Tumors with mutant BRAF are dependent on the RAF/MEK/ERK signaling pathway for their growth1-3. We found that ATP-competitive RAF inhibitors inhibit ERK signaling in cells with mutant BRAF, but unexpectedly enhance signaling in cells with wild-type BRAF. Here we demonstrate the mechanistic basis for these findings. We employed chemical genetic methods to show that drug-mediated transactivation of RAF dimers is responsible for paradoxical activation of the enzyme by inhibitors. Induction of ERK signaling requires direct binding of the drug to the ATP-binding site of one kinase of the dimer and is dependent on RAS activity. Drug binding to one member of RAF homo-(CRAF/CRAF) or heterodimers (CRAF/BRAF) inhibits one protomer, but results in transactivation of the drug-free protomer. In BRAFV600E tumors, RAS is not activated, thus transactivation is minimal and ERK signaling is inhibited in cells exposed to RAF inhibitors. These results imply that RAF inhibitors will be effective in tumors in which BRAF is mutated. Furthermore, since they do not inhibit ERK signaling in other cells, the model predicts that they would have a higher therapeutic index and greater antitumor activity than MEK inhibitors, but could also cause toxicity due to MEK/ERK activation. These predictions have been borne out strikingly in a recent clinical trial of the RAF inhibitor PLX40324-5. Finally, the model suggests that promotion of RAF dimerization by elevation of wild-type RAF expression or RAS activity could lead to drug resistance in mutant BRAF tumors. In agreement with this prediction, RAF inhibitors do not inhibit ERK signaling in cells that coexpress BRAFV600E and mutant RAS.
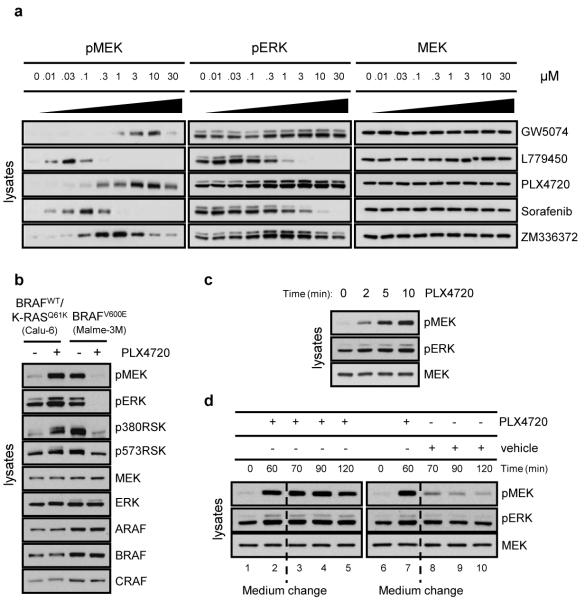
Six distinct ATP-competitive RAF inhibitors induced ERK activation in cells with wild-type BRAF, but inhibited signaling in mutant BRAFV600E cells (Fig. 1a, b, Supplementary Fig. 2a, b, Data Not Shown (DNS), structures of compounds shown in Supplementary Fig. 3, except that of PLX4032, which is unavailable). PLX47206, and its analog in clinical trial PLX4032, were studied in more detail. PLX4032 inhibited ARAF, BRAF and CRAF immunoprecipitated from 293H cells (Supplementary Fig. 4) and purified catalytic domains of BRAFV600E, wild-type BRAF and CRAF (IC50s: 35, 110 and 48nM) (Supplementary Table 1). PLX4032 was assayed against 62 additional kinases that span the kinome, and had IC50s of 1μM-10μM against eight of these and greater than 10μM against the rest (G.B., unpublished data). Induction of ERK signaling by PLX4720 was rapid (Fig. 1c), reversible (Fig. 1d), and associated with increased phosphorylation of the ERK substrate RSK (Fig 1b). MEK and ERK phosphorylation were induced at intermediate concentrations of RAF inhibitor, and inhibited at much higher doses (Fig. 1a).
Figure 1. RAF inhibitors rapidly activate MEK/ERK in cells with wild-type BRAF.
a, Calu-6 cells (BRAFwild-type/K-RASQ61K) were treated with increasing doses of the indicated RAF inhibitors and the effects on ERK signaling were determined by immunoblotting for pMEK and pERK. b, Cells with wild-type BRAF (Calu-6) or mutant BRAF (Malme-3M) were treated with vehicle or PLX4720 (1μM/1 hour). Phosphorylation and expression of the indicated proteins were assayed by immunoblotting. c, Calu-6 cells treated with 1μM PLX4720 for the indicated time points. d, Calu-6 cells were treated with 1μM PLX4720 for 60 minutes, then medium was replaced with medium containing 1μM PLX4720 (lanes 3-5) or vehicle (lanes 8-10) for the indicated time points.
Physiologic induction of ERK signaling depends on upstream activation of RAS by receptor-induced signaling7-8. PLX4032 induced ERK signaling in SKBR3 breast cancer cells, in which RAS activation is HER2-dependent9. The HER2 inhibitor Lapatinib abolished basal and PLX4032-induced ERK signaling in these cells (Supplementary Fig. 5a). In 293H cells, induction of MEK and ERK phosphorylation by either PLX4032 or PLX4720 was barely detectable (PLX will refer to data obtained with both compounds). HA-tagged wild-type RAS overexpression resulted in enhanced MEK/ERK activation by RAF inhibitor, which was more pronounced when mutant RAS was overexpressed (Fig. 2a and Supplementary Fig. 5b). The results suggest that RAS activity is required for MEK/ERK activation by RAF inhibitors. In contrast, in 293H cells expressing FLAG-tagged BRAFV600E, ERK signaling was inhibited by PLX4032 (Supplementary Fig. 5c). These results suggest that RAF inhibitors will inhibit the growth of tumors with mutant BRAF, but not those with wild-type BRAF, including those with RAS mutation. This is indeed the case: MEK-dependent tumors with RAS mutation are unaffected by PLX4032 (unpublished data).
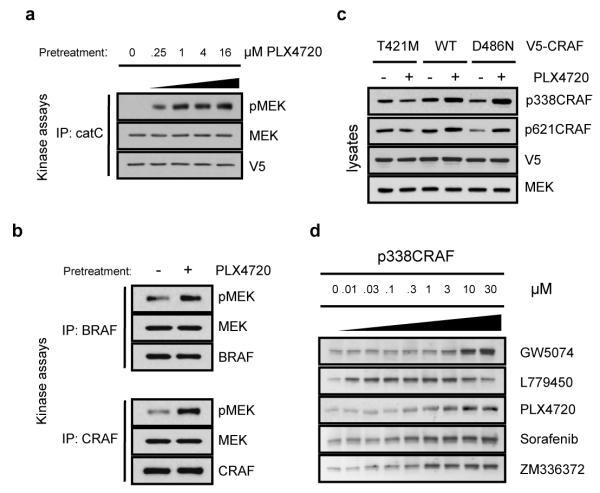
Figure 2. MEK/ERK activation requires binding of drug to the catalytic domain of RAF.
a, 293H cells transfected with EGFP (control), HA-tagged RASG12V, the catalytic domain of CRAF (V5-tagged catC) and catC carrying a mutation at the gatekeeper residue (V5-tagged catCT421M), treated with vehicle or PLX4720 (1μM/1 hour). Lysates were subjected to immunoblot analysis for pMEK and pERK. b, Wild-type (+/+), BRAF knock-out (BRAF −/−) or CRAF knock-out (CRAF −/−) mouse embryonic fibroblasts (MEFs) were treated with the indicated concentrations of PLX4720 for 1 hour. c, Sorafenib inhibits the gatekeeper mutant catCT421M protein in vitro (Supplementary Fig. 8c) and activates MEK/ERK in cells expressing it. 293H cells overexpressing catCT421M were treated with the indicated concentrations of sorafenib for 1 hour. Lysates were subjected to analysis for pMEK and pERK.
BRAF and CRAF kinases form homo- and heterodimers upon RAS activation10-12. PLX induced pronounced phosphorylation of MEK and ERK in wild-type MEFs and BRAF (−/−) MEFs. The response was diminished markedly in CRAF (−/−) MEFs (Fig. 2b, Supplementary Fig. 6a). Coexpression of CRAF and active RAS in CRAF (−/−) MEFs reconstituted the wild-type phenotype (Supplementary Fig. 6b, c). We conclude that BRAF is dispensable for MEK/ERK activation by PLX, and that CRAF expression is required for significant induction. We therefore investigated the mechanism of CRAF-dependent induction of ERK signaling in response to the drug.
Autoinhibition of RAF by its N-terminal domain13 is relieved upon binding to activated RAS7. We asked whether overexpression of an N-truncated form of CRAF would bypass the requirement for RAS activity. In 293H cells expressing the catalytic domain of CRAF (catC), PLX caused dramatic induction of MEK and ERK phosphorylation (Fig. 2a, Supplementary Fig. 7a). We focused mechanistic investigations on catC, in which PLX-induced MEK/ERK activation is RAS-independent. To test whether direct binding of PLX to CRAF is required for induction of signaling, we generated a catC carrying a mutation at the gatekeeper position (T421) in the kinase domain (mutations used and their properties are in Supplementary Fig. 1a). Structural studies6 predict that the T421M mutation should prevent drug binding and catCT421M was indeed resistant to inhibition by PLX in vitro (Supplementary Fig. 8a, b). ERK signaling was not induced by PLX in cells expressing catCT421M (Fig. 2a, Supplementary Fig. 7b). Thus, activation of MEK/ERK by PLX depends on its direct binding to the RAF kinase active site. Sorafenib inhibited catCT421M in vitro (Supplementary Fig. 8c) and induced ERK signaling in cells expressing catCT421M (Fig. 2c), demonstrating that this mutant is capable of inhibitor-induced MEK/ERK activation. Thus, direct binding of an ATP-competitive inhibitor to CRAF is required for induction of ERK signaling.
Recent work shows that binding of ATP-competitive inhibitors to AKT and PKC inhibits their activity, but induces the active, phosphorylated state of these kinases14-15. Washed catC immunoprecipitated from PLX-treated cells was more active than that isolated from untreated cells (Fig. 3a, Supplementary Fig. 9a). The same was true for endogenous BRAF and CRAF immunoprecipitated from treated Calu-6 cells (Fig. 3b, Supplementary Fig. 9b). Phosphorylation of CRAF at S338 and S621 has been correlated with its activation7. PLX caused increased phosphorylation of both sites on wild-type and kinase-dead CRAF in 293H cells. In contrast, it did not affect the phosphorylation of the PLX-resistant CRAFT421M mutant. (Fig. 3c, Supplementary Fig. 9c). All RAF inhibitors tested induced phosphorylation at p338 of endogenous CRAF (Fig. 3d). The data suggest that binding of PLX to CRAF induces activation of the enzyme and, subsequently, ERK signaling. The result seems paradoxical: binding of ATP-competitive inhibitors to the catalytic domain of CRAF activate its function.
Figure 3. RAF inhibitor induces the active, phosphorylated state of wild-type and kinase-dead RAF.
a, 293H cells over-expressing catC were treated with the indicated amounts of PLX4720 for 1 hour. Cells were lysed, catC was immunoprecipitated, washed extensively and subjected to kinase assay. Kinase activity was determined by immunoblotting for pMEK. b, Calu-6 cells were treated with PLX4720 (1μM/1 hour). Endogenous BRAF and CRAF were immunoprecipitated, washed and assayed for kinase activity. c, Treatment with RAF inhibitor results in elevated phosphorylation at activating phosphorylation sites on RAF. V5-tagged wild-type CRAF or kinase-dead CRAFD486N were overexpressed in 293H cells. After 24 hours cells were treated with vehicle or PLX4720 (5μM/1hour) and lysates were immunoblotted for p338CRAF and p621CRAF. The gatekeeper mutant CRAFT421M was used as negative control. d. Samples as in Fig. 1a, immunoblotted for pS338CRAF. Note that phosphorylation at S338 steadily increased, even when concentrations were reached that inhibited MEK/ERK.
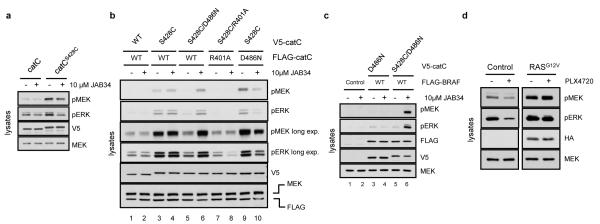
RAF isoforms form dimers in cells10-12,16. Since binding of both the drug and ATP to the catalytic domain would be required for activation and cannot occur simultaneously on the same molecule, we hypothesized that RAF inhibitors activate CRAF dimers in trans. (Supplementary Fig. 1b). To test this model, we generated mutant catCS428C that binds to quinazoylacrylamide-based inhibitors17, whereas catC does not. Two inhibitors JAB1317 and JAB34 (PD-168393)18 both inhibited catCS428C, but up to 30μM had no effect in vitro on catC (Supplementary Fig. 10a, b). JAB13 and JAB34 selectively affected ERK signaling in cells expressing catCS428C, and were inactive in those expressing catC (Supplementary Fig. 11). Like the other inhibitors (Fig. 1a), lower doses (40nM - 1μM) induced ERK signaling (Supplementary Fig. 11), whereas higher doses (10μM) inhibited (Fig. 4a). The specificity of this system allows us to test the dimer transactivation model. We coexpressed a V5-tagged, JAB-sensitive, kinase-dead catC, V5-catCS428C/D486N and FLAG-catC in 293H cells. V5-catCS428C/D486N is deficient in catalytic activity; it can bind to the inhibitor (JAB34) but cannot phosphorylate MEK, while FLAG-catC is catalytically active, but cannot bind JAB34. Treatment of cells expressing both constructs with a concentration of JAB34 that inhibited ERK signaling in cells expressing catCS428C alone (10μM JAB34, Fig. 4a) resulted in marked induction of ERK signaling (Fig. 4b, lanes 5-6). Thus, binding of JAB34 to kinase-dead, V5-catCS428C/D486N transactivated the catalytically competent FLAG-catC. When the catalytically active drug-binding mutant V5-catCS428C is coexpressed with catalytically inactive catC (FLAG-catCD486N), 10μM JAB34 inhibited, rather than activated ERK signaling. (Fig. 4b, lanes 9-10). When both constructs were insensitive to JAB, JAB34 had no effect on ERK signaling (Fig. 4b, lanes 1-2). When both constructs were catalytically active, we observed moderate MEK/ERK activation, likely resulting from inhibition of V5-catCS428C and transactivation of FLAG-catC (Fig. 4b, lanes 3-4).
Figure 4. MEK/ERK induction occurs via transactivation of RAF dimers.
a, Similarly to RAF inhibitors, JAB34 inhibits MEK/ERK at higher concentrations. 293H cells expressing V5-tagged catC or catCS428C were treated with either vehicle or 10μM JAB34 for 1 hour. b, Coexpression of drug-sensitive V5-tagged catC with drug-resistant FLAG-catC reveals that activation in the homodimer occurs in trans. 293H cells expressing the indicated mutants V5-tagged catC and FLAG-tagged catC were treated with a dose of JAB34 (10μM/1 hour) that inhibits catCS428C when expressed alone. c, Activation in the context of the heterodimer BRAF/CRAF occurs in trans. 293H cells co-expressing FLAG-tagged wild-type BRAF and V5- tagged kinase-dead catC (catCD486N) (lanes 3,4) or JAB34-sensitive/kinase-dead catC (catCS428C/D486N) (lanes 5,6) treated with vehicle or 10μM JAB34 for 1 hour. d, HT-29 cells (colorectal – BRAFV600E) were transfected with EGFP or HA-tagged N-RASG12V and treated with PLX4720 (1μM/1hour). Lysates were blotted for pMEK and pERK.
Transactivation from CRAF to BRAF can occur as well. JAB34 activated ERK signaling in cells coexpressing FLAG-BRAF with V5-catCS428C/D486N (Fig. 4c). Finally, JAB34 induced ERK activation in cells coexpressing full-length V5-CRAFS428C/D486N and wild-type FLAG-CRAF, confirming that our model is valid in the context of full-length CRAF (Supplementary Fig. 12).
Thus, activation of RAF by ATP-competitive inhibitors can be explained by transactivation: binding of drug to one RAF in the dimer activates the other. This is consistent with the enhancement of induction by active RAS, which promotes homo- and hetero-dimerization of BRAF and CRAF10,12. Our model suggests that transactivation will be dependent on formation of RAF dimers. A side-to-side dimer of the kinase domain is observed in crystal structures of BRAF11 and the residues at the dimer interface are conserved in all RAF isoforms. Based on the BRAF crystal structures, we identified a conserved Arg (R509) at the center of the dimer interface. Structural analysis predicts that mutation of R509 will diminish contacts between the two interacting proteins and reduce dimer formation, as also recently reported19. In that study, mutation of BRAF at R509 to Histidine resulted in dramatic loss of activity. The corresponding mutation in catC (R401H) results in severe loss of both expression and activity (DNS). We therefore mutated R401 to Alanine in V5-catCS428C and FLAG-catC. This mutation diminished dimerization (Supplementary Fig. 13), but retained expression and activity. In cells coexpressing these mutants, JAB34 failed to induce ERK signaling (Fig. 4b, lanes 7-8). Thus, a mutation that affects dimerization prevents transactivation.
The transactivation model explains the observation that inhibitors of RAF activate ERK signaling at low concentrations, but inhibit at higher concentrations in BRAFwild-type cells. Binding of an ATP-competitive inhibitor to one protomer within a RAF dimer results in both abolition of the catalytic activity of the inhibitor-bound RAF and transactivation of the other. Transactivation of RAF homo- and heterodimers is therefore responsible for induction of MEK/ERK phosphorylation by RAF inhibitors in cells with wild-type BRAF. Our model explains the paradoxical phenomenon of ERK activation by RAF inhibitors, previously reported by others20-22. Other kinases that exist in dimeric or multimeric complexes may behave in a similar manner. Recently, another model to explain these phenomena has been proposed23. They report that only selective BRAF inhibitors activate CRAF and ERK signaling, whereas pan-RAF inhibitors do not. Our data that all RAF inhibitors activate ERK signaling at low concentrations, that the phenomenon occurs in BRAF-null cells and that binding to CRAF activates CRAF and BRAF-dependent ERK signaling render that model unlikely.
Nevertheless, the clinical utility of these inhibitors depends on their inhibition of ERK signaling in tumor cells with BRAFV600E. Since transactivation of wild-type RAF requires dimerization and depends on RAS activity, we hypothesized that the levels of RAS activity in BRAFV600E mutant tumors may not be sufficient to support transactivation. If so, activation of RAS in BRAFV600E cells should prevent inhibition of ERK signaling by RAF inhibitors. In 293H cells overexpressing BRAFV600E and in HT29 tumor cells with endogenous BRAFV600E, ERK signaling was inhibited by either PLX or a MEK inhibitor. In contrast, when mutant RAS was coexpressed with BRAFV600E in either cell, ERK signaling became resistant to PLX, but remained sensitive to the MEK inhibitor (Fig. 4d, Supplementary Fig. 14a, b).
The data are consistent with the idea that RAF inhibitors suppress ERK signaling in BRAFV600E tumors because the level of RAS activation in these cells is insufficient to support transactivation of wild-type RAF and inhibition of BRAFV600E activity becomes the dominant effect of the drug. The findings suggest that increases in RAS activation or RAF dimerization may be sufficient to cause drug resistance.
The clinical implications of these findings are profound. BRAFV600E tumors and some with RAS mutation are dependent on ERK signaling. However, in clinic, a MEK inhibitor had only a 12% response rate in melanomas with BRAF mutation24. MEK inhibitors block ERK signaling in all tumor and normal cells and the dose of the drug that can be administered is limited by toxicity. RAF inhibitors and MEK inhibitors might have been expected to have similar biologic effects. Our findings show otherwise. RAF inhibitors will be useful for the treatment of tumors driven by BRAFV600E, but could have deleterious effects in some contexts due to ERK activation. However, the absence of ERK inhibition in normal cells may allow administration of high doses of RAF inhibitors and thus more complete inhibition of ERK signaling in BRAFV600E tumors, than is possible with MEK inhibitors.
The recent phase I clinical trial of PLX4032 in metastatic melanoma strikingly confirmed these predictions4-5. High serum levels of drug were achieved with modest toxicity and resulted in profound inhibition of ERK signaling in tumors. Tumor regression was observed in more than 90% of patients with BRAFV600E mutation, with 64% achieving a partial response by RECIST criteria. We believe that the remarkable activity of this drug, compared to that of MEK inhibitors, is due to its ability to inhibit ERK signaling in tumors more completely because of the absence of ERK inhibition in normal tissue.
Resistance to PLX4032 does develop, with a median time to disease progression of 8-9 months5. Potential mechanisms include gatekeeper mutations in BRAF and activating mutations in parallel signaling pathways. Our results suggest the possibility of novel mechanisms as well. Lesions that activate RAS or, as recently reported, overexpression of wild type RAF isoforms25 could result in inability of RAF inhibitors to suppress ERK signaling in the tumor and thus lead to resistance.
Methods Summary
Compounds and cell culture
PLX4032 and PLX4720 were obtained from Plexxikon, Inc. PD0325901 was synthesized in the MSKCC Organic Synthesis Core Facility by Dr. Ouathek Ouerfelli. Sorafenib was synthesized using published procedures26. JAB13, JAB34 were synthesized as previously described17. All other drugs were obtained from Calbiochem. Drugs were dissolved in DMSO and stored at −20°C. Cells were maintained in either DMEM or RPMI, supplemented with 2mM glutamine, antibiotics, and 10% fetal bovine serum. Wild-type, BRAF (−/−) and CRAF (−/−) MEFs were kindly provided by Dr. Manuela Baccarini, University of Vienna, Austria. 293H cells were from Invitrogen. All other cell lines were from ATCC.
Antibodies
Western blot analysis was performed as described1. The following antibodies were used: p217/p221MEK (pMEK), p202/p204ERK (pERK), p338CRAF, p380RSK, p573RSK, MEK, ERK, myc-tag (Cell Signaling), p621CRAF, V5-tag (Invitrogen), ARAF, BRAF (Santa Cruz Biotechnology), FLAG-tag (Sigma), CRAF (BD Transduction Laboratories), HA-tag (Covance). For immunoprecipitations of tagged proteins, the following reagents were used: Anti-V5 agarose affinity gel, Anti-FLAG M2 affinity gel, anti-c-myc agarose affinity gel (all from Sigma).
Plasmids
Plasmids encoding HA-tagged wild-type and mutant N-RAS were obtained from Biomyx. Plasmids for wild-type BRAF and BRAFV600E were kindly provided by Dr. Walter Kolch, (University of Glasgow, UK), and were used as template to create FLAG-tagged constructs. All other plasmids were created using standard cloning methods, with pcDNA3.1 (Invitrogen) as a vector. Mutations were introduced using site-directed Mutagenesis Kit (Stratagene). The catalytic domain of CRAF (catC) was created by truncating the first 305 amino-acids of CRAF.
Kinase assays
RAF kinase assays were conducted in the presence of 100μM ATP, at 30°C for 20 minutes. Recombinant inactive K97R MEK (MIllipore) was used as a substrate and kinase activity was estimated by immunoblotting for pMEK.
Transfections
Cells were seeded at 35mm or 100mm plates and transfected the following day using Lipofectamine 2000 (Invitrogen).
Methods
Compounds and cell culture
PLX4032 and PLX4720 were obtained from Plexxikon, Inc. PD0325901 was synthesized in the MSKCC Organic Synthesis Core Facility by Dr. Ouathek Ouerfelli. Sorafenib was synthesized using published procedures26. JAB13, JAB34 were synthesized as previously described17. All other drugs were obtained from Calbiochem. Drugs were dissolved in DMSO to yield 10 mM stock and stored at −20°C. Cells were maintained in DMEM (MEFs, 293H, NIH3T3 and Hela) or RPMI (all other cell lines), supplemented with 2mM glutamine, antibiotics, and 10% fetal bovine serum. Wild-type, BRAF (−/−) and CRAF (−/−) MEFs were kindly provided by Dr. Manuela Baccarini, University of Vienna, Austria. 293H cells were from Invitrogen. All other cell lines were from ATCC.
Antibodies
Western blot analysis was performed as described1. The following antibodies were used: p217/p221MEK (pMEK), p202/p204ERK (pERK), p338CRAF, p380RSK, p573RSK, MEK, ERK, myc-tag (Cell Signaling), p621CRAF, V5-tag (Invitrogen), ARAF, BRAF (Santa Cruz Biotechnology), FLAG-tag (Sigma), CRAF (BD Transduction Laboratories), HA-tag (Covance). For immunoprecipitations of tagged proteins, the following reagents were used: Anti-V5 agarose affinity gel, Anti-FLAG M2 affinity gel, anti-c-myc agarose affinity gel (all from Sigma).
Plasmids
Plasmids encoding HA-tagged wild-type and mutant N-RAS were obtained from Biomyx. Plasmids expressing myc-tagged wild-type BRAF and BRAFV600E were kindly provided by Dr. Walter Kolch, (University of Glasgow, UK), and were used as template to create FLAG-tagged constructs. All other plasmids were created using standard cloning methods, with pcDNA3.1 (Invitrogen) as a vector. Mutations were introduced using site-directed Mutagenesis Kit (Stratagene). The catalytic domain of CRAF (catC) was created by truncating the first 305 amino-acids of CRAF.
Immunoprecipitations and Kinase assays
Cells were lysed in lysis buffer (50 mM Tris (pH 7.5), 1% NP40, 150 mM NaCl, 10% glycerol, 1mM EDTA, supplemented with protease and phosphatase inhibitor cocktail tablets (Roche). Immunoprecipitations were carried out at 4°C for 4 hours, followed by 3 washes with lysis buffer and, in cases of subsequent kinase assay, one extra wash with kinase buffer (25mM Tris, pH 7.5, 10mM MgCl2). RAF kinase assays were conducted in the presence of 100μM ATP, at 30°C for 20 minutes. Recombinant inactive K97R MEK (MIllipore) was used as a substrate and the reaction was terminated with the addition of sample buffer and boiling. Kinase activity was estimated by immunoblotting for pMEK.
Transfections
Cells were seeded at 35mm or 100mm plates and transfected the following day using Lipofectamine 2000 (Invitrogen). 24 hours later, cells were collected for subsequent analysis.
Supplementary Material
Acknowledgements
We are grateful to Dr. Walter Kolch for the BRAF plasmids and Dr. Manuella Baccarini for the RAF knock-out MEFs. We thank Jimmy Blair for synthesis of JAB compounds and Arvin Dar, Sarat Chandarlapaty and David Solit for helpful discussions. This work has been funded by the Melanoma Research Alliance, the Starr Cancer Consortium, an NIH/NCI P01 grant (1P01CA129243-02) and by Joan’s Legacy: United Against Lung Cancer Foundation (PIP, NR). KMS would like to thank NIH-2R01EB001987, The Children’s Tumor Foundation and the Waxman Foundation for funding.
Footnotes
Author contributions: PIP and CZ designed research, performed experiments, analyzed data, and co-wrote the paper. GB provided reagents, analyzed data and co-wrote the paper. KMS and NR designed research, analyzed experiments and co-wrote the paper.
REFERENCES
- 1.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDermott U, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–41. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellbrock C, et al. V599EB-RAF is an oncogene in melanocytes. Cancer Res. 2004;64:2338–42. doi: 10.1158/0008-5472.can-03-3433. [DOI] [PubMed] [Google Scholar]
- 4.Chapman P, et al. Early efficacy signal demonstrated in advanced melanoma in a phase I trial of the oncogenic BRAF-selective inhibitor PLX4032 Eur. J. Cancer Suppl. 2009;7:5. [Google Scholar]
- 5.Flaherty K, et al. Phase I study of PLX4032: Proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J. Clin. Oncol. 2009;27 Abstract 9000. [Google Scholar]
- 6.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–6. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–85. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 8.Young A, et al. Ras signaling and therapies. Adv Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- 9.Konecny GE, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–9. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 10.Weber CK, Slupsky JR, Kalmes HA, Rapp UR. Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res. 2001;61:3595–8. [PubMed] [Google Scholar]
- 11.Wan PT, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 12.Rushworth LK, Hindley AD, O’Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26:2262–72. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cutler RE, Jr., Stephens RM, Saracino MR, Morrison DK. Autoregulation of the Raf-1 serine/threonine kinase. Proc Natl Acad Sci U S A. 1998;95:9214–9. doi: 10.1073/pnas.95.16.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okuzumi T, et al. Inhibitor hijacking of Akt activation. Nat Chem Biol. 2009;5:484–493. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron AJ, Escribano C, Saurin AT, Kostelecky B, Parker PJ. PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase activity. Nat Struct Mol Biol. 2009;16:624–30. doi: 10.1038/nsmb.1606. [DOI] [PubMed] [Google Scholar]
- 16.Karreth FA, DeNicola GM, Winter SP, Tuveson DA. C-Raf inhibits MAPK activation and transformation by B-Raf(V600E) Mol Cell. 2009;36:477–86. doi: 10.1016/j.molcel.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Blair JA, et al. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat Chem Biol. 2007;3:229–38. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, et al. Growth inhibition of nasopharyngeal carcinoma cells by EGF receptor tyrosine kinase inhibitors. Anticancer Res. 1999;19:919–24. [PubMed] [Google Scholar]
- 19.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–5. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 20.Hall-Jackson CA, et al. Paradoxical activation of Raf by a novel Raf inhibitor. Chem Biol. 1999;6:559–68. doi: 10.1016/s1074-5521(99)80088-x. [DOI] [PubMed] [Google Scholar]
- 21.King AJ, et al. Demonstration of a genetic therapeutic index for tumors expressing oncogenic BRAF by the kinase inhibitor SB-590885. Cancer Res. 2006;66:11100–5. doi: 10.1158/0008-5472.CAN-06-2554. [DOI] [PubMed] [Google Scholar]
- 22.Hoeflich KP, et al. Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression. Cancer Res. 2009;69:3042–51. doi: 10.1158/0008-5472.CAN-08-3563. [DOI] [PubMed] [Google Scholar]
- 23.Heidorn SJ, et al. Kinase-Dead BRAF and Oncogenic RAS Cooperate to Drive Tumor Progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dummer R, et al. AZD6244 (ARRY-142886) vs temozolomide (TMZ) in patients (pts) with advanced melanoma: An open-label, randomized, multicenter, phase II study. J. Clin. Oncol. 2008;26 Abstract 9033. [Google Scholar]
- 25.Montagut C, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–61. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankston D, et al. A Scaleable Synthesis of BAY 43-9006: A Potent Raf Kinase Inhibitor for the Treatment of Cancer. Organic Process Research and Development. 2002;6:777–781. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.