Abstract
Unconventional antigens, such as metals, stimulate T cells in a very specific manner. To delineate the binding landscape for metal-specific T cell recognition, alanine screens were performed on a set of beryllium (Be)-specific T cell receptors (TCR) derived from the lung of a chronic beryllium disease patient. These TCRs are HLA-DP2-restricted and express nearly identical TCR Vβ5.1 chains coupled with different TCR α-chains. Site-specific mutagenesis of all amino acids comprising the complementarity determining regions of the TCRA and TCRB genes showed a dominant role for Vβ5.1 residues in Be recognition, with little contribution from the TCR α-chain. Solvent-exposed residues along the α-helices of the HLA-DP2 α- and β-chains were also mutated to alanine. Two β-chain residues, located near the proposed Be binding site of HLA-DP2, played a dominant role in T cell recognition with no contribution from the HLA-DP2 α-chain. These findings suggest that Be-specific T cells recognize antigen using an unconventional binding topology, with the majority of interactions contributed by TCR Vβ5.1 residues and the HLA-DP2 β1-chain. Thus, unusual docking topologies are not exclusively used by autoreactive T cells, but also for the recognition of unconventional metal antigens, such as Be.
Keywords: Human, T cells, MHC, Lung, T cell receptors
Introduction
A conventional immune response is initiated by αβ T cell receptor (TCR) recognition of a major histocompatibility complex (MHC) molecule with a bound peptide derived from a foreign protein (1). Immune-mediated clearance of unconventional antigens, such as metals, is poorly understood, especially in the context of interactions required for TCR recognition. Yet, understanding these interactions are critical since metal-induced diseases, such as chronic beryllium disease (CBD) and nickel-induced contact hypersensitivity, occur in a significant number of exposed individuals, leading to morbidity and, in the case of CBD, mortality. Metal binding to an MHC/peptide complex adds a third component of complexity for recognition by T cells, with all three components required to elicit T cell activation. For example, in the case of nickel-induced hypersensitivity, the nickel-specific TCR interacts with an antigenic complex formed by nickel, an unknown peptide, and HLA-DR52c (2).
Chronic beryllium disease (CBD) is a granulomatous lung disorder characterized by the accumulation of beryllium (Be)-responsive CD4+ T cells in the lung (3, 4). These Be-specific T cells express a polarized Th1 phenotype and recognize Be in an HLA-DP restricted manner (5, 6). Genetic susceptibility to Be-induced disease has been linked to MHC class II (MHCII) molecules. In particular, HLA-DP alleles possessing a glutamic acid at the 69th position of the β-chain (βGlu69) are strongly linked to disease susceptibility (7–11). The HLA-DP molecules that present Be to T cells match those implicated in the genetic susceptibility, suggesting that the HLA contribution to disease is based on the ability of those molecules to bind and present Be to T cells (6, 12, 13). We have recently crystallized HLA-DP2, and its unique structural features, including surface exposure of βGlu69, provide an explanation of the genetic linkage between βGlu69-containing HLA-DP alleles and Be-induced disease (14).
Of the approximately 20 TCR-pMHC structures solved to date, analyses show a distinction in T cell recognition of foreign- versus self-derived antigens. The TCR generally uses a conserved binding topology for recognition of foreign-derived antigens, including a diagonal orientation, conserved polarity, and a centered position over the peptide/MHC complex (15, 16). This conserved polarity places the TCR β-chain in close contact with the α1 helix of MHC and the TCR α-chain over the α2 helix of MHCI or β1 helix of MHCII. Essential contacts are made with both α-helices of the MHC, generating optimal binding interactions between the T cell and the antigen-presenting cell. In contrast, unconventional binding interactions have been reported for the majority of human autoimmune complexes solved to date (17–19). The docking footprint for these TCRs is shifted laterally or tilted over the peptide/MHC complex, creating suboptimal binding interactions. It has been postulated that these unusual binding features may allow these low affinity self-reactive T cells to escape negative selection in the thymus (20). In this regard, it remains unknown how Be-specific TCRs recognize this unconventional antigen in the context of HLA-DP2/peptide.
Using a set of Be-specific Vβ5+ TCRs derived from the lung of a subject with CBD, we show that these TCRs recognize the HLA-DP2/peptide/Be complex using an unusual binding topology. The atypical docking footprint for recognition of the Be antigen was characterized by residues in the Vβ5 chain playing a dominant role in Be antigen recognition, with no contribution from germline-derived CDR1 and CDR2 residues of the TCR α-chain. Selected mutations of solvent-exposed residues of the HLA-DP2 α-helices confirmed these findings and showed little contribution from the α1 helix of HLA-DP2. Taken together, our findings suggest that the Be-specific TCRs are centered directly over the putative Be binding site of HLA-DP2, with residues of the TCR Vβ domain and the β1 helix of DP2 contributing to binding and recognition of the Be antigen.
Materials and Methods
Study population
Experiments performed in the current study used CD4+ T cells derived from the lung of patients with CBD. The diagnosis of CBD was established using previously defined criteria (11, 21), including a history of Be exposure, the presence of granulomatous inflammation on lung biopsy, and a positive proliferative response of BAL T cells to BeSO4 in vitro. Informed consent was obtained from these subjects, and the protocol was approved by the Human Subject Institutional Review Board at the University of Colorado Denver and National Jewish Health.
Immunofluorescence staining and analysis of intracellular cytokine expression
BAL-derived, Be-specific T cell lines were derived as previously described (6, 12) and maintained in culture by cycles of restimulation (every 2–3 weeks) with 100 µM BeSO4-pulsed autologous EBV-transformed lymphoblastoid cell lines (LCLs) and further expanded with rIL-2. For stimulation of cytokine secretion, the CD4+ T cell line (2.5 × 105 cells) was stimulated with either medium or 100 µM BeSO4 using autologous LCLs (2.5 × 105 cells) as antigen-presenting cells. Cells were incubated for 6 hours at 37° C in a humidified 5% CO2 atmosphere with 10 µg/ml brefeldin A added after the first hour of stimulation (5, 22). After stimulation, cells were washed and stained with mAbs directed against CD3, CD4 (all from BD Bioscience) and anti-TCR Vβ5.1 (clone LC4) (23). Cells were fixed, permeabilized and stained with an anti- IFN-γ mAb (BD Bioscience) for 30 minutes at 4°C. The lymphocyte population was identified using forward and 90° light scatter patterns, and fluorescence intensity was analyzed using a FACSCaliber flow cytometer (BD Biosciences) (5, 22). Data files were analyzed using FlowJo software (Tree Star).
Analysis of TCRBV5S1 gene expression of BAL CD4+ T cell clones
Since the majority of Be-responsive CD4+ T cells in this line expressed Vβ5.1, we sorted CD4+Vβ5.1+ T cells using a FACSAria flow cytometer (BD Immunocytometry Systems) and cloned by limiting dilution. Cells from confluent cultures were harvested, and RNA was isolated using a QIAGEN RNeasy kit according to manufacturer’s instructions. cDNA was prepared, and the TCRB gene fragments were amplified using a TCRBV5S1 primer (5’-ATACTTCAGTGAGACACAGAGAAAC-3’) and TCRBC primer (5’-TTCTGATGGCTCAAACAC-3’). PCR products were purified using a DNA binding membrane spin column (QIAGEN) and sequenced using a TCRBC sequencing primer (5’-CGACCTCGGGTGGGAACA-3’). The corresponding TCRAV gene segment for each CD4+ T cell clone was determined using a complete set of TCRAV primers. This method was also used to characterize the T cell clone designated 1041-3.3, which expresses a Vα1/Vβ3 TCR and was derived from lung T cells isolated from CBD subject 1041. The dengue virus-specific, Vα11/Vβ23+ TCR was derived from a T cell clone, designated JK34, which was kindly provided by Dr. Ennis’ laboratory (24).
Cloning of TCR and MHC constructs into retroviral vectors
Full length chimeric TCRs were cloned into a Murine Stem Cell Virus (MSCV) plasmid for retroviral transduction of murine T cell hybridoma line, 5KC (25). PCR fragments encoding the extracellular variable domains of the TCR α- and β-chains of each T cell clone were cloned into MSCV plasmids that encode either a murine Cα or Cβ domain, an internal ribosomal entry site (IRES), and GFP reporter for selection. Thus, these chimeric TCRs were composed of human variable and mouse constant domains. Although it is possible that this could influence the docking of the TCR with the MHCII molecule, this possibility is unlikely since antigen specificity of the TCR is dictated by the CDRs of the variable domain.
For the Vβ5+ Be-specific TCRs, the MSCV plasmids encoding the full length TCR constructs, containing human variable and mouse constant domains, were used as templates to introduce single-site alanine substitutions at each CDR using the QuickChange Site-Directed Mutagenesis Kit (Stratagene). In addition, genes for human HLA-DPA1*0103 and HLA-DPB1*0201 were cloned into separate MSCV plasmids encoding GFP or hygromycin for selection. Alanine or glutamine mutations were introduced at solvent-exposed amino acid positions within the α-helices of both the DPA and DPB genes as described above.
Expression of TCR variants on the murine T cell hybridoma
Full length TCRA and TCRB constructs were packaged as retrovirus by transient transfection of Phoenix 293T cells with MSCV plasmids (25). Phoenix cells were plated at 4.5 × 105 cells/well in 6-well plates (Corning) coated with 100 µg/ml poly D lysine (Sigma). After 24 hours, wells were washed with PBS and replenished with IMDM-glutaMAX (Invitrogen) culture medium without FBS. Cells were transfected with Lipofectamine 2000 (Invitrogen) preincubated with 6 µg MSCV and 1.5 µg pCL-Eco packaging vector, with FBS added after 4 hours at 37°C. After 24 hours, the cells were replenished with medium containing 10% FBS and incubated for 24 hours before collecting retrovirus-containing supernatants.
For expression of human TCRs, the parental TCRα−β− murine T cell hybridoma, 5KC.73.8.20, was used (26). This line was first retrovirally transduced with human CD4 using a Ψ2 CD4 packaging cell line (27). Transduced T cell hybridomas were stained with an anti-human CD4 mAb (BD Biosciences) and cells expressing the highest levels of human CD4 (designated 5KC-9C6) were used in the current study. 5KC-9C6 cells were transduced with filtered viral supernatant using a spin-infection protocol as previously described (25). Positively-staining cells were sorted using either a MoFLo (Cytomation) or FACSAria flow cytometer (BD Biosciences).
Fibroblast expression of HLA-DP2 variants
Wild-type and HLA-DP2 variants were packaged as retrovirus, as described for the TCR variants, and the retrovirus was used to transduce the murine fibroblast line, B6DK10, which had previously been transfected with murine B7 and ICAM. Briefly, fibroblasts (5 × 105) were adhered to a 6-well plate (Corning) in cell culture medium (IMDM-Glutamax, Invitrogen) and 10% FBS for 1 hr at 37°C. Media was removed and 1 ml of filtered viral supernatant and 8 µg/ml polybrene (Millipore) were added. For the spin-infection, plates were centrifuged at 1200 rpm for 1 hr at room temperature and incubated for 2–4 hrs at 37°C. Viral supernatant was removed by washing with PBS and cultures were replenished with medium containing FBS. After 2–4 days, cell surface expression of HLA-DP was measured using a conformation-dependent anti-HLA-DP mAb, B7.21 (28). Fibroblasts were stained and sorted as described above, and data analysis was performed using FlowJo software (Tree Star).
T cell hybridoma activation assays
T cell hybridoma cells (1 × 105) and murine fibroblasts transfected with wild-type or variant HLA-DP2 were incubated overnight at 37°C with various concentrations of BeSO4 or dengue virus peptide. Supernatants were harvested, and IL-2 measured using the mouse IL-2 Ready-Set-Go ELISA kit (eBioscience). Activation curves were generated by plotting percentage of maximal IL-2 release, (A450 (sample) - A450 (control)) / (Max A450 (sample) - A450 (control)) × 100, against antigen concentration. The concentration of BeSO4 or dengue virus peptide required for half-maximal IL-2 release, or EC50 value, was determined using non-linear regression (sigmoidal-fit, GraphPad Prism) of the activation curves. Assays utilizing the murine fibroblast line B6DK10 expressing wild-type or HLA-DP2 variants required 1–2 × 105 antigen-presenting cells per well, whereas for the HLA-DP2 transfected DAP.3L fibroblasts, 5 × 104 cells were used.
Results
Characterization of Be-responsive T cell clones derived from the lung of a CBD patient
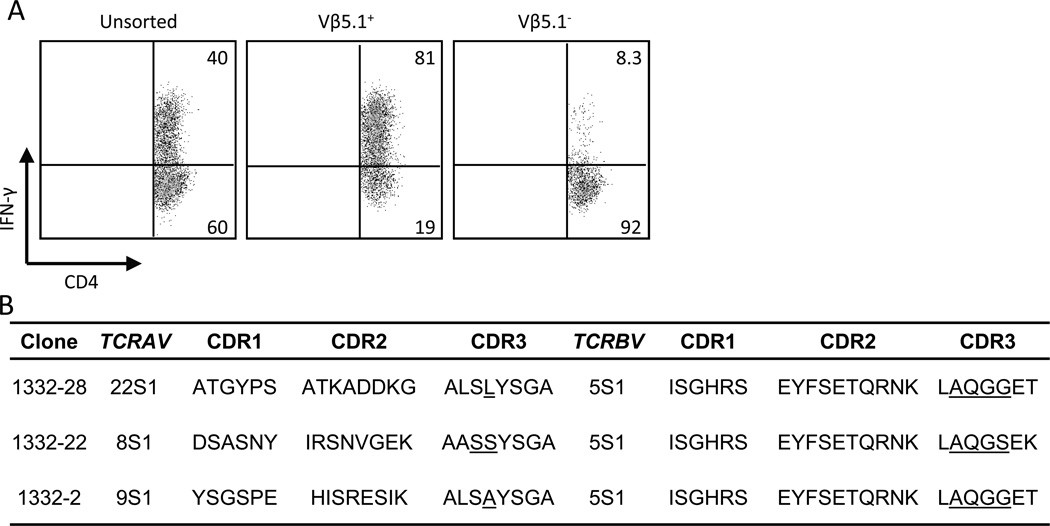
We have previously identified a large number of Be-responsive CD4+ T cells compartmentalized to the bronchoalveolar lavage (BAL) of CBD patients (5). With repeated cycles of BeSO4 stimulation, a Be-responsive CD4+ T cell line was developed from one CBD patient (designated 1332). Using intracellular cytokine staining as a measure of T cell response, 40% of the CD4+ T cells were Be-responsive and expressed IFN-γ (Fig. 1A). Interestingly, the predominant TCR Vβ expansion comprising this line was Vβ5.1 (data not shown), and the vast majority of Vβ5.1-expressing CD4+ T cells were Be-specific (81% in the density plot shown compared to 8.3% in the Vβ5.1− T cell subset) (Fig. 1A). In order to determine whether this expansion of Vβ5.1-expressing CD4+ T cells was composed of a single T cell clone or related clones, we sorted CD4+Vβ5.1+ T cells and cloned via limiting dilution. RNA and cDNA were prepared from each clonal isolate, and the TCRB genes were amplified and sequenced.
FIGURE 1.
Deduced amino acid sequences of a set of Be-responsive, Vβ5.1+CD4+ T cells. A, Intracellular IFN-γ staining of a Be-responsive T cell line derived from the BAL of a CBD patient is shown. The cells were stimulated with either medium alone or BeSO4 and subsequently stained for CD4, Vβ5.1 and intracellular IFN-γ expression. The number in the upper right quadrant of each density plot indicates the percentage of CD4+ T cells expressing IFN-γ. B, Analysis of deduced TCRA and TCRB CDR1, CDR2 and CDR3 amino acid sequences expressed by the Be-responsive T cell clones derived from the T cell line in A. Underlined amino acids indicate those encoded by non-germline nucleotides. These sequence data are available from GenBank under accession numbers JF834314 - JF834319 http://www.ncbi.nlm.nih.gov/genbank/.
We identified three Be-specific T cell clones that express TCRBV5S1 and nearly identical CDR3 sequences (Fig. 1B). Two of the T cell clones, 1332-28 and 1332-2, utilized different nucleotide combinations in the nBDn region and TCRBJ2S5 to encode identical amino acids. The third T cell clone, 1332-22, utilized TCRBJ1S4 resulting in a two amino acid difference in the TCRB gene sequence (serine at position 99 and lysine at position 101). Interestingly, the three T cell clones utilized different TCRAV genes coupled with TCRAJ28. As a result, these TCRs have different germline-derived CDR1α and CDR2α despite encoding nearly identical somatically-rearranged CDR3α (except for two amino acid differences; one encoded by germline and the other by an N-region modification) (Fig. 1B). These findings raise the possibility that the TCR Vβ domain of the Be-specific TCR is playing the dominant role in recognition of the HLA-DP2/peptide/Be complex, whereas the contribution of the Vα domain is limited to CDR3α.
Generation of T cell hybridomas expressing Be-specific TCRs
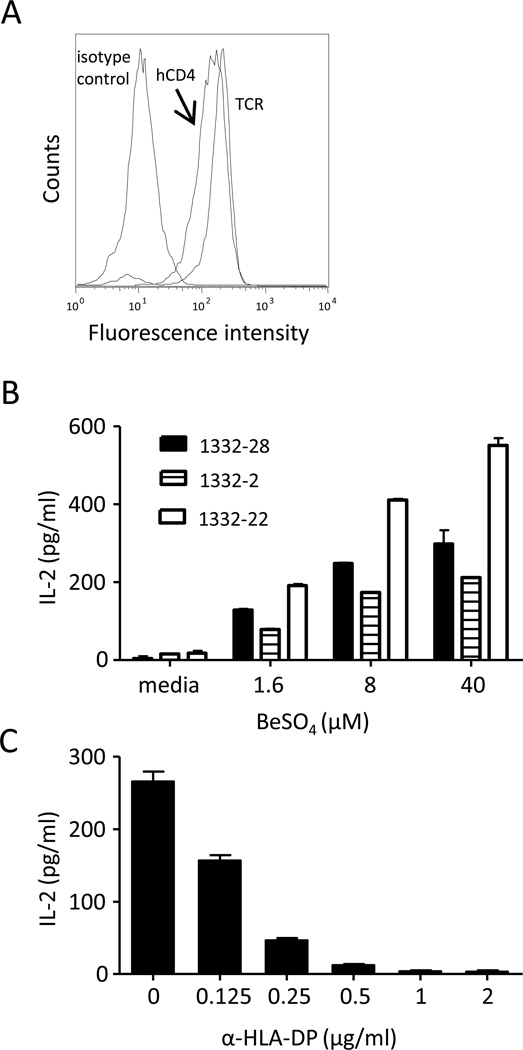
Because it is difficult to perpetuate BAL-derived, Be-specific T cell clones indefinitely, we generated T cell hybridomas that express the TCR of these Be-specific T cell clones. Hybridomas have several advantages compared to the T cell clones from which their TCRs are derived. They can be repeatedly grown to large numbers, and their stimulation is independent of the functional state of the parental T cell clone. For 1332-28, -22, and -2, gene fragments of the variable domains of each clones’ TCRA and TCRB genes were cloned into retroviral vectors encoding murine TCR constant domains and expressed on the surface of the murine T cell hybridoma line, 5KC (25). Thus, these chimeric TCRs consist of human variable and murine constant domains. The 5KC T cell line no longer expresses murine CD4 and has been transduced with human CD4 (Fig. 2A). Fig. 2B shows the ability of these three Be-specific hybridomas to secrete IL-2 in response to various concentrations of BeSO4 presented by murine DAP3.L cells transfected with HLA-DP2. This antigen-specific response was completely blocked by the addition of an anti-HLA-DP-specific mAb (Fig. 2C).
FIGURE 2.
Characterization of Be-specific T cell hybridomas. A, Expression of TCR and human CD4 on the 1332-28 T cell hybridoma. Constructs encoding the variable domain of the TCR α- and β-chains of Be-specific T cell clones were transduced into the TCR− murine T cell hybridoma, 5KC. The resultant T cell hybridomas were stained with anti-human CD4 and anti-TCR mAbs and analyzed on a FACSCaliber flow cytometer. B, Stimulation of T cell hybridomas, designated 1332-28, -22, and -2, with varying BeSO4 concentrations presented by HLA-DP2-transfected murine fibroblasts. The mean ± SEM IL-2 (pg/ml) is shown, and data are representative of three independent experiments. C, Inhibition of Be-induced IL-2 secretion by the addition of various concentrations of the anti-DP mAb, B7.21, is shown.
Mapping T cell recognition of the Be antigen in the context of HLA-DP2
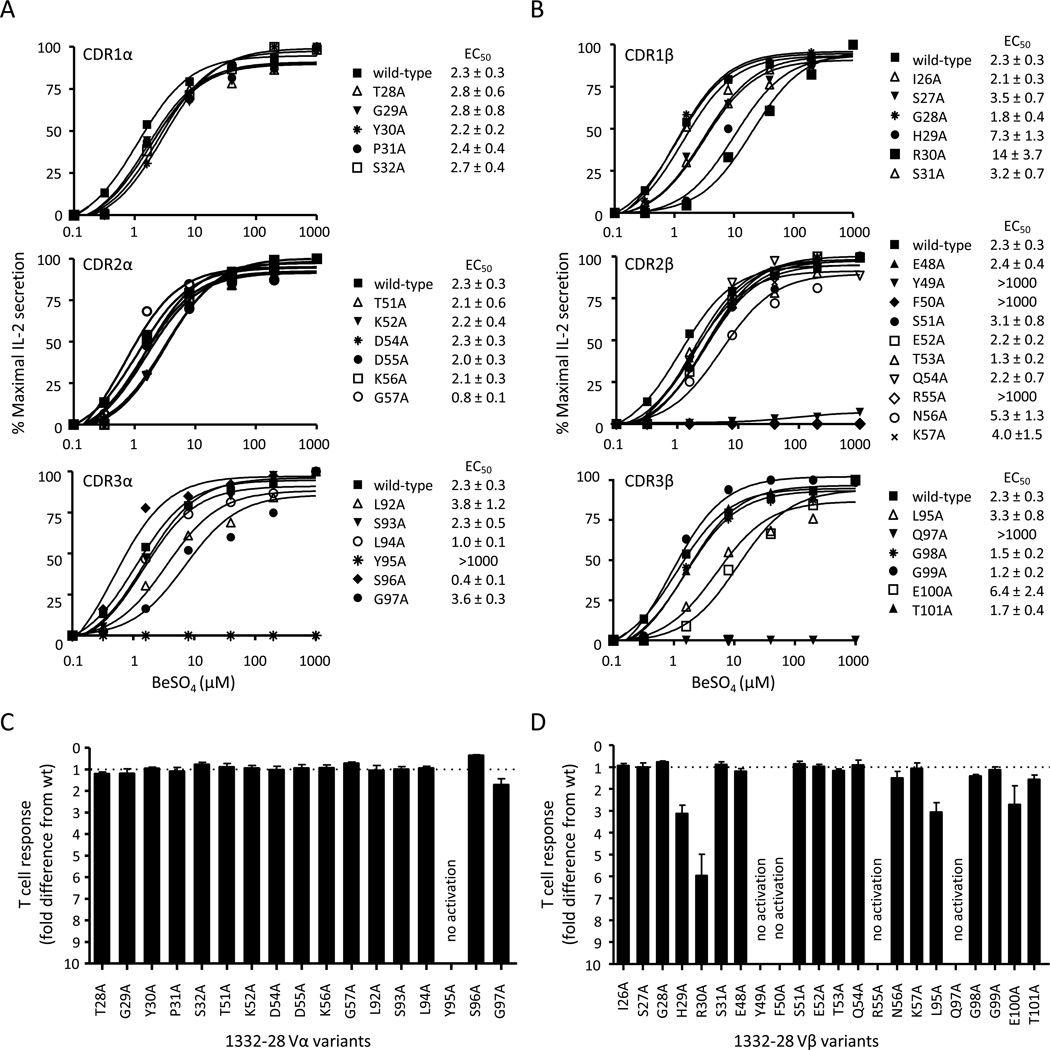
To determine whether TCRs recognize Be in a similar manner as other antigen-specific TCRs, we mutated TCR residues of the 1332-28 T cell clone at potential antigen-contact sites, with the aim of mapping T cell recognition of the Be antigen. We initially focused on 1332-28 since a crystal structure of an autoimmune Vα22/Vβ5.1 TCR (same as that expressed by 1332-28) bound to a myelin basic protein (MBP) self-peptide in the context of HLA-DR2a exists (DRA1*0101/DRB5*0101; PDB ID code 1ZGL) (19). Each TCRA and TCRB CDR residue was separately mutated to alanine using site-directed mutagenesis, and each mutant TCR was expressed on the surface of 5KC. Using a mouse anti-TCR Cβ mAb, similar levels of TCR expression were documented for each cell line (Supplemental Fig. 1). The T cell hybridoma response to BeSO4 was measured using IL-2 secretion, and activation curves were generated by plotting percentage of maximal IL-2 release against BeSO4 concentration (Fig. 3A, 3B). Surprisingly, none of the alanine mutations in the germline-derived CDR1α and CDR2α of 1332-28 had any effect on Be-induced IL-2 secretion, with EC50 values (defined as the BeSO4 concentration required for a half-maximal T cell hybridoma response) similar to the wild-type TCR (Fig. 3A, top and middle panels). In contrast, mutation of the tyrosine at position 95 of the somatically rearranged CDR3α to alanine (Y95A) abolished Be-induced T cell activation (Fig. 3A, bottom panel). This tyrosine is derived from the TCRAJ28 gene segment. In order to quantitatively display the EC50 differences of the individual mutant TCRs, the overall EC50 fold-change difference (mean ± SEM) for each TCR compared to the wild-type TCR for three separate experiments is shown (Fig. 3C). Thus, only one amino acid of the TCR α-chain is critical for functional recognition of the HLA-DP2/peptide/Be complex.
FIGURE 3.
Contact sites of the Be-responsive TCR, 1332-28, with the HLA-DP2/peptide/Be complex. Be-specific response of T cell hybridoma 1332-28 expressing wild-type TCR and TCR variants with single-site alanine mutations at each amino acid position of the CDR1 (upper panel), CDR2 (middle panel) and CDR3 (lower panel) of Vα22 (A) and Vβ5.1 (B). TCRs were expressed on 5KC and stimulated with various concentrations of BeSO4 presented by HLA-DP2-transfected murine fibroblasts. IL-2 secretion was measured by ELISA and plotted as % maximal IL-2 secretion against BeSO4 concentration. Activation curves are representative of three independent experiments, and EC50 values (the concentration of BeSO4 required for a half-maximal T cell hybridoma response) were determined by non-linear regression of the activation curves using Graph Pad Prism software. The mean ± SEM EC50 values for the wild-type TCR and each of the variants are shown. The overall EC50 fold-change difference (mean ± SEM) for the TCR α-chain (C) and β-chain (D) variants compared to the wild-type TCR is shown. The dotted line at y = 1 represents the wild-type TCR response.
For the TCR β-chain analysis of 1332-28, several mutations resulted in either reduced or abolished T cell hybridoma response compared to the wild-type TCR (Fig. 3B, 3D). For example, four mutations including Y49A, F50A, R55A in CDR2β and Q97A in CDR3β abolished the Be-induced T cell hybridoma response (EC50 values greater than 1000 µM). Several other mutations in the β-chain moderately reduced (increased EC50 values 2–6 fold compared to wild-type), but did not abolish, IL-2 secretion in response to Be (H29A and R30A in CDR1β and L95A and E100A in CDR3β) (Fig. 3B). Similar to the α-chain mutants, the EC50 values were plotted as a fold-difference compared to the wild-type TCR response (Fig. 3D). Overall, these data show that the Vβ domain of 1332-28 TCR plays the dominant role in Be recognition, with no contribution from the germline-derived regions of the Vα22 chain.
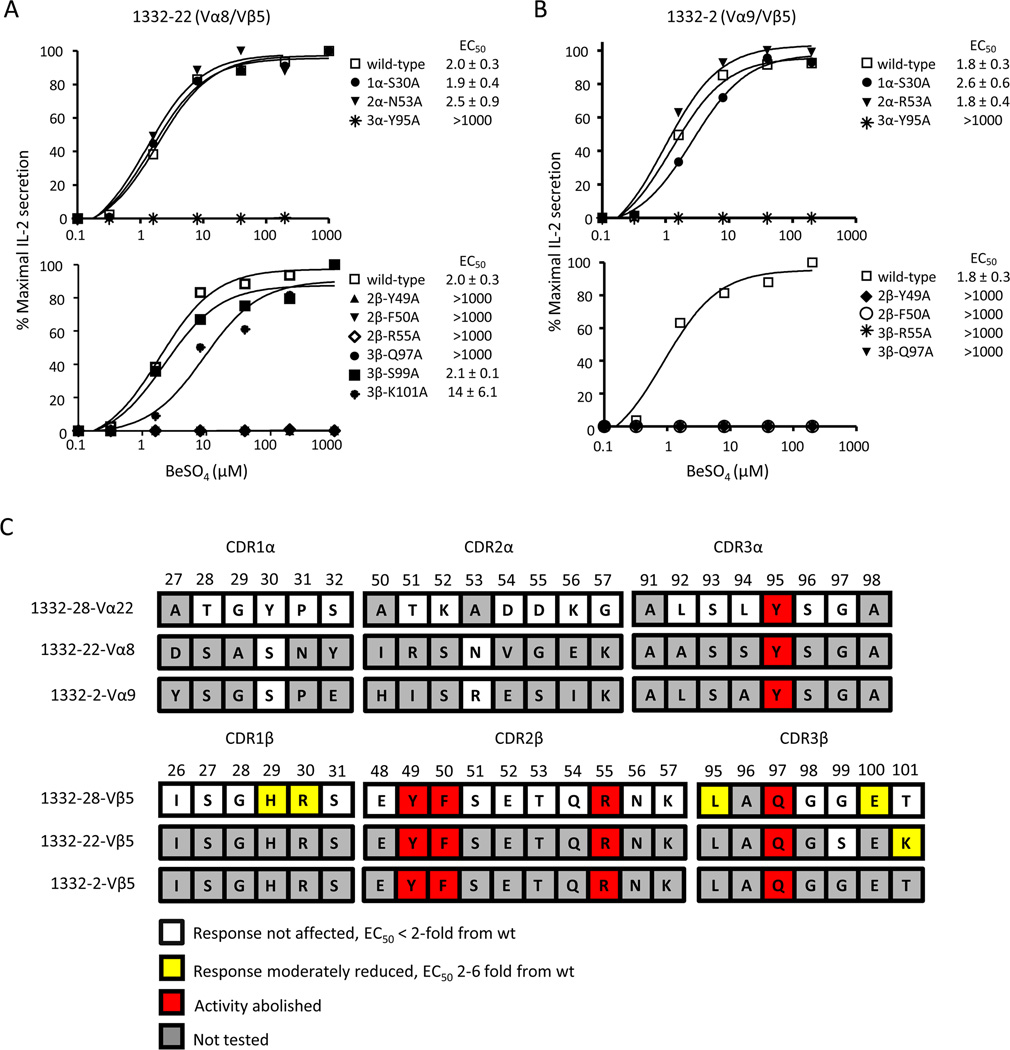
In order to determine whether the other Be-specific Vβ5+ T cell clones, 1332-22 and -2, recognize antigen in a similar manner, we performed site-directed mutagenesis on selected CDR α- and β-chain residues that abolished IL-2 secretion in the 1332-28 TCR. The 1332-22 and -2 TCR variants were expressed on 5KC, and the IL-2 response to various BeSO4 concentrations presented by HLA-DP2-expressing fibroblasts was compared to the wild-type 1332-22 and -2 TCRs. Of note, the three Be-specific TCRs had similar EC50 values, ranging from 1.8 to 2.3 µM BeSO4 (compare Figs. 3 and 4). Identical to 1332-28, the same five mutations (Y95A in CDR3α, Y49A, F50A and R55A in CDR2β, and Q97A in CDR3β) in the TCRs of 1332-22 and -2 abolished Be-induced IL-2 secretion (Fig. 4A, 4B). As controls, we also mutated germline-derived CDR1α and CDR2α residues of 1332-22 (S30A in CDR1α and N53A in CDR2α) and 1332-2 (S30A in CDR1α and R53A in CDR2α) at positions that had no effect on Be recognition in 1332-28. As expected, these mutations in the TCRAV gene segments of 1332-22 and -2 also had no effect on Be-induced IL-2 secretion compared to the wild-type TCR (Fig. 4A, 4B, top panels).
FIGURE 4.
Vβ5+ TCRs recognize the HLA-DP2/peptide/Be complex using identical contact sites. A and B Stimulation of Be-specific T cell hybridomas, 1332-22 and -2, expressing selected mutations in the CDRs of the TCR α- and TCR β-chains with HLA-DP2-expressing DAP.3 fibroblasts and various concentration of BeSO4. T cell secretion of IL-2 was measured by ELISA and plotted as described in Figure 3. Activation curves are representative of three independent experiments. The mean ± SEM EC50 values for the wild-type TCR and each of the variants are shown. C, A color-coded summary of T cell activation data for the stimulation of T cell hybridomas 1332-28, -22, and -2. TCR CDR amino acid sequences for each of the three T cell hybridomas are aligned with the amino acid sequence number shown above. Residues are highlighted based on their effect when mutated to alanine, in comparison to the wild-type response. Residues highlighted in white indicate that an alanine mutation had no effect on T cell response, with EC50 values identical to the wild-type response. Residues highlighted in yellow indicate a mutation that had a moderate effect on T cell hybridoma response, 2–6-fold reduction in response compared to wild-type. Residues highlighted in red indicate the absence of a hybridoma response when mutated to alanine.
Even though the Vβ5.1+ T cell clones express nearly identical TCR β-chains, minor differences exist between 1332-28 and -22, with variations at positions 99 and 101 of CDR3β (see Fig. 1B). To determine if the S99 and K101 in the CDR3β of 1332-22 have an influence on Be recognition, we also mutated these residues to alanine. Similar to 1332-28, the CDR3β S99A mutation had no effect on the 1332-22 T cell hybridoma response (Fig. 4A, bottom panel). In contrast to mutation of the threonine (T) residue at position 101 of 1332-28 which had no effect on Be-induced T cell activation, a moderate decrease was seen in the 1332-22 response with the CDR3β K101A mutation (Fig. 4A, bottom panel). The EC50 fold-differences for each variant compared to the wild-type response are shown in Supplemental Fig. 2. To facilitate interpretation of the results, we used a color-coded scheme to depict the substitutions and their respective effects on the T cell hybridoma response (Fig. 4C). Collectively, these data show that the three Be-specific TCRs utilize identical β-chain amino acid residues, for either direct or indirect recognition of the HLA-DP2/peptide/Be complex, despite the expression of different TCRAV gene segments.
Function of the 1332-28 TCR expressing a non-cognate TCRA gene
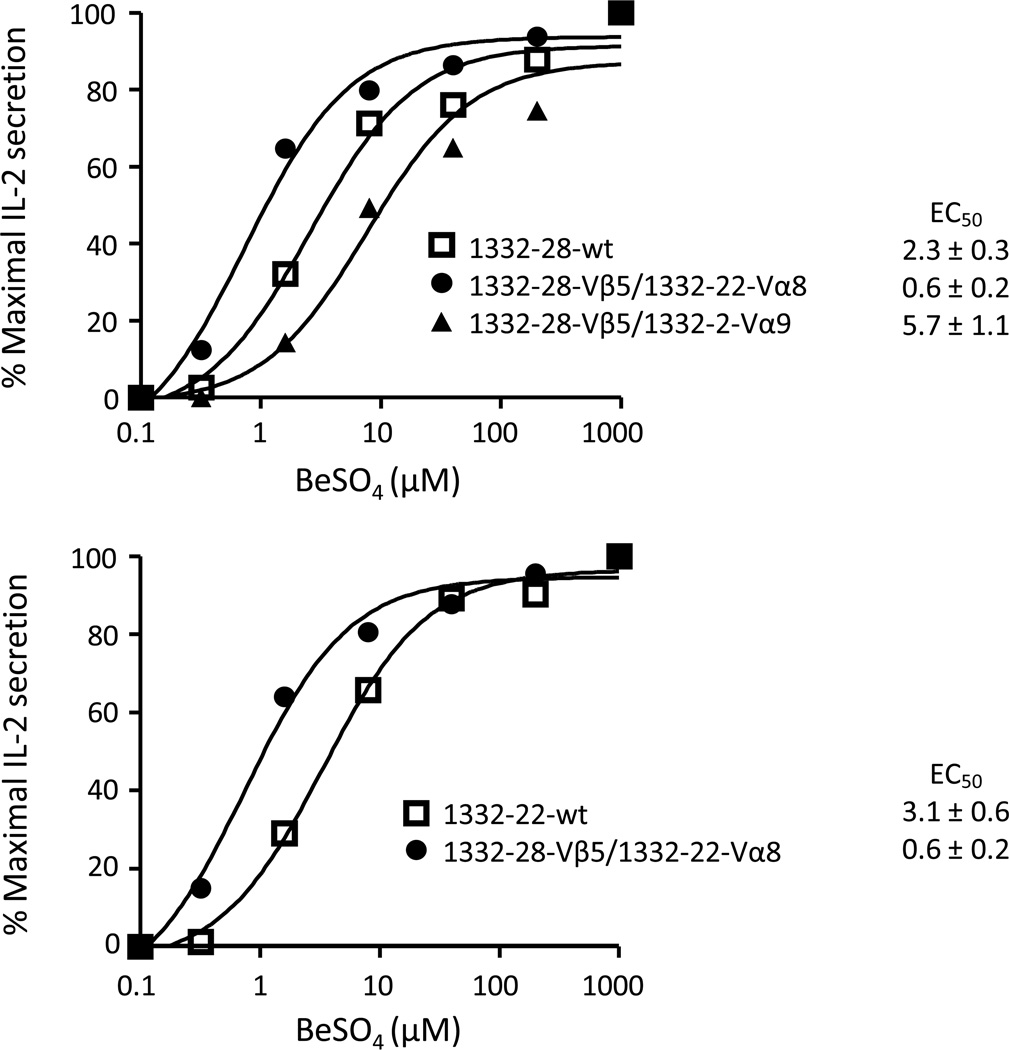
To confirm that the TCR β-chain of the Be-specific TCRs plays the dominant role in antigen recognition, the variable domain of the native 1332-28 TCRA gene was substituted with non-cognate TCRA genes. The TCRB gene of 1332-28 was paired with either the TCRA gene of 1332-22 (AV8S1) or 1332-2 (AV9S1) and expressed on the surface of 5KC with similar TCR levels (data not shown). T cell hybridomas were stimulated with BeSO4 using HLA-DP2-expressing fibroblasts as antigen-presenting cells, and activation curves were generated as described above. As shown in Fig. 5A, switching the native 1332-28 Vα22 chain to either Vα8 or Vα9 had minimal effects on the Be-specific T cell hybridoma response, with a 4-fold reduction in EC50 with Vα8 and a 2.5-fold increase with Vα9.
FIGURE 5.
Activation of Be-responsive, Vβ5.1+ TCRs is independent of CDR1α and CDR2α. Stimulation of Be-specific T cell hybridomas expressing noncognate TCRAV genes. The native TCRA gene of the 1332-28 TCR was switched to non-cognate 1332-22 TCRAV8 or 1332-2 TCRAV9 genes, and the resultant hybridomas were stimulated as in Figure 3. Data are plotted as % maximal IL-2 secretion against concentration of beryllium and are representative of three independent experiments. The mean ± SEM EC50 values for the wild-type TCR and the variants are shown.
Since the β-chains of 1332-28 and -22 differ at positions 99 and 101, we queried whether these amino acids would affect Be-specific T cell activation. Thus, the Be-specific response of the 1332-22-Vα8/1332-28-Vβ5.1 TCR was also compared to wild-type 1332-22 TCR. As shown in Fig. 5A (bottom panel), the mutant TCR had a slightly increased Be-induced response with a 5-fold decrease in the EC50 value compared to the wild-type TCR. These differences are likely the result of a neutral threonine versus a positively-charged lysine at position 101 of the β-chain of 1332-28 and 1332-22, respectively. Although the changes were subtle, the optimal combination of α- and β-chains for Be-induced T cell activation was the Vβ5.1 chain from 1332-28 and the Vα8.1 chain from 1332-22. Taken together, these findings confirm our alanine scan of three Be-specific TCRs, showing that the β-chain of the TCR plays the dominant role in binding to the HLA-DP2/peptide/Be complex and that coupling with any of the TCR Vα chains will result in Be-induced T cell activation as long as the tyrosine (Y) at position 95 of the CDR3α is present.
Effect of HLA-DP2 variants on the Be-induced T cell response
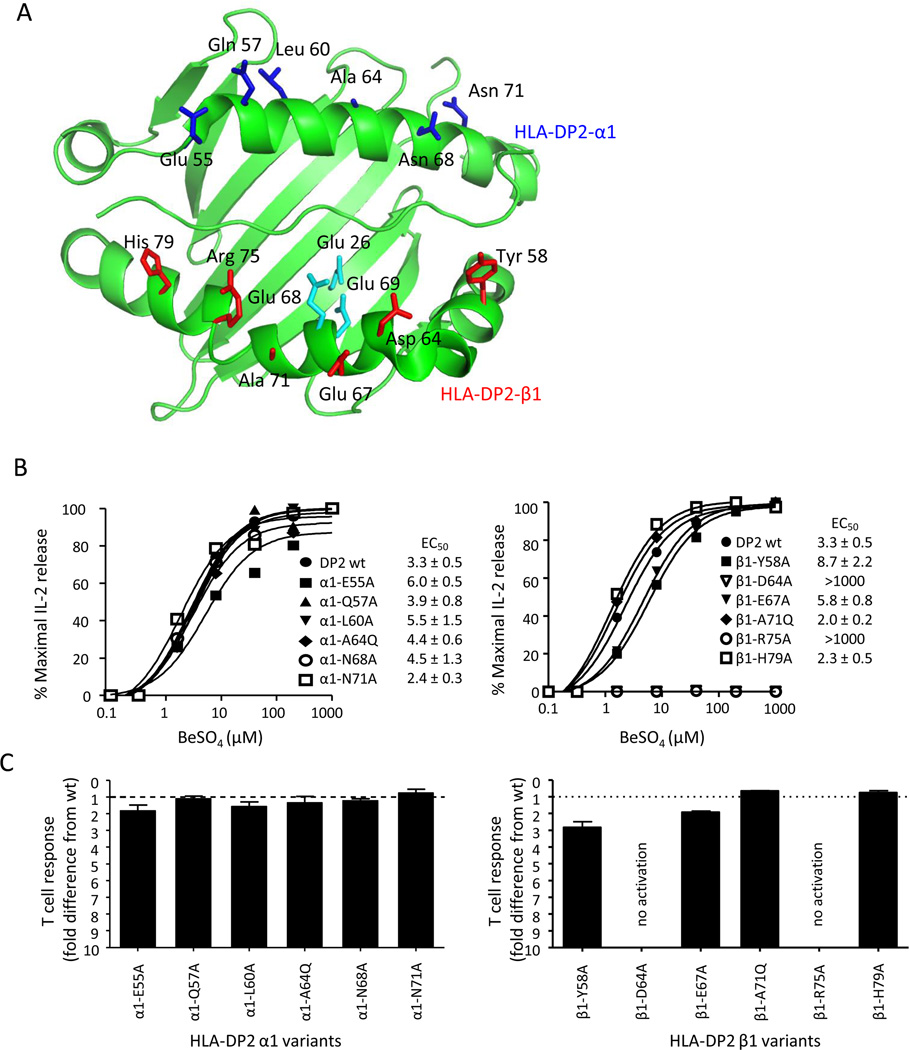
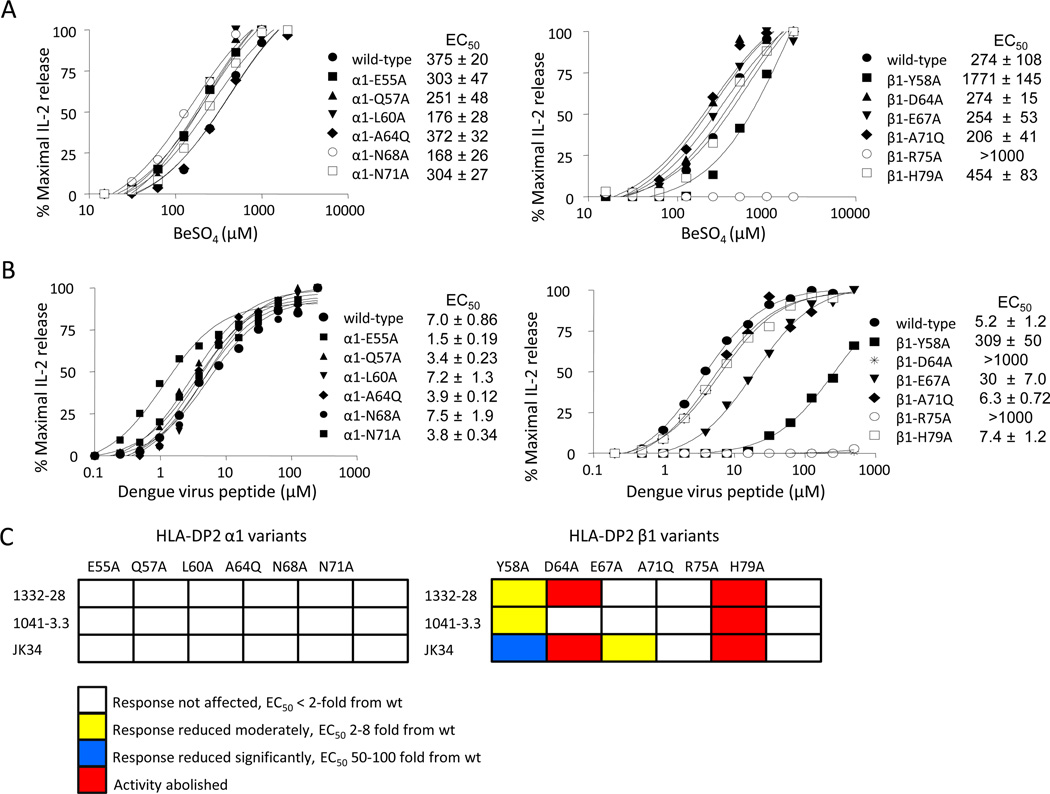
The predominant role of the TCR β-chain in Be recognition with a limited requirement for the Vα domain suggest an unconventional binding topology for these Be-specific TCRs. To further characterize T cell recognition of the Be antigen in the context of HLA-DP2, we performed site-directed mutagenesis on HLA-DP2. We have recently crystallized HLA-DP2 (PDB ID code 3LQZ) (14) and used this structure to delineate solvent-exposed residues most likely to impact T cell recognition when mutated and not affect stability or expression of the MHCII molecule. Amino acid residues that spanned the length of HLA-DP2 α1 and β1 helices were chosen for mutagenesis, and their location on the α-helices of HLA-DP2 is shown in Fig. 6A. The following mutations were introduced in the HLA-DP2 α1 helix: E55A, Q57A, L60A, A64Q, N68A, N71A, and β1 helix: Y58A, D64A, E67A, A71Q, R75A, H79A. We chose to replace the alanine at positions 64 of the α1 helix and 71 of the β1 helix with a glutamine in order to disrupt TCR binding through steric hindrance (29). Wild-type and variants of HLA-DP2 were expressed on the surface of the murine fibroblast line, B6DK10, and all antigen-presenting cell lines expressed similar levels of HLA-DP2 as shown by staining with a conformation dependent anti-HLA-DP mAb (B7.21) (data not shown). The Be-specific T cell hybridoma 1332-28 was stimulated with various concentrations of BeSO4 added to the fibroblasts expressing the HLA-DP2 variants, and activation curves were generated as described above (Fig. 6B). Surprisingly, not a single mutation in the HLA-DP2 α1 helix reduced the T cell hybridoma response as indicated by similar EC50 values of the variants compared to wild-type DP2 (Fig. 6B, left panel). In contrast, several mutations in the α-helix of the DP2 β-chain either abolished or reduced the 1332-28 T cell hybridoma response. For example, two DP2 β1 helix mutations (D64A and R75A) abolished 1332-28 hybridoma response, as indicated by no IL-2 secretion in response to up to 1000 µM BeSO4 (Fig. 6B, right panel). In addition, β1-Y58A moderately reduced the response as indicated by a 2.6-fold increase in the EC50 compared to wild-type HLA-DP2 (Fig. 6B, right panel). A summary of the EC50 fold differences for the 12 variants compared to wild-type DP2 is shown in Fig. 6C, with only two of these upward facing, solvent-exposed residues of HLA-DP2 representing major sites for either direct or indirect contact with the Be-specific 1332-28 TCR.
FIGURE 6.
Identification of HLA-DP2 contacts for Be-specific, Vβ5.1+ T cells. A, Crystal structure of HLA-DP2 with a self-derived HLA-DRα peptide (PBD ID code 3LQZ). The α1- and β1-domains of HLA-DP2, along with the peptide, are shown in green. Solvent-exposed, upward-facing residues of the HLA-DP2 α1 and β1 α-helices subjected to mutagenesis are shown in blue and red, respectively. Cyan residues denote three glutamic acids proposed to contribute to Be binding. B, HLA-DP2 molecules with single-site mutations introduced along both α-helices were expressed on the surface of murine fibroblasts and used to stimulate 1332-28 TCR. Representative activation curves for the α-chain (left panel) and β-chain (right panel) variants are plotted as % maximal IL-2 release against BeSO4 concentration. Data are representative of at least three separate experiments. C, Shown is EC50 fold-change differences (mean ± SEM) for hybridoma 1332-28 induced by the HLA-DP2 variants compared to wild-type HLA-DP2 for three separate experiments. The dotted line at y = 1 represents the wild-type TCR response.
Effect of HLA-DP2 variants on the response of additional HLA-DP2 restricted TCRs
In order to determine whether the abnormal binding topology shown for the three Vβ5+ TCRs was present in other Be-specific TCRs, we investigated a T cell hybridoma (designated 1041-3.3) expressing a Be-specific TCR derived from the lung of an additional CBD patient. The TCR of the parental T cell clone utilizes TCRAV1 and TCRBV3 gene segments (Supplemental Fig. 3) and is dissimilar to the CDR3α and CDR3β sequences of the Vβ5.1+ T cell clones (compare Figs. 1B and Supplemental Fig. 3). The 1041-3.3 T cell hybridoma was generated in a similar manner as described for the Vβ5.1+ T cell hybridomas and had a >100-fold increase in the Be-induced EC50 value compared to 1332-28, suggesting a significantly decreased binding affinity of this TCR for the HLA-DP2/peptide/Be complex (compare Figs. 3 and 7A). Fibroblasts expressing the HLA-DP2 α1 helix mutations had no effect on the Be-induced IL-2 response of this Vα1/Vβ3 T cell hybridoma (Fig. 7A, left panel). Conversely, two DP2 β1 helix mutations either reduced (Y58A) or abolished (R75A) the Be-induced IL-2 response (Fig. 7A, right panel). These findings suggest that this hybridoma contacts the HLA-DP2/peptide/Be complex in the same unconventional manner as the three related Be-specific TCRs despite expressing distinctly different TCR genes and having a much lower binding affinity for antigen.
FIGURE 7.
Effect of HLA-DP2 variants on the response of other DP2-restricted TCRs. A, Stimulation of a T cell hybridoma expressing a Be-specific, Vα1/Vβ3+ TCR derived from the lung of CBD patient 1041 with HLA-DP2 variants. The T cell hybridoma, designated 1041-3.3, was stimulated with fibroblasts expressing wild-type and α-chain (left panel) and β-chain (right panel) variants of HLA-DP2 and varying BeSO4 concentrations. Activation curves and EC50 values are representative of three independent experiments. B, Stimulation of cells expressing a dengue virus specific Vα11/Vβ23 TCR, designated JK34, with fibroblasts expressing HLA-DP2 α-chain (left panel) and β-chain (right panel) variants and presenting a dengue virus peptide. Representative activation curves from three independent experiments are plotted and used to determine EC50 values. C, A color-coded summary shows the effects of HLA-DP2 variants on various DP2-restricted T cell hybridomas, expressing beryllium-specific TCRs 1332-28 and 1041-3.3 and a dengue-virus-specific JK34 TCR. Residues are colored based upon their effect when mutated to alanine, in comparison to the wild-type response. Residues highlighted in white indicate that an alanine mutation had no effect on T cell hybridoma response, with EC50 values identical to the wild-type response. Residues highlighted in yellow and blue indicate a mutation that either had a moderate (2–8 fold) or large (50–100-fold reduction) effect on the hybridoma response. Residues highlighted in red abolished the IL-2 response when mutated to alanine.
In order to verify that the abnormal binding topology observed for these Be-specific TCRs is unique for this unconventional antigen, we determined how a T cell recognizes a conventional (viral) antigen in the context of HLA-DP2. Thus, we expressed on 5KC a Vα11/Vβ23 TCR derived from a T cell clone specific to an epitope of the NS3 protein of dengue virus (amino acids 251–264; HTGREIVDLMCHAT) (24). Upon exposure to the dengue virus peptide, hybridoma cells expressing the dengue-specific TCR (designated JK34) secreted IL-2, with an EC50 value ranging between 5.2 and 7.0 µM dengue virus peptide (Fig. 7B). Using fibroblasts expressing single-site variants of HLA-DP2 to map antigen recognition of this dengue virus-specific TCR, the HLA-DP2 α1 helix variants had no affect on the JK34 T cell hybridoma response, as indicated by identical EC50 values for the α1 helix variants compared to the wild-type TCR (Fig. 7B, left panel). Conversely, alanine mutations at positions 64 and 75 (D64A and R75A) of the HLA-DP2 β1-chain abolished the dengue viral-specific T cell response (Fig. 7B, right panel). In addition, two other HLA-DP2 β1-chain mutations (Y58A and E67A) markedly reduced the dengue viral-specific hybridoma response. In Fig. 7C, we used a color-coded scheme to depict the amino acid substitutions and their respective effects on the T cell hybridoma response. Taken together, these findings suggest that these HLA-DP2-restricted TCRs predominantly contact the HLA-DP2 β1 chain with little to no interactions with the α1 chain, raising the possibility that all DP2-restricted TCRs display a binding footprint distinct from those seen with other MHCII-restricted TCRs. Interestingly, the β1 R75A mutation abolished the response for all three T cell hybridomas (Fig. 7C), suggesting that this amino acid is a critical contact site for HLA-DP2 restricted TCRs.
Discussion
CD4+ T cells are intimately tied to the pathogenesis of CBD (4). Progression from beryllium sensitization to disease is characterized by the presence of granulomatous inflammation and the accumulation of beryllium-specific, Th1-polarized CD4+ T cells in the lung (30–33). These lung T cells are composed of oligoclonal TCR Vβ expansions that include Vβ5.1 (34, 35). These expansions are specific for CBD patients, compartmentalized to lung, and persist at high frequency in patients with active disease (34). Thus, it was of interest when a large population of Be-responsive CD4+ T cells isolated from the lung of an HLA-DP2-expressing CBD patient was shown to express nearly identical TCR Vβ5.1 chains associated with different Vα usage. In the present study, we used site-directed mutagenesis to determine how this set of related, Be-responsive TCRs recognizes Be in the context of HLA-DP2/peptide. Our findings suggest that these Vβ5.1+ TCRs utilize an abnormal binding footprint for the recognition of the HLA-DP2/peptide/Be complex.
A crystal structure exists of an autoimmune TCR (3A6) expressing the same Vα22/Vβ5.1 pairing as our Be-specific TCR 1332-28. The 3A6 TCR is specific for a MBP self-peptide presented by HLA-DR2a (PDB ID code 1ZGL) and has a CDR footprint that is shifted toward the N-terminus of the bound peptide and the MHCII β1 helix (19). Our alanine scan of the CDRs of 1332-28 showed a dominant role for the TCR Vβ5.1 chain in the recognition of Be/DP2, with several mutations in the CDR1β, CDR2β, and CDR3β abolishing the T cell hybridoma response while only one mutation in the Vα domain (Y95A in CDR3α) affected recognition of Be/DP2. Importantly, these findings were corroborated in two other Vβ5.1-expressing, Be-specific TCRs that used different TCR Vα chains. In addition, the Vβ5.1+ TCRs maintained Be specificity even in the presence of a noncognate α-chain as long as the tyrosine (Y) at position 95 was present. Consistent with our findings that the CDR2α of 1332-28, -2 and -22 had no significant interactions with Be/DP2, the CDR2α of 3A6 also made no contact with the MBP/DR2a, being too short to interact with either DR2a or MBP (16, 19). Whereas the highly conserved tyrosine residue at position 30 of CDR1α of 1332-28 TCR played no role in binding to Be/DP2, this residue in 3A6 strongly interacts with numerous DR2a β1 helix residues (16, 19). Comparing the TCR Vα chain contacts of 3A6 and 1332-28, eight Vα residues of 3A6 interact with MHCII compared with one for 1332-28. Accordingly, our findings suggest that the docking footprint of this set of Be-specific TCRs is more profoundly shifted than that described for 3A6, using mainly Vβ5.1 residues for the recognition of Be/DP2 with no contribution from germline-derived CDR1α or CDR2α.
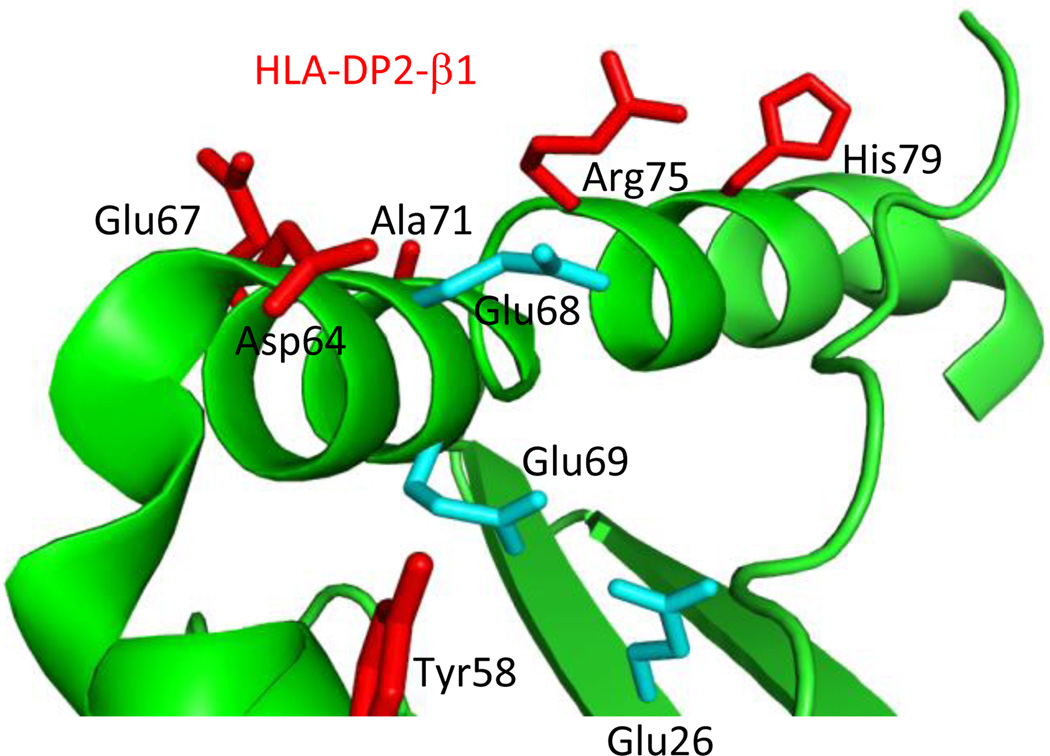
Previous mutagenesis and structural studies show that both α-helices of the MHC contribute to TCR recognition. Germline-derived CDR1 and CDR2 residues from both TCR α- and β-chains of the murine 2C TCR contributed energetically to recognition of MHCI Ld and Kb, which was confirmed by the solved structures (36–38). In addition, residues spanning both α-helices of murine I-Ek contributed to the recognition by the moth cytochrome C-specific TCR, 2B4 (39). Similar results were shown with a variety of T cells specific for 3K/I-Ab, with mutations along both α-helices of I-Ab reducing TCR binding affinity (29). Conversely, the present findings show that the α1 helix of HLA-DP2 plays a minor role in interacting with the Be-specific TCRs, with the β1 helix contributing the majority of the contact sites. Interestingly, the two HLA-DP2 β-chain mutations that abolished the Be-specific T cell hybridoma response (D64A and R75A in the β1 helix) surround the putative Be binding site on HLA-DP2, involving glutamic acid residues at positions 26, 68, and 69 (Fig. 8). Site-directed mutagenesis of each of these three negatively-charged amino acids was previously shown to abrogate T cell recognition of Be/DP2 (14). Thus, the docking footprint used by Be-specific TCRs for the recognition of antigenic HLA-DP2 is skewed, being dominated by the TCR β-chain and β1 helix residues of DP2. To date, there is only one reported case in which an unconventional binding topology has been used by a TCR for the recognition of a foreign-derived antigen (40). Since we do not know the peptide(s) that complete the Be-specific TCR ligand for this set of TCRs, it remains unknown how peptide contributes to TCR binding.
FIGURE 8.
Structural features of the HLA-DP2 β-chain highlighting the putative Be-binding pocket. Crystal structure of HLA-DP2 with a self-derived HLA-DR α-chain peptide as depicted with PyMol software (PDB ID code, 3LQZ). A cartoon representation of the DR α-chain peptide and the β-chain of HLA-DP2 are shown in green. HLA-DP2 β1 helix residues, subjected to single-site mutagenesis in the current study, are highlighted red. The three glutamic acid residues that make up the putative Be-binding site are highlighted in cyan.
Although defined criteria have been used to characterize T cell recognition of foreign-derived antigen, these rules do not apply to human autoantigen-specific TCRs that exhibit atypical binding interactions, as discussed above for TCR 3A6. The present study suggests that the unconventional binding topologies observed for autoimmune TCRs can also be used for recognition of unconventional antigens such as Be and even HLA-DP2-restricted, anti-viral TCRs. There are several interpretations of the unusual binding mechanism adopted by the Be-specific TCRs. For example, the docking footprint of the Be-specific TCRs may be shifted towards the β1 helix of MHCII, with CDR1α and CDR2α making no significant contacts with HLA-DP2 (Supplemental Fig. 4A). This shift would position the TCR Vβ domain directly over the putative Be-binding site of HLA-DP2, located between the peptide backbone and β1 α-helix. To a lesser degree, similar findings were reported for human TCRs Ob.1A12 and 3A6, which each recognize multiple sclerosis-derived autoimmune complexes (18, 19). Alternatively, the Be-specific TCRs may recognize antigen using a tilted and reversed polarity, with the Vβ chain contacting the β1 α-helix of MHC and the Vα-chain hovering over the α1 helix of MHC, yet making no contacts (Supplemental Fig. 4B). As supporting evidence, the docking footprint for the human multiple sclerosis-derived TCR Hy.1B11 is tilted with interactions between TCR Vβ and the MHC α1 helix dominating, while maintaining a conserved docking polarity (17). The former mechanism seems more plausible since a reversed polarity has not been reported to date. Ultimate proof requires a structure of TCR-DP2/peptide/Be complex, which has not been feasible without knowing which peptide(s) completes the Be-specific TCR ligand.
The binding polarity between TCR and MHC is suggested to be a function of evolutionarily conserved interactions between these two molecules. However, whether conserved docking topologies are a consequence of evolutionarily conserved interactions between germline-derived TCR regions (CDR1 and CDR2) and α-helix residues of MHC is still unresolved (41). Supporting evidence from analyses of TCR/pMHC crystal structures show that conserved pair-wise interactions exist for related TCRs with certain MHC alleles (16, 42, 43). We suggest that the Be-antigen generates an alternative binding landscape for recognition by T cells even though evolutionarily conserved interactions exist. To illustrate, the majority of MHCII molecules express conserved amino acids which lack bulky side chains (e.g., alanine) at the center of each α-helix. These small residues are thought to create a “cup” that is used to pivot the TCR for a diagonal binding orientation (16, 43). For all of the DP2-restricted TCRs tested here, substitution of a large bulky side chain at conserved alanine residues (A64Q in α1 helix and A71Q in β1 helix; Fig. 6) of HLA-DP2 had no effect on the T cell response. Even though conserved α-helical residues exist in HLA-DP2, the Be-specific Vβ5.1+ TCRs lack conserved tyrosine residues in CDR2α and CDR1β, which have been suggested to play a role in how TCR recognizes peptide/MHC (16). As mentioned, even though 1332-28 expresses a conserved tyrosine in CDR1α, an alanine mutation at this position did not affect Be-specific T cell activation. It is noteworthy that the mutated CDR2β-Tyr49 residue abolished the Be-specific T cell response, since many other TCRs express a conserved Tyr in this region.
From the structural analysis of a Vα22/Vβ5.1 TCR (3A6) and our TCR mutagenesis studies, it was expected that the Be-specific Vβ5.1+ TCR, 1332-28, would respond in a similar manner to Be presented by either wild-type DP2 or DP2 with alanine mutations in selected solvent-exposed α-helix residues. However, it was surprising that two other DP2-restricted TCRs (a Be-specific Vα1/Vβ3 TCR derived from the lung of another CBD patient and a dengue virus-specific Vα11/Vβ22 TCR derived from the blood of a subject infected with dengue virus) displayed similar results. These findings raise the possibility that, unlike interactions with other MHCII molecules, the majority of HLA-DP2-restricted TCRs utilize a distinct binding topology where most of the contact sites involve the β1 helix of DP2. One possible reason for this atypical footprint may lie in the unusual structural features of the HLA-DP2 molecule. For example, HLA-DP2 possesses a widened distance between the peptide backbone and β1 helix compared to most other MHCII molecules (14). A recent modeling study of HLA-DR1 with and without a bound peptide suggested that this region of the β-chain α-helix may be flexible (44), contributing to the variation in the width of this part of the binding groove and possibly to differing TCR footprints.
In conclusion, the present work suggests that the Be-antigen (HLA-DP2/peptide/Be) generates a unique landscape for recognition by Be-specific TCRs, dominated by TCR β-chain and β1 α-helix residues of DP2. Our findings provide additional evidence that unusual binding topologies are not used exclusively by T cells to recognize autoimmune complexes, but also for T cell recognition of unconventional antigens, such as Be, and DP2-restricted anti-viral TCRs.
Supplementary Material
Acknowledgments
We thank Dr. Francis Ennis for providing the dengue-virus specific T cell clone JK34.
Non-standard abbreviations used
- Be
beryllium
- CBD
chronic beryllium disease
Footnotes
This work is supported by the following NIH grants: HL62410, HL92997, and ES011810 (to APF), and the Clinical Translational Research Center (UL1 RR025780) from the National Center for Research Resources.
References
- 1.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Lu L, Vollmer J, Moulon C, Weltzien HU, Marrack P, Kappler J. Components of the ligand for a Ni++ reactive human T cell clone. J. Exp. Med. 2003;197:567–574. doi: 10.1084/jem.20021762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falta MT, Bowerman NA, Dai S, Kappler JW, Fontenot AP. Linking genetic susceptibility and T cell activation in beryllium-induced disease. Proc. Am Thorac Soc. 2010;7:126–129. doi: 10.1513/pats.201002-022RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontenot AP, Maier LA. Genetic susceptibility and immune-mediated destruction in beryllium-induced disease. Trends Immunol. 2005;26:543–549. doi: 10.1016/j.it.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Fontenot AP, Canavera SJ, Gharavi L, Newman LS, Kotzin BL. Target organ localization of memory CD4+ T cells in patients with chronic beryllium disease. J. Clin. Invest. 2002;110:1473–1482. doi: 10.1172/JCI15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontenot AP, Torres M, Marshall WH, Newman LS, Kotzin BL. Beryllium presentation to CD4+ T cells underlies disease-susceptibility HLA-DP alleles in chronic beryllium disease. Proc. Natl. Acad. Sci. U S A. 2000;97:12717–12722. doi: 10.1073/pnas.220430797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maier LA, McGrath DS, Sato H, Lympany P, Welsh K, Du Bois R, Silveira L, Fontenot AP, Sawyer RT, Wilcox E, Newman LS. Influence of MHC class II in susceptibility to beryllium sensitization and chronic beryllium disease. J. Immunol. 2003;171:6910–6918. doi: 10.4049/jimmunol.171.12.6910. [DOI] [PubMed] [Google Scholar]
- 8.Richeldi L, Kreiss K, Mroz MM, Zhen B, Tartoni P, Saltini C. Interaction of genetic and exposure factors in the prevalence of berylliosis. Am. J. Ind. Med. 1997;32:337–340. doi: 10.1002/(sici)1097-0274(199710)32:4<337::aid-ajim3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 9.Richeldi L, Sorrentino R, Saltini C. HLA-DPB1 glutamate 69: a genetic marker of beryllium disease. Science. 1993;262:242–244. doi: 10.1126/science.8105536. [DOI] [PubMed] [Google Scholar]
- 10.Rosenman KD, Rossman M, Hertzberg V, Reilly MJ, Rice C, Kanterakis E, Monos D. HLA class II DPB1 and DRB1 polymorphisms associated with genetic susceptibility to beryllium toxicity. Occup. Environ. Med. 2010 doi: 10.1136/oem.2010.055046. In press. [DOI] [PubMed] [Google Scholar]
- 11.Rossman MD, Kern JA, Elias JA, Cullen MR, Epstein PE, Preuss OP, Markham TN, Daniele RP. Proliferative response of bronchoalveolar lymphocytes to beryllium. A test for chronic beryllium disease. Ann. Intern. Med. 1988;108:687–693. doi: 10.7326/0003-4819-108-5-687. [DOI] [PubMed] [Google Scholar]
- 12.Bill JR, Mack DG, Falta MT, Maier LA, Sullivan AK, Joslin FG, Martin AK, Freed BM, Kotzin BL, Fontenot AP. Beryllium presentation to CD4+ T cells is dependent on a single amino acid residue of the MHC class II β-chain. J. Immunol. 2005;175:7029–7037. doi: 10.4049/jimmunol.175.10.7029. [DOI] [PubMed] [Google Scholar]
- 13.Lombardi G, Germain C, Uren J, Fiorillo MT, du Bois RM, Jones-Williams W, Saltini C, Sorrentino R, Lechler R. HLA-DP allele-specific T cell responses to beryllium account for DP- associated susceptibility to chronic beryllium disease. J. Immunol. 2001;166:3549–3555. doi: 10.4049/jimmunol.166.5.3549. [DOI] [PubMed] [Google Scholar]
- 14.Dai S, Murphy GA, Crawford F, Mack DG, Falta MT, Marrack P, Kappler JW, Fontenot AP. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Proc. Natl. Acad. Sci. U S A. 2010;107:7425–7430. doi: 10.1073/pnas.1001772107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 16.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu. Rev. Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sethi DK, Schubert DA, Anders AK, Heroux A, Bonsor DA, Thomas CP, Sundberg EJ, Pyrdol J, Wucherpfennig KW. A highly tilted binding mode by a self-reactive T cell receptor results in altered engagement of peptide and MHC. J. Exp. Med. 2011;208:91–102. doi: 10.1084/jem.20100725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat. Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 21.Newman LS, Kreiss K, King TE, Jr, Seay S, Campbell PA. Pathologic and immunologic alterations in early stages of beryllium disease. Re-examination of disease definition and natural history. Am. Rev. Respir. Dis. 1989;139:1479–1486. doi: 10.1164/ajrccm/139.6.1479. [DOI] [PubMed] [Google Scholar]
- 22.Fontenot AP, Gharavi L, Bennett SR, Canavera SJ, Newman LS, Kotzin BL. CD28 costimulation independence of target organ versus circulating memory antigen-specific CD4+ T cells. J. Clin. Invest. 2003;112:776–784. doi: 10.1172/JCI18317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yssel H, Blanchard D, Boylston A, De Vries JE, Spits H. T cell clones which share T cell receptor epitopes differ in phenotype, function and specificity. Eur. J. Immunol. 1986;16:1187–1193. doi: 10.1002/eji.1830161002. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto Y, Kurane I, Leporati AM, Ennis FA. Definition of the region on NS3 which contains multiple epitopes recognized by dengue virus serotype-cross-reactive and flavivirus-cross-reactive, HLA-DPw2-restricted CD4+ T cell clones. J. Gen. Virol. 1998;79:697–704. doi: 10.1099/0022-1317-79-4-697. [DOI] [PubMed] [Google Scholar]
- 25.Scott-Browne JP, Matsuda JL, Mallevaey T, White J, Borg NA, McCluskey J, Rossjohn J, Kappler J, Marrack P, Gapin L. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat. Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- 26.White J, Pullen A, Choi K, Marrack P, Kappler JW. Antigen recognition properties of mutant Vβ3+ T cell receptors are consistent with an immunoglobulin-like structure for the receptor. J. Exp. Med. 1993;177:119–125. doi: 10.1084/jem.177.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Littman DR. A kinase-independent function of Lck in potentiating antigen-specific T cell activation. Cell. 1993;74:633–643. doi: 10.1016/0092-8674(93)90511-n. [DOI] [PubMed] [Google Scholar]
- 28.Watson AJ, DeMars R, Trowbridge IS, Bach FH. Detection of a novel human class II HLA antigen. Nature. 1983;304:358–361. doi: 10.1038/304358a0. [DOI] [PubMed] [Google Scholar]
- 29.Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat. Immunol. 2006;7:1191–1199. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- 30.Kreiss K, Newman LS, Mroz M, Campbell PA. Screening blood test identifies subclinical beryllium disease. J. Occup. Med. 1989;31:603–608. doi: 10.1097/00043764-198907000-00011. [DOI] [PubMed] [Google Scholar]
- 31.Mroz MM, Kreiss K, Lezotte DC, Campbell PA, Newman LS. Reexamination of the blood lymphocyte transformation test in the diagnosis of chronic beryllium disease. J. Allergy Clin. Immunol. 1991;88:54–60. doi: 10.1016/0091-6749(91)90300-d. [DOI] [PubMed] [Google Scholar]
- 32.Rossman MD, Kern JA, Elias JA, Cullen MR, Epstein PE, Preuss OP, Markham TN, Daniele RP. Proliferative response of bronchoalveolar lymphocytes to beryllium. Ann. Intern. Med. 1988;108:687–693. doi: 10.7326/0003-4819-108-5-687. [DOI] [PubMed] [Google Scholar]
- 33.Fontenot AP, Canavera SJ, Gharavi L, Newman LS, Kotzin BL. Target organ localization of memory CD4+ T cells in patients with chronic beryllium disease. J. Clin. Invest. 2002;110:1473–1482. doi: 10.1172/JCI15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fontenot AP, Falta MT, Freed BM, Newman LS, Kotzin BL. Identification of pathogenic T cells in patients with beryllium-induced lung disease. J. Immunol. 1999;163:1019–1026. [PubMed] [Google Scholar]
- 35.Fontenot AP, Kotzin BL, Comment CE, Newman LS. Expansions of T-cell subsets expressing particular T-cell receptor variable regions in chronic beryllium disease. Am. J. Respir. Cell Mol. Biol. 1998;18:581–589. doi: 10.1165/ajrcmb.18.4.2981. [DOI] [PubMed] [Google Scholar]
- 36.Manning TC, Schlueter CJ, Brodnicki TC, Parke EA, Speir JA, Garcia KC, Teyton L, Wilson IA, Kranz DM. Alanine scanning mutagenesis of an αβ T cell receptor: mapping the energy of antigen recognition. Immunity. 1998;8:413–425. doi: 10.1016/s1074-7613(00)80547-6. [DOI] [PubMed] [Google Scholar]
- 37.Lee PU, Churchill HR, Daniels M, Jameson SC, Kranz DM. Role of 2CT cell receptor residues in the binding of self- and allo-major histocompatibility complexes. J. Exp. Med. 2000;191:1355–1364. doi: 10.1084/jem.191.8.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 39.Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–556. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 40.Gras S, Burrows SR, Kjer-Nielsen L, Clements CS, Liu YC, Sullivan LC, Bell MJ, Brooks AG, Purcell AW, McCluskey J, Rossjohn J. The shaping of T cell receptor recognition by self-tolerance. Immunity. 2009;30:193–203. doi: 10.1016/j.immuni.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Jerne NK. The somatic generation of immune recognition. Eur. J. Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- 42.Sim BC, Zerva L, Greene MI, Gascoigne NR. Control of MHC restriction by TCR Valpha CDR1 and CDR2. Science. 1996;273:963–966. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- 43.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'. Nat. Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 44.Painter CA, Cruz A, Lopez GE, Stern LJ, Zavala-Ruiz Z. Model for the peptide-free conformation of class II MHC proteins. PLoS One. 2008;3:e2403. doi: 10.1371/journal.pone.0002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.