Abstract
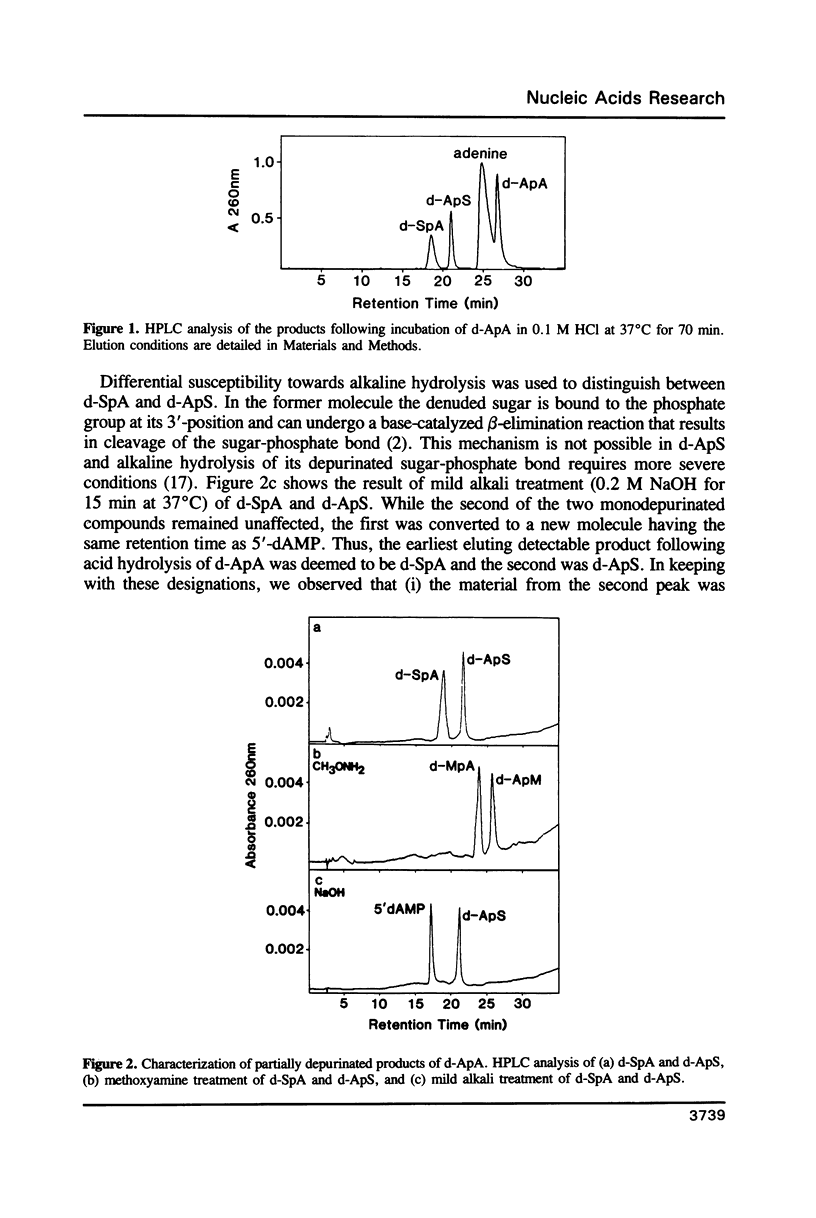
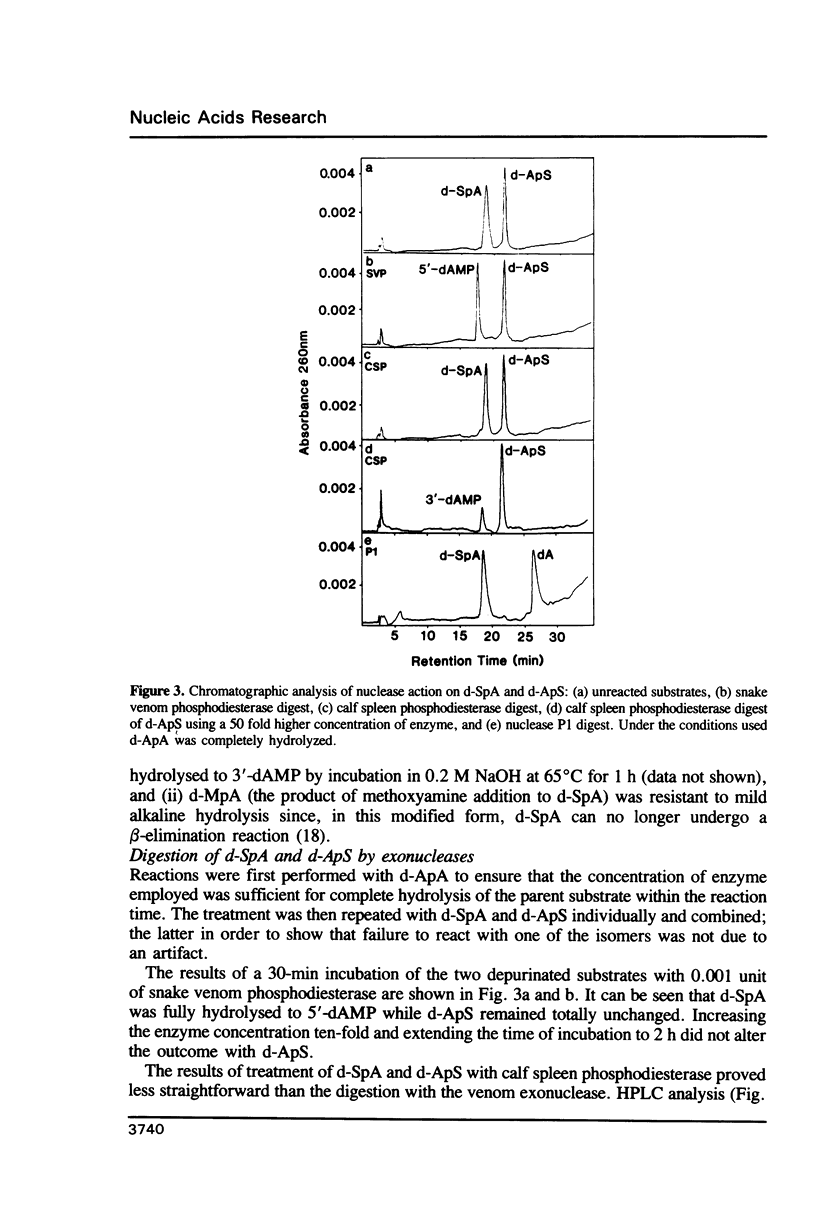
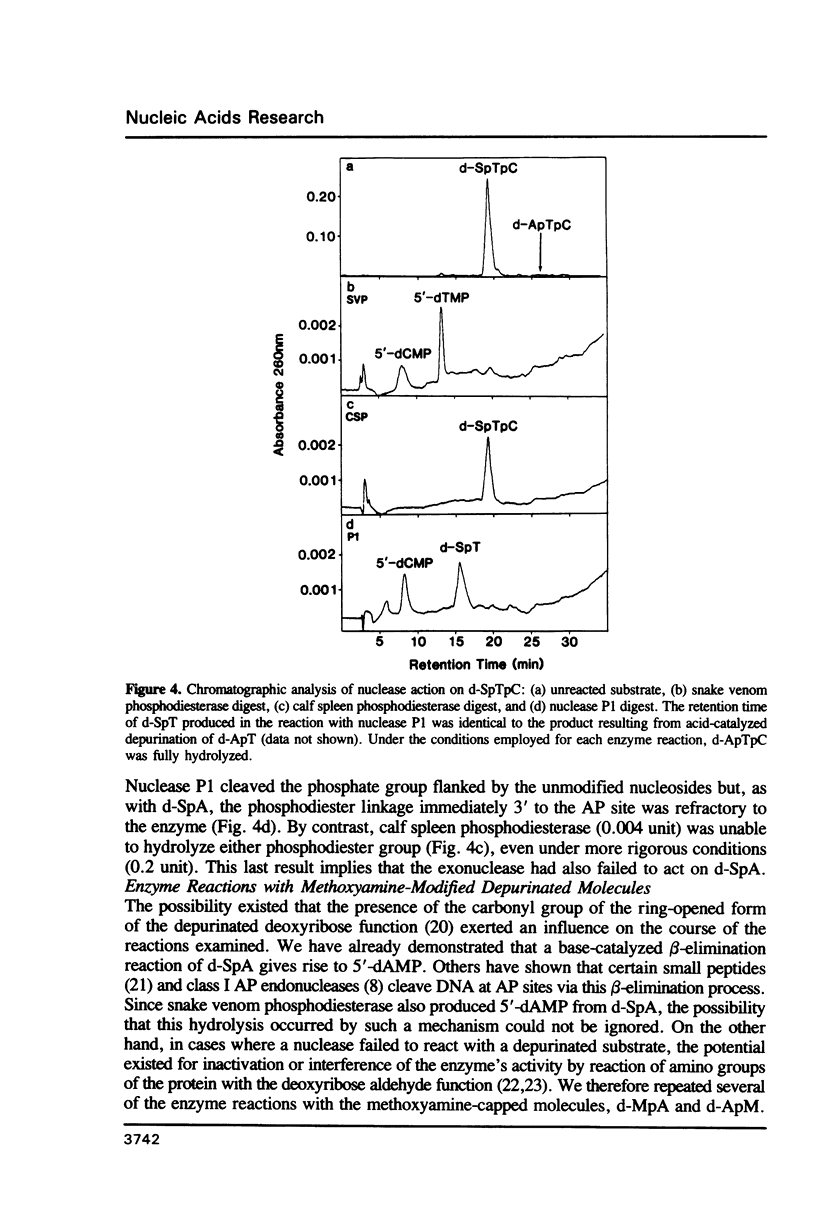
Partial depurination of d-ApA produced two UV260nm-absorbing isomers, d-SpA and d-ApS (where S represents the depurinated deoxyribose sugar), that provided simple model compounds with which to examine, by HPLC, the response of nucleases to phosphodiester bonds flanked 3' or 5' by an apurinic site. The structural identity of each compound was established by (i) reaction with methoxyamine to confirm the presence of an abasic deoxyribose group, and (ii) degradation of d-SpA under mild alkaline conditions to distinguish it from d-ApS. At an enzyme concentration which led to complete hydrolysis of d-ApA, snake venom phosphodiesterase readily cleaved d-SpA to 5'-dAMP but had no discernible effect on d-ApS. Calf spleen phosphodiesterase also failed to act on one isomer, in this instance d-SpA, but additionally reacted at a much slower rate (approximately 100 fold) with d-ApS than with d-ApA. Three single-strand specific endonucleases, nuclease P1, nuclease S1 and mung bean nuclease, all responded in an identical manner, hydrolysing d-ApS but not d-SpA. The possibility that the aldehyde group at the AP sites might be responsible for some of these observations was rejected after repeating the enzyme digestions with the methoxyamine-capped molecules and observing no differences from the reactions with d-SpA and d-ApS.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailly V., Verly W. G. Escherichia coli endonuclease III is not an endonuclease but a beta-elimination catalyst. Biochem J. 1987 Mar 1;242(2):565–572. doi: 10.1042/bj2420565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V., Verly W. G. The excision of AP sites by the 3'-5' exonuclease activity of the Klenow fragment of Escherichia coli DNA polymerase I. FEBS Lett. 1984 Dec 10;178(2):223–227. doi: 10.1016/0014-5793(84)80605-5. [DOI] [PubMed] [Google Scholar]
- Behmoaras T., Toulmé J. J., Hélène C. A tryptophan-containing peptide recognizes and cleaves DNA at apurinic sites. Nature. 1981 Aug 27;292(5826):858–859. doi: 10.1038/292858a0. [DOI] [PubMed] [Google Scholar]
- Beranek D. T., Weis C. C., Swenson D. H. A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis. 1980 Jul;1(7):595–606. doi: 10.1093/carcin/1.7.595. [DOI] [PubMed] [Google Scholar]
- Bernardi A., Cantoni G. L. Action of spleen exonuclease on transfer ribonucleic acid. J Biol Chem. 1969 Mar 25;244(6):1468–1476. [PubMed] [Google Scholar]
- Coombs M. M., Livingston D. C. Reaction of apurinic acid with aldehyde reagents. Biochim Biophys Acta. 1969 Jan 21;174(1):161–173. doi: 10.1016/0005-2787(69)90239-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Goffin C., Bailly V., Verly W. G. Nicks 3' or 5' to AP sites or to mispaired bases, and one-nucleotide gaps can be sealed by T4 DNA ligase. Nucleic Acids Res. 1987 Nov 11;15(21):8755–8771. doi: 10.1093/nar/15.21.8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondal-Zocchi G., Verly W. G. Deoxyribonuclease IV from rat liver chromatin and the excision of apurinic sites from depurinated DNA. Biochem J. 1985 Jan 15;225(2):535–542. doi: 10.1042/bj2250535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman L., Grafstrom R. AP sites and AP endonucleases. Biochimie. 1982 Aug-Sep;64(8-9):577–580. doi: 10.1016/s0300-9084(82)80090-4. [DOI] [PubMed] [Google Scholar]
- Jennette K. W., Jeffrey A. M., Blobstein S. H., Beland F. A., Harvey R. G., Weinstein I. B. Nucleoside adducts from the in vitro reaction of benzo[a]pyrene-7,8-dihydrodiol 9,10-oxide or benzo[a]pyrene 4,5-oxide with nucleic acids. Biochemistry. 1977 Mar 8;16(5):932–938. doi: 10.1021/bi00624a019. [DOI] [PubMed] [Google Scholar]
- LIVINGSTON D. C. DEGRADATION OF APURINIC ACID BY CONDENSATION WITH ALDEHYDE REAGENTS. Biochim Biophys Acta. 1964 Jul 22;87:538–540. doi: 10.1016/0926-6550(64)90136-7. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Liuzzi M., Talpaert-Borlé M. A new approach to the study of the base-excision repair pathway using methoxyamine. J Biol Chem. 1985 May 10;260(9):5252–5258. [PubMed] [Google Scholar]
- Loeb L. A., Preston B. D. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Margison G. P., O'Connor P. J. Role of apurinic sites in the resistance of methylated oligodeoxyribonucleotides to degradation by spleen exonuclease. Biochem J. 1975 Nov;151(2):249–256. doi: 10.1042/bj1510249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T., Brutlag D. Addition of homopolymers to the 3'-ends of duplex DNA with terminal transferase. Methods Enzymol. 1979;68:41–50. doi: 10.1016/0076-6879(79)68005-9. [DOI] [PubMed] [Google Scholar]
- Niewiarowski W., Uznanski B. Substrate specificity and stereospecificity of calf spleen phosphodiesterase towards deoxyribonucleosidyl 3'-(4-nitrophenyl phosphates) and phosphorothioates. Eur J Biochem. 1985 Nov 15;153(1):145–153. doi: 10.1111/j.1432-1033.1985.tb09280.x. [DOI] [PubMed] [Google Scholar]
- Ogilvie K. K., Hruska F. H. Affect of spleen and snake venom phosphodiesterases on nucleotides containing nucleosides in the syn conformation. Biochem Biophys Res Commun. 1976 Jan 26;68(2):375–378. doi: 10.1016/0006-291x(76)91155-4. [DOI] [PubMed] [Google Scholar]
- RAZZELL W. E., KHORANA H. G. Studies on polynucleotides. III. Enzymic degradation; substrate specificity and properties of snake venom phosphodiesterase. J Biol Chem. 1959 Aug;234(8):2105–2113. [PubMed] [Google Scholar]
- Randerath K., Reddy M. V., Gupta R. C. 32P-labeling test for DNA damage. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6126–6129. doi: 10.1073/pnas.78.10.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. V., Randerath K. Nuclease P1-mediated enhancement of sensitivity of 32P-postlabeling test for structurally diverse DNA adducts. Carcinogenesis. 1986 Sep;7(9):1543–1551. doi: 10.1093/carcin/7.9.1543. [DOI] [PubMed] [Google Scholar]
- SETLOW R. B., CARRIER W. L., BOLLUM F. J. NUCLEASE-RESISTANT SEQUENCES IN ULTRAVIOLET-IRRADIATED DEOXYRIBONUCLEIC ACID. Biochim Biophys Acta. 1964 Nov 15;91:446–461. doi: 10.1016/0926-6550(64)90075-1. [DOI] [PubMed] [Google Scholar]
- Singer B., Kröger M., Carrano M. O2- and O4-alkyl pyrimidine nucleosides: stability of the glycosyl bond and of the alkyl group as a function of pH. Biochemistry. 1978 Apr 4;17(7):1246–1250. doi: 10.1021/bi00600a018. [DOI] [PubMed] [Google Scholar]
- Stuart G. R., Chambers R. W. Synthesis and properties of oligodeoxynucleotides with an AP site at a preselected position. Nucleic Acids Res. 1987 Sep 25;15(18):7451–7462. doi: 10.1093/nar/15.18.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talpaert-Borlé M., Liuzzi M. Reaction of apurinic/apyrimidinic sites with [14C]methoxyamine. A method for the quantitative assay of AP sites in DNA. Biochim Biophys Acta. 1983 Sep 9;740(4):410–416. doi: 10.1016/0167-4781(83)90089-1. [DOI] [PubMed] [Google Scholar]
- Tomasz M., Chowdary D., Lipman R., Shimotakahara S., Veiro D., Walker V., Verdine G. L. Reaction of DNA with chemically or enzymatically activated mitomycin C: isolation and structure of the major covalent adduct. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6702–6706. doi: 10.1073/pnas.83.18.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIGLER P. W. The kinetics of snake venom phosphodiesterase, with a new type of substrate, 3-pyridyl thymidine 5'-phosphate. J Biol Chem. 1963 May;238:1767–1771. [PubMed] [Google Scholar]
- Zemlicka J. Synthesis of dicytidylyl-(3'-5')-1,2-di(adenosin-N6-yl)ethane and dicytidylyl-(3'-5')-1,4-di(adenosin-N6-yl)butane: covalently joined terminals of two transfer ribonucleic acids and their behavior toward snake venom phosphodiesterase. Biochemistry. 1980 Jan 8;19(1):163–168. doi: 10.1021/bi00542a025. [DOI] [PubMed] [Google Scholar]


