Abstract
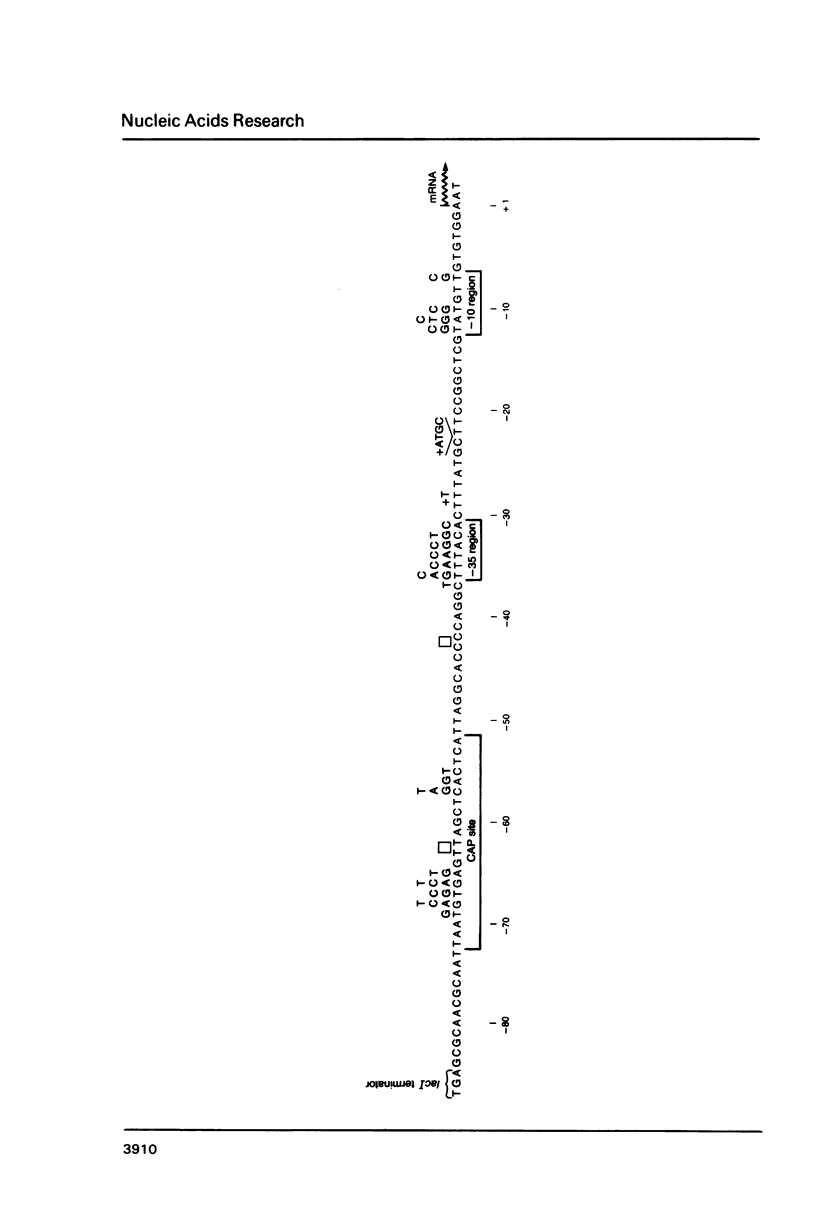
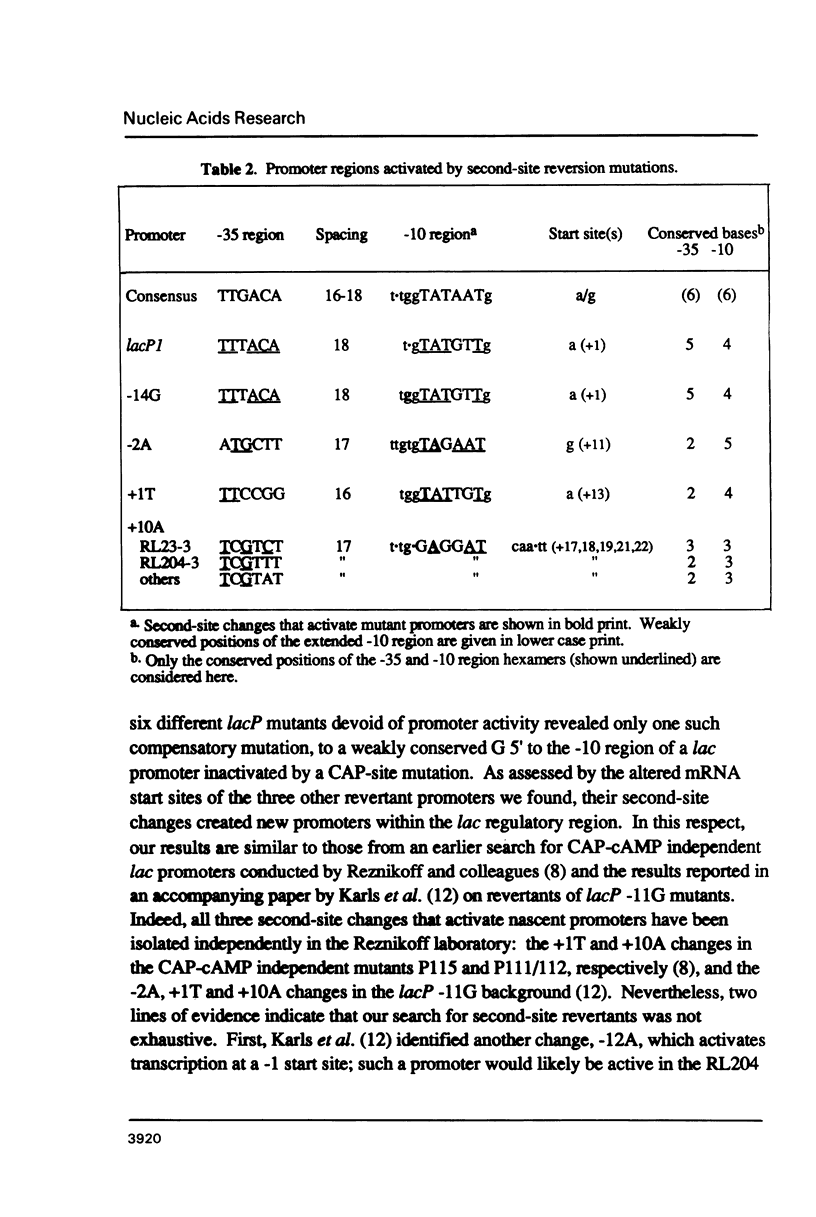
Second-site mutations that restored activity to severe lacP1 down-promoter mutants were isolated. This was accomplished by using a bacteriophage f1 vector containing a fusion of the mutant E. coli lac promoters with the structural gene for chloramphenicol acetyltransferase (CAT), so that a system was provided for selecting phage revertants (or pseudorevertants) that conferred resistance of phage-infected cells to chloramphenicol. Among the second-site changes that relieved defects in mutant lac promoters, the only one that restored lacP1 activity was a T----G substitution at position -14, a weakly conserved site in E. coli promoters. Three other sequence changes, G----A at -2, A----T at +1, and C----A at +10, activated nascent promoters in the lac regulatory region. The nascent promoters conformed to the consensus rule, that activity is gained by sequence changes toward homology with consensus sequences at the -35 and -10 regions of the promoter. However, the relative activities of some promoters cannot be explained solely by consideration of their conserved sequence elements.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman M. L., Landy A. Promoter mutations in the transfer RNA gene tyrT of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4303–4307. doi: 10.1073/pnas.76.9.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina-Stein M., Thoren M., Salzman N., Thomspon J. A. Rapid sequence determination of late simian virus 40 16S mRNA leader by using inhibitors of reverse transcriptase. Proc Natl Acad Sci U S A. 1979 Feb;76(2):731–735. doi: 10.1073/pnas.76.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham A. H., Ponnambalam S., Chan B., Busby S. Mutations that reduce expression from the P2 promoter of the Escherichia coli galactose operon. Gene. 1986;41(1):67–74. doi: 10.1016/0378-1119(86)90268-4. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Vovis G. F., Zinder N. D. Insertion mutant of bacteriophage f1 sensitive to EcoRI. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2699–2702. doi: 10.1073/pnas.76.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Busby S., Aiba H., de Crombrugghe B. Mutations in the Escherichia coli operon that define two promoters and the binding site of the cyclic AMP receptor protein. J Mol Biol. 1982 Jan 15;154(2):211–227. doi: 10.1016/0022-2836(82)90061-4. [DOI] [PubMed] [Google Scholar]
- Busby S., Truelle N., Spassky A., Dreyfus M., Buc H. The selection and characterisation of two novel mutations in the overlapping promoters of the Escherichia coli galactose operon. Gene. 1984 May;28(2):201–209. doi: 10.1016/0378-1119(84)90257-9. [DOI] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Molecular basis of DNA sequence recognition by the catabolite gene activator protein: detailed inferences from three mutations that alter DNA sequence specificity. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7274–7278. doi: 10.1073/pnas.81.23.7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feramisco J. R., Smart J. E., Burridge K., Helfman D. M., Thomas G. P. Co-existence of vinculin and a vinculin-like protein of higher molecular weight in smooth muscle. J Biol Chem. 1982 Sep 25;257(18):11024–11031. [PubMed] [Google Scholar]
- Godson G. N., Vapnek D. A simple method of preparing large amounts of phiX174 RF 1 supercoiled DNA. Biochim Biophys Acta. 1973 Apr 11;299(4):516–520. doi: 10.1016/0005-2787(73)90223-2. [DOI] [PubMed] [Google Scholar]
- Graña D., Youderian P., Susskind M. M. Mutations that improve the ant promoter of Salmonella phage P22. Genetics. 1985 May;110(1):1–16. doi: 10.1093/genetics/110.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Inouye M. Up-promoter mutations in the lpp gene of Escherichia coli. Nucleic Acids Res. 1985 May 10;13(9):3101–3110. doi: 10.1093/nar/13.9.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilty S., Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987 May 5;262(13):6389–6395. [PubMed] [Google Scholar]
- Kunkel T. A., Alexander P. S. The base substitution fidelity of eucaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J Biol Chem. 1986 Jan 5;261(1):160–166. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClerc J. E., Istock N. L., Saran B. R., Allen R., Jr Sequence analysis of ultraviolet-induced mutations in M13lacZ hybrid phage DNA. J Mol Biol. 1984 Dec 5;180(2):217–237. doi: 10.1016/s0022-2836(84)80001-7. [DOI] [PubMed] [Google Scholar]
- LeClerc J. E., Istock N. L. Specificity of UV mutagenesis in the lac promoter of M13lac hybrid phage DNA. Nature. 1982 Jun 17;297(5867):596–598. doi: 10.1038/297596a0. [DOI] [PubMed] [Google Scholar]
- Mandecki W., Goldman R. A., Powell B. S., Caruthers M. H. lac Up-promoter mutants with increased homology to the consensus promoter sequence. J Bacteriol. 1985 Dec;164(3):1353–1355. doi: 10.1128/jb.164.3.1353-1355.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L. E., Reznikoff W. S. lac Promoter mutation Pr115 generates a new transcription initiation point. J Mol Biol. 1980 May 25;139(3):551–556. doi: 10.1016/0022-2836(80)90146-1. [DOI] [PubMed] [Google Scholar]
- Maquat L. E., Thornton K., Reznikoff W. S. lac Promoter mutations located downstream from the transcription start site. J Mol Biol. 1980 May 25;139(3):537–549. doi: 10.1016/0022-2836(80)90145-x. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., McClure W. R. Analysis of the occurrence of promoter-sites in DNA. Nucleic Acids Res. 1986 Jan 10;14(1):109–126. doi: 10.1093/nar/14.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Reznikoff W. S. Lactose promoter mutation Pr115 activates an overlapping promoter within the lactose control region. J Mol Biol. 1985 Oct 5;185(3):525–533. doi: 10.1016/0022-2836(85)90069-5. [DOI] [PubMed] [Google Scholar]
- Piette J., Cunin R., Boyen A., Charlier D., Crabeel M., Van Vliet F., Glansdorff N., Squires C., Squires C. L. The regulatory region of the divergent argECBH operon in Escherichia coli K-12. Nucleic Acids Res. 1982 Dec 20;10(24):8031–8048. doi: 10.1093/nar/10.24.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnambalam S., Webster C., Bingham A., Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal -35 region sequences. J Biol Chem. 1986 Dec 5;261(34):16043–16048. [PubMed] [Google Scholar]
- Rosen E. D., Hartley J. L., Matz K., Nichols B. P., Young K. M., Donelson J. E., Gussin G. N. DNA sequence analysis of prm-mutations of coliphage lambda. Gene. 1980 Nov;11(3-4):197–205. doi: 10.1016/0378-1119(80)90060-8. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D., Shimatake H., Brady C., Wulff D. L. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature. 1978 Mar 30;272(5652):414–423. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Treisman R., Proudfoot N. J., Shander M., Maniatis T. A single-base change at a splice site in a beta 0-thalassemic gene causes abnormal RNA splicing. Cell. 1982 Jul;29(3):903–911. doi: 10.1016/0092-8674(82)90452-4. [DOI] [PubMed] [Google Scholar]
- Wickner W., Brutlag D., Schekman R., Kornberg A. RNA synthesis initiates in vitro conversion of M13 DNA to its replicative form. Proc Natl Acad Sci U S A. 1972 Apr;69(4):965–969. doi: 10.1073/pnas.69.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. M., Reznikoff W. S. Deletion analysis of RNA polymerase interaction sites in the Escherichia coli lactose operon regulatory region. J Mol Biol. 1986 Apr 20;188(4):545–553. doi: 10.1016/s0022-2836(86)80004-3. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]



