an HFD for 180 days. PGC-1α and PEPCK mRNA levels were measured by quantitative RT-PCR in liver (A and B) and WAT (C and D) from WT and Nrf2-KO mice (n = 8 for each genotype) fed an HFD or an SD. The RT-PCR was performed in triplicate wells for each sample. Bars show means ± SEM. *P < 0.05, **P < 0.001.
Nrf2 overexpression reduces FGF21 mRNA expression levels and suppresses FGF21 promoter activity.
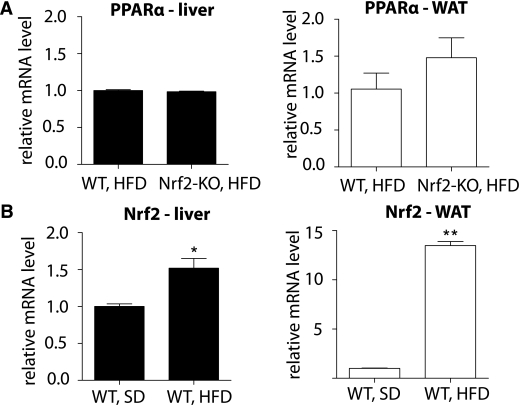
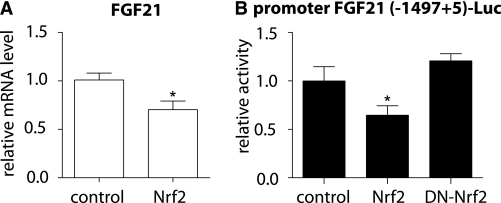
To investigate the mechanism underlying the differential induction of FGF21 in Nrf2-KO and WT mice, the mRNA level of peroxisome proliferator–activated receptor (PPAR)-α was compared between the two genotypes, because PPAR-α is known to be involved in the upregulation of FGF21 in mice during the response to starvation (32). However, no difference was found between the two genotypes in either the liver or the WAT (Fig. 5A). To investigate whether Nrf2 might be directly involved in the differential induction of FGF21, the mRNA levels of Nrf2 were quantified in WT mice on an HFD or SD for 180 days. The Nrf2 mRNA levels in HFD-fed mice were ∼50% higher in the liver (P < 0.01) and ∼14 times higher in the WAT (P < 0.0001) compared with the SD-fed mice (Fig. 5B). Thus, the compromised induction of FGF21 observed in WT mice compared with Nrf2-KO mice correlated with an induction of Nrf2; this result suggested that Nrf2 induction might repress FGF21. To test this hypothesis, the ST-2 cells were used, because they express both functional Nrf2 (28,30) and FGF21 mRNA (Fig. 6A). Nrf2 was overexpressed in ST-2 cells, and FGF21 mRNA levels were measured using quantitative RT-PCR. Nrf2 overexpression led to an ∼40% decrease in FGF21 mRNA levels (P < 0.001) (Fig. 6A). To examine the possible involvement of Nrf2 in the repression of the FGF21 promoter, ST-2 cells were transiently transfected with an FGF21 promoter luciferase reporter plasmid along with WT Nrf2 or dominant-negative Nrf2 (DN-Nrf2). Consistent with the repression of FGF21 mRNA (Fig. 6A), the overexpression of WT Nrf2 lead to a corresponding decrease (∼40%, P < 0.001) in FGF21 promoter activity (Fig. 6B). On the other hand, overexpression of DN-Nrf2 had no effect on FGF21 promoter activity (Fig. 6B). These results indicate that Nrf2 can inhibit FGF21 expression by repressing its cognate promoter.
FIG. 5.
A: PPARα mRNA levels in liver and WAT are similar between WT and Nrf2-KO mice on a long-term HFD. PPARα mRNA levels were measured by quantitative RT-PCR in liver and WAT of WT and Nrf2-KO mice (n = 8 for each genotype) fed an HFD for 180 days. The RT-PCR was performed in triplicate wells for each sample. Bars show means ± SEM. B: Nrf2 mRNA levels are elevated in the liver and WAT of mice fed an HFD for 180 days compared with an SD. Nrf2 mRNA levels were measured by quantitative RT-PCR in liver and WAT of WT mice fed an HFD or SD (n = 8 for each diet group). The RT-PCR was performed in triplicate wells for each sample. Bars show means ± SEM. *P < 0.01, **P < 0.0001.
FIG. 6.
Overexpression of Nrf2 reduces FGF21 mRNA levels and suppresses FGF21 promoter activity. A: Nrf2 was overexpressed in ST-2 cells in a 60-mm plate, and FGF21 mRNA level was measured by quantitative RT-PCR 48 h after transfection. The RT-PCR was performed in triplicate wells for each sample. Bars show means ± SEM of three independent experiments. B: (−1497+5) promoter FGF21-Luc was transiently transfected in ST-2 cells in 24-well plates, and luciferase activity was measured after cotransfection with pcDNA 3.1 Hygro(–)-mNrf2, pcDNA 3.1 Hygro(–)-DN-Nrf2, or the empty vector. The luciferase activity was normalized to β-galactosidase activity. In each experiment, the luciferase activity was measured in 12 technical replicates. Bars show means ± SEM of three independent experiments. *P < 0.001.
DISCUSSION
The current study investigated the role of Nrf2 in a mouse model of obesity induced by long-term (180 days) HFD. Lack of Nrf2 was found to partly protect male mice of a C57BL6 J background against obesity. Consistent with this finding, a protective role of Nrf2 was recently shown in two shorter-term (90 days) studies of HFD-induced obesity, one of which used female C57BL6J mice (23); the other used male mice of a mixed C57BL6;129SV background (27). Taken together, these observations suggest that deletion of Nrf2 is a protective factor against HFD-induced obesity independent of the animals’ sex and genetic background; this protection is sustained over several months on an obesogenic diet.
It is noteworthy that, on the HFD regimen, the Nrf2-KO mice initially gained weight at about the same rate as WT mice and later reached a plateau, while the WT mice continued to gain weight. Given that food consumption was not found to differ between the two genotypes, a plausible hypothesis for the difference in weight gain is that Nrf2-KO mice have increased energy expenditure. Increased locomotor activity or increased fat excretion cannot be excluded but are highly improbable because an independent study did not find such differences in mice on an HFD for 3 months (27). Nrf2-KO mice are known to have increased levels of oxidative stress in their tissues (43); this could stimulate mitochondrial biogenesis (44) and activate uncoupling proteins in mitochondria leading to thermogenesis (45). The increased mitochondrial activity in tissues such as white and brown adipose tissue and muscle could be responsible for the increased energy expenditure and consequently for the lower weights of Nrf2-KO mice. Further research is needed to investigate these metabolic parameters in Nrf2-KO mice under an HFD.
Obesity is associated with impaired glucose tolerance and reduced insulin sensitivity. Consistent with their lower weights, Nrf2-KO mice were more glucose tolerant and more insulin sensitive than WT mice after 180 days on an HFD. Interestingly, Nrf2-KO mice were more glucose tolerant than WT mice, even before their weights diverged significantly (30 days). This finding (which, to the best of our knowledge, has not been described before) indicates that the improved glucose tolerance of Nrf2-KO mice is not wholly attributable to their leaner body habitus. Notably, there was no difference in glucose tolerance or insulin sensitivity between the genotypes before the initiation of HFD. Another study reported that Nrf2-KO mice have impaired glucose tolerance at baseline (46). Although the reason(s) for this discrepancy is not known, in the current study, no differences in glucose tolerance were seen between the two genotypes, even after 180 days on an SD (data not shown). The present results suggest that the lack of Nrf2 leads to a more insulin-sensitive phenotype under HFD.
Previous work has attributed the protection of Nrf2-KO mice against obesity to impaired adipogenesis, acknowledging, however, that additional mechanisms are probably involved (27). To identify such novel mechanisms, the metabolic profiles of Nrf2-KO and WT animals were compared. Significant difference was observed in the plasma levels of FGF21, a recently discovered hormone that has been implicated in metabolic control. FGF21 improves glucose tolerance and leads to weight loss in obese mice (10). It was proposed that obesity is a state of FGF21 resistance, in which plasma FGF21 levels are elevated and administration of exogenous FGF21 has attenuated signaling effects in obese mice compared with lean (35). Accordingly, lower levels of circulating FGF21 were anticipated in the leaner Nrf2-KO mice compared with the more obese WT mice after an HFD. Unexpectedly, however, while plasma FGF21 levels were increased in both genotypes by the obesogenic diet as expected (35), they were significantly higher in the Nrf2-KO mice.
Consistent with the increased plasma levels of FGF21 in Nrf2-KO mice on an HFD, significantly higher FGF21 mRNA expression levels were found in the liver and WAT compared with WT mice. In accordance with these findings, it was recently shown that a diet deficient in methionine and choline (a model of fatty liver) induced higher FGF21 mRNA expression in the liver of Nrf2-KO mice compared with WT mice (47). Thus, Nrf2 may exert a negative effect on FGF21 that is important in various pathological states. The increased FGF21 mRNA and plasma levels observed in the Nrf2-KO mice were accompanied by increased mRNA expression of specific genes that have been previously suggested to be regulated by FGF21 (12). Under an SD, there was no difference between the two genotypes in the mRNA levels of PGC-1α, a transcriptional coactivator involved in energy metabolism, and PEPCK, a PGC-1α−regulated rate-limiting enzyme in gluconeogenesis (37) in either liver or WAT. After HFD feeding, both PGC-1α and PEPCK mRNA levels were downregulated in both genotypes (Fig. 4), in accordance with previous studies (38–42). The significantly higher FGF21 levels in Nrf2-KO mice could at least partially account for the smaller decrease of PEPCK and PGC-1α levels in Nrf2-KO mice after HFD feeding. Given that Nrf2-KO mice on HFD weigh less and are more insulin sensitive than WT mice, and because insulin is a negative regulator of PEPCK mRNA (48), one might predict PEPCK mRNA levels to be decreased in Nrf2-KO mice compared with WT mice on an HFD. However, the regulation of PEPCK mRNA levels is likely complex, with insulin and FGF21 exerting effects in opposite directions. Similar to the present findings, transgenic mice with liver-specific overexpression of FGF21 have also been found to have increased insulin sensitivity and elevated liver PEPCK mRNA levels (11,36).
In WT mice fed an HFD for 180 days, Nrf2 mRNA is induced in both liver and WAT. Another study also using C57BL6 J mice showed similar findings after 90 days on an HFD (49). It should be noted that Nrf2 mRNA levels were reported to be reduced after 30 days on an HFD (26); this discrepancy may be due to the shorter duration of HFD feeding. After a long-term HFD, the induction of Nrf2 may partially explain the lower FGF21 levels of WT mice compared with Nrf2-KO mice. It is also noteworthy that the increase of FGF21 mRNA levels in Nrf2-KO mice is more pronounced in the WAT than in the liver. This might be explained by the fact that HFD induces Nrf2 mRNA 10-fold more in the WAT than in the liver (Fig. 5B). Thus, the effect of withdrawing the Nrf2-mediated repression on FGF21 should be more prominent in the WAT.
The notion that Nrf2 represses FGF21 mRNA expression was further tested in transiently transfected ST-2 cells, a cell line of mouse bone marrow stroma origin (50) that expresses both functional Nrf2 and FGF21. Importantly, the overexpression of Nrf2 significantly lowered the mRNA expression levels of FGF21; moreover, Nrf2 suppressed the activity of a FGF21 promoter reporter. These findings indicate that Nrf2 can repress FGF21 mRNA expression in a cell-autonomous manner (i.e., without requiring interactions of different tissues, endocrine mediators, neuronal effects, or other indirect mechanisms). The finding that overexpression of DN-Nrf2 has no effect on FGF21 promoter reporter activity is not due to technical reasons, since a sufficient level of DN-Nrf2 expression was obtained (see the Supplementary Data). The lack of effect of DN-Nrf2 on FGF21 promoter reporter activity could reflect weak repression of FGF21 by Nrf2 under basal conditions. Work is in progress to investigate whether endogenous Nrf2 directly affects FGF21 expression by binding to specific antioxidant response element sequences that are present on the native FGF21 promoter; four such potential binding sites were identified by bioinformatics analyses (data not shown).
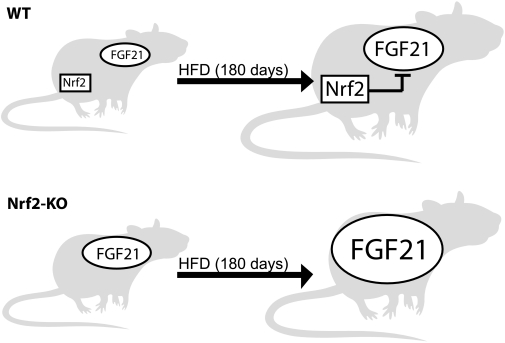
On the basis of the present findings, we propose the following model regarding the relationship between Nrf2 and FGF21 during prolonged HFD (Fig. 7). In WT mice, FGF21 as well as Nrf2 are induced after HFD. The increased Nrf2 expression after HFD leads to a partial repression of FGF21 expression, which accounts for the attenuated increase in FGF21 levels in WT mice. On the other hand, the absence of Nrf2-mediated FGF21 repression in Nrf2-KO mice permits a larger increase in FGF21 levels after HFD. We suggest that this phenomenon is at least in part responsible for the altered metabolic phenotype of the Nrf2 knockout mice.
FIG. 7.
Model depicting the repressive role of Nrf2 on the induced expression of FGF21 during HFD. WT and Nrf2-KO mice have similar low basal FGF21 levels. After HFD feeding, FGF21 levels increase in both genotypes, but to different extents. In WT mice, Nrf2 is induced by HFD and compromises the increase of FGF21 levels. In contrast, the lack of Nrf2 in the KO mice abolishes its inhibitory effect on FGF21, which leads to the significantly higher induction of FGF21 in Nrf2-KO mice.
In conclusion, the current study identified Nrf2 as a novel regulator of FGF21 gene expression and circulating FGF21 levels in response to an obesogenic diet. Increased FGF21 is a potential mediator of the protection of Nrf2-KO mice against obesity and insulin resistance induced by long-term HFD. The crosstalk between Nrf2 and FGF21 may be important under stress conditions such as HFD-induced obesity, and the underlying mechanisms of FGF21 regulation by Nrf2 warrant further elucidation. Because FGF21 is a potential new target, identifying the factors that regulate its expression may be important in understanding the development of obesity and the metabolic syndrome.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by a grant from the Hellenic Endocrine Society to I.G.H.
No potential conflicts of interest relevant to this article were reported.
D.V.C., G.P.S., and I.G.H. researched data and wrote the manuscript. P.G.Z. researched data. A.I.P., A.G.P., and V.E.K. contributed to discussion and reviewed the manuscript.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0112/-/DC1.
REFERENCES
- 1.Itoh N, Ornitz DM. Functional evolutionary history of the mouse Fgf gene family. Dev Dyn 2008;237:18–27 [DOI] [PubMed] [Google Scholar]
- 2.Goetz R, Beenken A, Ibrahimi OA, et al. Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol Cell Biol 2007;27:3417–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishimura T, Nakatake Y, Konishi M, Itoh N. Identification of a novel FGF, FGF-21, preferentially expressed in the liver. Biochim Biophys Acta 2000;1492:203–206 [DOI] [PubMed] [Google Scholar]
- 4.Muise ES, Azzolina B, Kuo DW, et al. Adipose fibroblast growth factor 21 is up-regulated by peroxisome proliferator-activated receptor gamma and altered metabolic states. Mol Pharmacol 2008;74:403–412 [DOI] [PubMed] [Google Scholar]
- 5.Kharitonenkov A, Dunbar JD, Bina HA, et al. FGF-21/FGF-21 receptor interaction and activation is determined by betaKlotho. J Cell Physiol 2008;215:1–7 [DOI] [PubMed] [Google Scholar]
- 6.Kurosu H, Choi M, Ogawa Y, et al. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J Biol Chem 2007;282:26687–26695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki M, Uehara Y, Motomura-Matsuzaka K, et al. betaKlotho is required for fibroblast growth factor (FGF) 21 signaling through FGF receptor (FGFR) 1c and FGFR3c. Mol Endocrinol 2008;22:1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa Y, Kurosu H, Yamamoto M, et al. BetaKlotho is required for metabolic activity of fibroblast growth factor 21. Proc Natl Acad Sci U S A 2007;104:7432–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito S, Kinoshita S, Shiraishi N, et al. Molecular cloning and expression analyses of mouse betaklotho, which encodes a novel Klotho family protein. Mech Dev 2000;98:115–119 [DOI] [PubMed] [Google Scholar]
- 10.Kliewer SA, Mangelsdorf DJ. Fibroblast growth factor 21: from pharmacology to physiology. Am J Clin Nutr 2010;91:254S–257S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kharitonenkov A, Shiyanova TL, Koester A, et al. FGF-21 as a novel metabolic regulator. J Clin Invest 2005;115:1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coskun T, Bina HA, Schneider MA, et al. Fibroblast growth factor 21 corrects obesity in mice. Endocrinology 2008;149:6018–6027 [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Lloyd DJ, Hale C, et al. Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes 2009;58:250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kharitonenkov A, Wroblewski VJ, Koester A, et al. The metabolic state of diabetic monkeys is regulated by fibroblast growth factor-21. Endocrinology 2007;148:774–781 [DOI] [PubMed] [Google Scholar]
- 15.Kharitonenkov A, Shanafelt AB. FGF21: a novel prospect for the treatment of metabolic diseases. Curr Opin Investig Drugs 2009;10:359–364 [PubMed] [Google Scholar]
- 16.Calkins MJ, Johnson DA, Townsend JA, et al. The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxid Redox Signal 2009;11:497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul 2006;46:113–140 [DOI] [PubMed] [Google Scholar]
- 18.Sykiotis GP, Bohmann D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci Signal 2010;3:re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 2007;47:89–116 [DOI] [PubMed] [Google Scholar]
- 20.Lee JM, Li J, Johnson DA, et al. Nrf2, a multi-organ protector? FASEB J 2005;19:1061–1066 [DOI] [PubMed] [Google Scholar]
- 21.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med 2004;10:549–557 [DOI] [PubMed] [Google Scholar]
- 22.Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway: identification of novel gene clusters for cell survival. J Biol Chem 2003;278:8135–8145 [DOI] [PubMed] [Google Scholar]
- 23.Shin S, Wakabayashi J, Yates MS, et al. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur J Pharmacol 2009;620:138–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sykiotis GP, Habeos IG, Samuelson AV, Bohmann D. The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr Opin Clin Nutr Metab Care 2011;141:41–48 [DOI] [PMC free article] [PubMed]
- 25.Yates MS, Tran QT, Dolan PM, et al. Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis 2009;30:1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka Y, Aleksunes LM, Yeager RL, et al. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J Pharmacol Exp Ther 2008;325:655–664 [DOI] [PubMed] [Google Scholar]
- 27.Pi J, Leung L, Xue P, et al. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J Biol Chem 2010;285:9292–9300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Habeos IG, Ziros PG, Chartoumpekis D, Psyrogiannis A, Kyriazopoulou V, Papavassiliou AG. Simvastatin activates Keap1/Nrf2 signaling in rat liver. J Mol Med (Berl) 2008;86:1279–1285 [DOI] [PubMed] [Google Scholar]
- 29.Hsieh CH, Rau CS, Hsieh MW, et al. Simvastatin-induced heme oxygenase-1 increases apoptosis of Neuro 2A cells in response to glucose deprivation. Toxicol Sci 2008;101:112–121 [DOI] [PubMed] [Google Scholar]
- 30.Chartoumpekis D, Ziros PG, Psyrogiannis A, Kyriazopoulou V, Papavassiliou AG, Habeos IG. Simvastatin lowers reactive oxygen species level by Nrf2 activation via PI3K/Akt pathway. Biochem Biophys Res Commun 2010;396:463–466 [DOI] [PubMed] [Google Scholar]
- 31.Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 1997;236:313–322 [DOI] [PubMed] [Google Scholar]
- 32.Inagaki T, Dutchak P, Zhao G, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab 2007;5:415–425 [DOI] [PubMed] [Google Scholar]
- 33.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a cap’n’collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem 1999;274:26071–26078 [DOI] [PubMed] [Google Scholar]
- 34.Shin S, Wakabayashi N, Misra V, et al. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol 2007;27:7188–7197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher FM, Chui PC, Antonellis PJ, et al. Obesity is an FGF21 resistant state. Diabetes 2010;59:2781–2789 [DOI] [PMC free article] [PubMed]
- 36.Potthoff MJ, Inagaki T, Satapati S, et al. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci U S A 2009;106:10853–10858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon JC, Puigserver P, Chen G, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 2001;413:131–138 [DOI] [PubMed] [Google Scholar]
- 38.Sajan MP, Standaert ML, Nimal S, et al. The critical role of atypical protein kinase C in activating hepatic SREBP-1c and NFkappaB in obesity. J Lipid Res 2009;50:1133–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheema SK, Clandinin MT. Dietary fat-induced suppression of lipogenic enzymes in B/B rats during the development of diabetes. Lipids 2000;35:421–425 [DOI] [PubMed] [Google Scholar]
- 40.Sutherland LN, Capozzi LC, Turchinsky NJ, Bell RC, Wright DC. Time course of high-fat diet-induced reductions in adipose tissue mitochondrial proteins: potential mechanisms and the relationship to glucose intolerance. Am J Physiol Endocrinol Metab 2008;295:E1076–E1083 [DOI] [PubMed] [Google Scholar]
- 41.Sparks LM, Xie H, Koza RA, et al. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes 2005;54:1926–1933 [DOI] [PubMed] [Google Scholar]
- 42.Arçari DP, Bartchewsky W, dos Santos TW, et al. Antiobesity effects of yerba maté extract (Ilex paraguariensis) in high-fat diet-induced obese mice. Obesity (Silver Spring) 2009;17:2127–2133 [DOI] [PubMed] [Google Scholar]
- 43.Beyer TA, Xu W, Teupser D, et al. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J 2008;27:212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda S, Foncea R, Guerrero J, Leighton F. Oxidative stress and upregulation of mitochondrial biogenesis genes in mitochondrial DNA-depleted HeLa cells. Biochem Biophys Res Commun 1999;258:44–49 [DOI] [PubMed] [Google Scholar]
- 45.Echtay KS, Roussel D, St-Pierre J, et al. Superoxide activates mitochondrial uncoupling proteins. Nature 2002;415:96–99 [DOI] [PubMed] [Google Scholar]
- 46.Aleksunes LM, Reisman SA, Yeager RL, Goedken MJ, Klaassen CD. Nuclear factor erythroid 2-related factor 2 deletion impairs glucose tolerance and exacerbates hyperglycemia in type 1 diabetic mice. J Pharmacol Exp Ther 2010;333:140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang YK, Yeager RL, Tanaka Y, Klaassen CD. Enhanced expression of Nrf2 in mice attenuates the fatty liver produced by a methionine- and choline-deficient diet. Toxicol Appl Pharmacol 2010;245:326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puigserver P, Rhee J, Donovan J, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 2003;423:550–555 [DOI] [PubMed] [Google Scholar]
- 49.Kim S, Sohn I, Ahn JI, Lee KH, Lee YS, Lee YS. Hepatic gene expression profiles in a long-term high-fat diet-induced obesity mouse model. Gene 2004;340:99–109 [DOI] [PubMed] [Google Scholar]
- 50.Ding J, Nagai K, Woo JT. Insulin-dependent adipogenesis in stromal ST2 cells derived from murine bone marrow. Biosci Biotechnol Biochem 2003;67:314–321 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.