Abstract
Nuclear xenobiotic receptor CAR activates transcription of the CYP2B6 gene by directly binding to the distal enhancer PBREM. This CAR-mediated activation is synergized by transcription factors EGR1 and HNF4α that bind to the proximal element OAREKI [1]. Two additional EGR1 binding sites have now been found just downstream from PBREM. Internal deletion of EGR 1 sites within the context of the −1.8 kb CYP2B6 promoter, which contains both PBREM and OAREKI, revealed that the distal and proximal EGR1 sites are essential for EGR1 to synergize CAR-mediated transcription. Chromatin conformation capture 3C assays demonstrated that ERG1 may loop the distal PBREM towards the proximal OAREKI so that together, CAR and HNF4α synergistically activate the CYP2B6 promoter.
INTRODUCTION
Hepatocytes have genes that encode xenobiotic metabolizing enzymes and transporters which are induced following exposure to xenobiotics including therapeutic drugs. Induction of these genes helps to defend the body against the toxicity and carcinogenicity of these xenobiotics, and also affects drug efficacy and can cause drug-drug interactions. Human cytochrome P450 2B6 (CYP2B6), one such enzyme, is clinically important as it metabolizes many of the commonly used therapeutics and activates anti-cancer pro-drugs such as cyclophosphamid and ifosfamide [2]. Numerous drugs, such as phenobarbital, phenytoin and rifampicin, induce transcription of the CYP2B6 gene, an activity in which the nuclear xenobiotic receptors CAR and PXR perform the essential roles [3]. For example, phenobarbital specifically translocates CAR from the cytoplasm into the nucleus [4], leading the direct binding of CAR to the distal sequence called “phenobarbital responsive enhance module” (PBREM) to trans-activate the CYP2B6 promoter [5]. While the distal PBREM is the primary CAR binding site required for drug activation of the CYP2B6 promoter, our recent study has shown that CAR also requires the 53-bp composite regulatory element OAREKI located in the proximal promoter region to activate the CYP2B6 gene [1,6]. The molecular mechanism by which the CYP2B6 promoter connects the function of the distal PBREM with that of OAREKI to regulate its activity, however, still remains elusive.
The OAREKI consists of the DR1, E-box and CACCC motifs, but dose not contain a CAR binding sequence. HNF4α, acting as the constitutive factor, always occupies the DR1 site, whereas early growth regulation 1 (EGR1) is the cellular signal-mediated inducible factor that binds to the CACCC site [1]. All of these three transcription factors are essential for the CYP2B6 promoter to be activated: CAR as the drug activated factor, EGR1 as the signal-induced factor and HNF4α as the constitutive factor. Intriguingly, our previous experiments including ChIP analysis have revealed that CAR indirectly interacts with the OAREKI as binding to CACCC site increases. These observations lead us to hypothesize that EGR1 may facilitate the interaction of CAR with the HNF4α on OAREKI to activate the CYP2B6 promoter. Here we have identified and characterized two additional EGR1 binding sites in the distal promoter region and performed chromatin conformation capture (3C) assays to investigate the molecular mechanism by which EGR1 facilitates CAR interaction with OAREKI.
MATERIALS and METHODS
Reagents
TCPOBOP and DMSO were purchased from Sigma Chemical Co. (St. Lois, MO); poly(dI-dC)·(dI-dC) from GE Healthcare Bio-Science Corp. (Piscataway, NJ). The plasmid –pGL3basic and pcDNA3.1-V5-His-TOPO were obtained from Promega (Madison, WI) and Invitrogen (Carlsbad, CA), respectively. Normal Rabbit IgG and antibodies against EGR1 {(588)X} were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); V5 antibody from Invitrogen.
Plasmids
The −1.8-kb CYP2B6 promoter was previously cloned into a basic firefly luciferase reporter plasmid pGL3-basic [6]. Deletions were constructed in the context of the −1.8-kb CYP2B6 promoter using the Quick Change® site-directed mutagenesis kit (Stratagene, TX) and proper primers: ΔCA1 (−269/−244), ΔCA2 (−1477/−1468), ΔCA3 (−1519/−1511) and ΔCA123 (−269/−244 and −1519/−1468). The promoters used for 3C assays were amplified to add a XhoI site at both the 5′ and 3′ ends and were cloned into a pGL3-basic plasmid. Human EGR1 cDNA was amplified from Caco2 RNA and was cloned into pcDNA3.1-V5-His-TOPO plasmids (Invitrogen). Various deletion mutants of EGR1 were likewise constructed using the Quick Change® site-directed mutagenesis kit: pcDNA3.1- EGR1 Δ1–283, pcDNA3.1- EGR1Δ3–283, pcDNA3.1-EGR1 Δ270–306 pcDNA3.1- EGR1 Δ324–377, pcDNA3.1- EGR1 Δ367–393, pcDNA3.1- EGR1 Δ422–543. All other plasmids used were previously described in our laboratory.
Cells and transfection
HepG2 and Ym17 cells were cultured in minimal essential medium supplemented with 10% (v/v) fetal bovine serum, antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin) and 2 mM glutamine. Ym17 cells are a mCAR-expressing stable cell line derived from HepG2 cells (6). For Luc reporter assays, cells were cultured on a 24-well plate at a density of 3.5 × 105 cells per well for 24 h prior to transfection with a given plasmid or a combination of multiple plasmids using FuGENE6 (Roche Diagnostics, Indianapolis, IN). After 24 h transfection, the cells were treated with TCPOBOP (250 nM), OA (10 nM) or CITCO (250 nM) for an additional 48h and were harvested for Luciferase assays using the Dual-Luciferase reporter assay system (Promega).
Chromosome conformation capture (3C)
3C assays were performed as described previously with some modifications [7]. Briefly, 100 μg of nuclear extract prepared from Ym17 cells were shaken with 50 ng of a given promoter at room temperature for 45 min and were incubated with formaldehyde to crosslink protein-DNA complexes. After glycines being added to stop cross linking, complexes were precipitate by ethanol at −20 °C., Precipitates were dissolved in 80 μl of TE, digested by XhoI at 37 °C for 2h and were incubated for 20 min at 70 °C to inactivate the enzyme activity of XhoI. Digests were diluted in ligation buffer not to exceed DNA concentration of over 0.5 ng/μl, from which 20 ng of DNA were ligated by T4 DNA ligase (2000 units, Promega) for 4h at 16 °C or by Quick ligase (Roche) for 5 min at room temperature. After ligation, protein DNA complexes were de-cross linked by adding SDS and NaCl up to 1% and 0.3 M, respectively and treated overnight at 55 °C and then treated with proteinase K for additional 1 hr at 65 °C, from which DNA was purified by PCR purification kit (QIAGEN). Purified DNA was subjected to amplification: the −1769/−31 and −682/−618 regions for the looped and linear DNA structures, respectively. For analysis by regular PCR, DNA was amplified by LA Taq (Takara), resolved by 1.5% agarose gel and visualized by ethylene bromide. For real-time PCR analysis using the 7900HT Fast Real-Time PCR System (Applied Biosystems) using two primer sets (forward: AGGATAAAAGGCCCAGTTGGA, reverse: CTAAGATTGGGTGCTCATTGCA; for circulating, forward: GGCCAGGATGGTCTCGAA, reverse: CCAGCACGTTGGGAAGCT for internal control).
Gel shift
Gel shift assays were performed with nuclear extracts prepared from cells harvested from 4 confluent 145 cm2 dishes for each drug treatment using the procedures described previously [1,8].
RESULTS AND DISCUSSION
Requirement of both DNA binding and activation domain
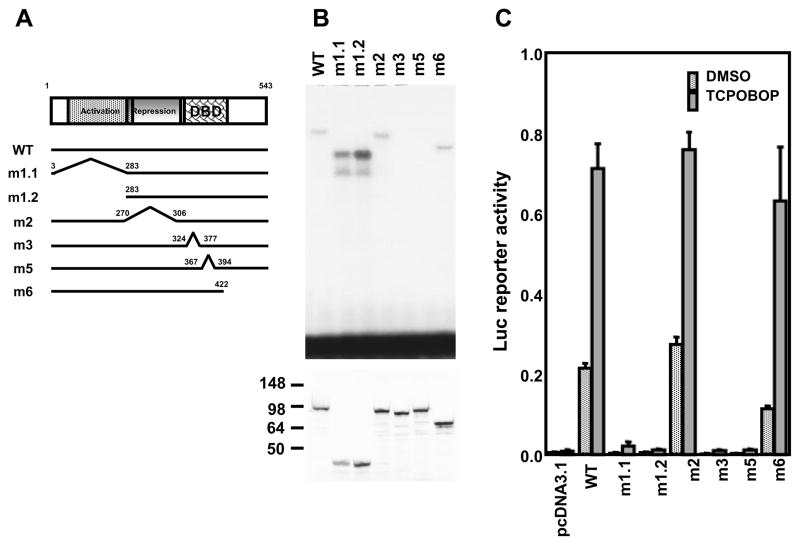
EGR1 molecule consists of multiple domains such as the activation, repression and DNA binding domains (Fig. 1A). Domain-based deletion mutants were constructed to delineate the region of EGR1 responsible for the synergistic activation of the CYP2B6 promoter. Deletion of either the activation (m1)[9], repression (m2)[10] or C-terminal (m6)[11] domain did not affect EGR1 binding to the CACCC motif of OAREKI in gel shift assays (Fig. 1B). As expected, two different deletions (m3 and m5) of the DNA binding domain [11] resulted in the loss of binding ability. These mutants were co-transfected into COS1 cells to examine their capability to synergize the CAR-HNF4α mediated activation of the CYP2B6 promoter. The m1, m3 and m5 mutants lost this capability, while the m2 and m6 mutants fully retained it as observed with wild type EGR1 (Fig. 1C). Thus, these results indicated that EGR1 requires its activation and DNA binding domains to synergize the CAR-HNF4α mediated transcription.
Figure 1. Structural domain-based function of EGR1.
(A) Deletion map of EGR1 molecule. Domains of activation, repression and DNA binding (DBD) are shown in boxes. Numeric numbers indicate residues beginning with and ending with for each deletion. (B) Gel shift assays to show binding of EGR1 and its deletion mutants to CA rich region within CYP2B6 OAREKI. Double-stranded 32P-end-labeled CA rich probe (−253/−242) was incubated with in vitro translated EGR1 (WT), its mutants (m1-m6). m1-1, 1–2, 2, 3, 5, and 6: EGR1 deletion mutants lacking the activation domain (1–283) (3–283)[9], repression domain (270–306)[10], DNA binding domain (324–377)[11], DNA binding domain (367–393)[11] and C-terminal region, respectively (422/543)[11]. Numbers indicate amino acid residues. As a negative control, empty vector (pcDNA3.1) was mixed with an in vitro translation cocktail and incubated with labeled probe. Autoradiography below is to show an equality of ERG1 proteins used for gel shift assays. EGR1 and its mutants were produced in vitro translated proteins in TNT coupled reticulocyte lysate system (Promega) and [35S] Methionine (Amersham Biosciences, Piscataway, NJ), were separated on a 11% SDS-PAGE gel and were subjected to autoradiography. Numeric numbers indicate sizes of molecular markers (C) Transient transfection assays to show EGR1 activity. Ym17 cells were co-transfected by the −1.8 kb promoter of CYP2B6 or a given deletion promoter with phRL-SV40 cells and were subsequently treated with either DMSO, or 250 nM TCPOBOP. Reporter activity was calculated by dividing firefly luciferase activity with renilla luciferase activity.
Requirement of both distal and proximal CACC motifs
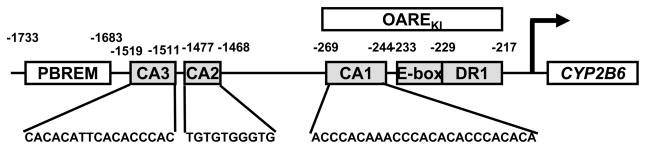
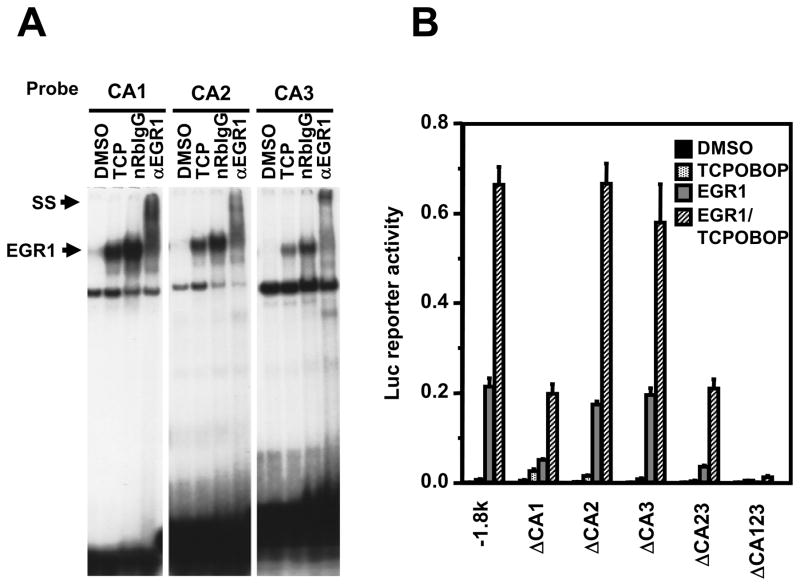
The CYP2B6 promoter is regulated by the two essential composite DNA sequences: the distal PBREM and the proximal OAREKI (Fig. 2). Our previous study showed that EGR1 depends on its direct interaction with the proximal CACCC motif (−269/−244) to synergistically activate the −1.8 kb CYP2B6 promoter [1]. This proximal CACCC motif (now designated CA1) is located at the 5′-end of the 53 bp OAREKI (−269/−217) of the CYP2B6 promoter. Sequence-based motif search of the −1.8 kb CYP2B6 promoter found two additional CACCC motifs (CA2 and CA3) at just down stream of the proximal PBREM (−1733/−1683): −1519CACACATTCACACCCAC−1511 and −1477TGTGTGGGTG−1468, respectively. Given this finding, the role of these distal CA2 and CA3 motifs in the EGR1 mediated synergistic activation was investigated. Gel shift and super-shift assays were performed using the CA1, CA2 and CA3 sequences as probes, confirming that both CA2 and CA3 bound to EGR1 in a similar manner to the CA1 (Fig. 3A). EGR1 appeared to bind to all three probes: CA1, CA2 and CA3. Within the context of the −1.8 kb CYP2B6 promoter, the CA1, CA2 and CA3 were internally deleted singularly or in combinations. Subsequently, these deletion promoters were co-transfected with an EGR1 expression plasmid into Ym17 cells so that their ability to be activated by EGR1 could be examined. The CA1 deletion decreased activation by EGR1 approximately 5-fold. Although the single deletion of either CA2 or CA3 resulted in the fully retention of the EGR1 activation, the simultaneous deletion of both CA2 and CA3 decreased the promoter activity to the same level as observed with the CA1 deletion (Fig. 3B). The triple deletion of CA1, CA2 and CA3 completely abrogated the ability of EGR1 to activate the −1.8 kb CYP2B6 promoter. These results suggested that both the proximal and distal CACC motifs are required for EGR1 to activate the −1.8 kb CYP2B6 promoter. With respect to the two distal motifs, only one of the distal CA2 and CA3 motifs was necessary for EGR1 activation in the presence of the proximal CA1 motif.
Figure 2. Motif map of CYP2B6 promoter.
The −1.8 kb CYP2B6 promoter contains three CA rich regions: promoter of CYP2B6: CA1 (−269/−244), CA2 (−1477/1468) and CA3 (−1519/−1511), which are positioned relative to PBREM (−1733/−1683) and OAREKI (−269/−217). The OAREKI sequence can be further divided into CA1, E-box (−233/−228) and DR1 (−229/−217) regions.
Figure 3. Role of CA motifs in trans-activation activity of ERG1.
(A) Gel shift assays to show binding of EGR1 to three CA rich regions within CYP2B6 promoter. A given double-stranded 32P-end-labeled CA rich probes (CA1, −253/−242; CA2, −1477/1468; CA3, −1519/−1511) was incubated with nuclear extracts prepared from Ym17 cells treated with DMSO or TCPOBOP plus PMA (TCP). For supershifts, anti-EGR1 antibody and normal rabbit IgG (nRbIgG) were co-incubated. (B) Transient transfections to show that the three CA-rich regions are critical to activate CYP2B6 promoter by EGR1. Ym17cells were co-transfected by −1.8 kb promoter construct or were its deletion mutants (−1.8 kb ΔCA1, −1.8 kb ΔCA2, −1.8 kb ΔCA3, −1.8 kb ΔCA23, −1.8 kb ΔCA123) with phRL-SV40, pcDNA3.1or pcDNA3.1-EGR1. The cells were first treated with DMSO or 250 mM TCPOBOP and were harvested for measuring luciferase activity. Reporter activity was calculated by dividing firefly luciferase with Renilla luciferase activities.
Looping distal PBREM close to proximal OAREKI
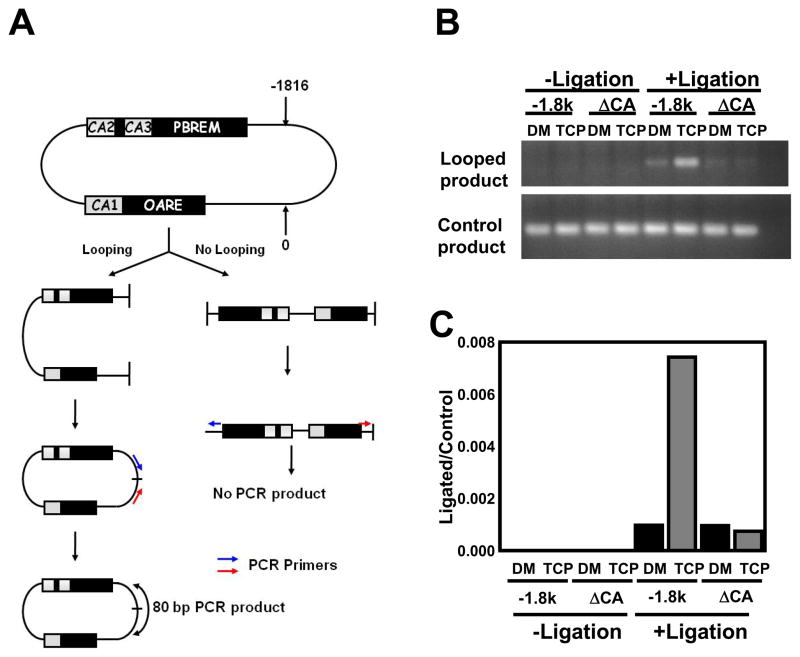
Requiring both the distal and proximal CA sites for EGR1 to synergize CAR-mediated transcription of CYP2B6 promoter led us to hypothesize the presence of a looping mechanism that enables physical access of CAR on the distal PBREM to HNF4α on the proximal OAREKI. Chromosome conformation capture (3C) assays [12,13] were employed to explore this looping mechanism. A recently developed in vitro version of 3C technique is an assay for demonstrating DNA looping, in which two distant cis-acting elements in a head-to-head orientation are designed to be PCR amplified as a single strand DNA in a head-to-tail orientation after ligation when protein bindings enable them to come close one to another [7]. 3C technique has already been employed to successfully show that the glucocorticoid receptor activates two promoters that are looped close to the single glucocoroticoid response element [12]. In our 3C assay with the −1.8 kb CYP2B6 promoter, if looping occurs, PCR reaction should produce the 80 bp fragment that contains a region of the sequence connecting PBREM and OAREKI in head-to-tail orientation (Fig. 4A). When the −1.8 kb CYP2B6 promoter was incubated with nuclear proteins prepared from Ym17 cells co-treated with TCPOBOP and PMA, a dramatic amplification of the 80 bp fragments was observed and compared with the corresponding assay with nuclear proteins from DMSO-treated Ym17 cells (Fig. 4B). No such increase of the amplification was detected when all three CA sites were deleted from the −1.8 kb CYP2B6 promoter. In addition to the regular PCR reaction, real time PCR was also utilized in our 3C assays, showing approximately 8-fold increase of amplification with the −1.8 kb CYP2B6 promoter by the nuclear extracts from the co-treated Ym17 cells and no increase with the promoter lacking all three CA site (Fig. 4C). These results are consistent with the hypothesis that EGR1, by binding to the CA sites, induces the looping of the distal PBREM closer to the proximal OAREKI within the CYP2B6 promoter.
Figure 4. Chromatin conformation capture.
(A) Schematic representation of the 3C experimental procedure. The numeric numbers 0 and −1816 indicate the XhoI cutting sites. Arrows indicate the position of PCR primers and double arrow indicates the position of the PCR amplicon. (B) The −1.8kb CYP2B6 (−1.8kb) and ΔCA123 CYP2B6 (ΔCAs) promoters were incubated with nuclear extracts prepared from Ym17 cells treated with DMSO (DM) or TCPOBOP plus PMA (TCP). After incubation, the reaction mixture was divided into two parts, of which one was ligated, while the other was unligated. Degrees of ligation were estimated by PCR amplification using two sets of primers for looped and linear products, respectively on an ethydium bromide-stained 1.5% agarose gel. (C) The same ligated and unligated samples used in Fig. 4A were subjected for real time PCR.
In conclusion, we have now uncovered a novel looping mechanism, whereby the distal PBREM close to the proximal OAREKI, EGR1 enables CAR to cross talk with HNF4α to synergistically activate CYP2B6 promoter. Multiple steps of protein-DNA and -protein interactions expected to occur during this DNA looping still remain a future subject of intense investigations. Functional cross talk of CAR with HNF4α is well known its role in the transcriptional regulation of SULT2A1and various CYP genes [14–16], for one of which indirect interactions of CAR with HNF4α are suggested [15]. Although the molecular mechanism by which EGR1 bends DNA to loop the CYP2B6 promoter is not yet understood, it is reasonable to think that ERG1 proteins at the distal and proximal CA sites may directly or indirectly interact to loop PBREM close to OAREKI within the CYP2B6 promoter. A presumed interaction of ERG1 with CAR and/or HNF4α and/or the CAR interaction with HNF4α may simultaneously undergo in order for EGR1 to bend DNA. The concept of DNA looping is widely accepted as a principle mechanism by which two distantly separated cis-acting elements interact to regulate transcription, another example of which is a possible EGR1 involvement in looping the RUSH-1α gene promoter for activation [17]. CYP2B6 is one of the human P450 enzymes that metabolize numerous therapeutic drugs. CAR mediates induction of CYP genes by therapeutics, affecting drug efficacy and causing drug-drug interactions. Since CAR is not able to activate the CYP2B6 gene without HNF4α and EGR1, understanding the molecular mechanism of their cross talk should provide us with various ways to control CYP2B6 activities with clinical implications.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program (Z01ES71005-01) of the NIH and NIEHS.
Abbreviations used
- CAR
nuclear constitutive active/androstane receptor
- PXR
pregnane X receptor
- CYP
cytochrome P450
- EGR1
early growth response 1
- HNF4α
hepatocyte-enriched nuclear factor 4α
- OAREKI
OA response element KI
- PB
phenobarbital
- PBREM
PB responsive enhancer module
- TCPOBOP
1,4-bis-[2-(3,5-dichloropyridyloxy)]benzen
- PMA
phorbol 12-myristate 13-acetate
- PKC
(PMA)-activated protein kinase C
- DMSO
dimethylsulfoxide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Inoue K, Negishi M. Nuclear receptor CAR requires early growth response 1 to activate the human cytochrome P450 2B6 gene. J Biol Chem. 2008;283:10425–32. doi: 10.1074/jbc.M800729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 pharmacogenetics and cancer. Oncogene. 2006;25:1679–91. doi: 10.1038/sj.onc.1209377. [DOI] [PubMed] [Google Scholar]
- 3.Sueyoshi T, Negishi M. Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol. 2001;41:123–43. doi: 10.1146/annurev.pharmtox.41.1.123. [DOI] [PubMed] [Google Scholar]
- 4.Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol Cell Biol. 1999;19:6318–22. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol. 1998;18:5652–8. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swales K, Kakizaki S, Yamamoto Y, Inoue K, Kobayashi K, Negishi M. Novel CAR-mediated mechanism for synergistic activation of two distinct elements within the human cytochrome P450 2B6 gene in HepG2 cells. J Biol Chem. 2005;280:3458–66. doi: 10.1074/jbc.M411318200. [DOI] [PubMed] [Google Scholar]
- 7.Babu DA, Chakrabarti SK, Garmey JC, Mirmira RG. Pdx1 and BETA2/NeuroD1 participate in a transcriptional complex that mediates short-range DNA looping at the insulin gene. J Biol Chem. 2008;283:8164–72. doi: 10.1074/jbc.M800336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274:6043–6. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 9.Gashler AL, Swaminathan S, Sukhatme VP. A novel repression module, an extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol Cell Biol. 1993;13:4556–71. doi: 10.1128/mcb.13.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russo MW, Matheny C, Milbrandt J. Transcriptional activity of the zinc finger protein NGFI-A is influenced by its interaction with a cellular factor. Mol Cell Biol. 1993;13:6858–65. doi: 10.1128/mcb.13.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matheny C, Day ML, Milbrandt J. The nuclear localization signal of NGFI-A is located within the zinc finger DNA binding domain. J Biol Chem. 1994;269:8176–81. [PubMed] [Google Scholar]
- 12.Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–33. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 13.Gondor A, Rougier C, Ohlsson R. High-resolution circular chromosome conformation capture assay. Nat Protoc. 2008;3:303–13. doi: 10.1038/nprot.2007.540. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Kissling G, Negishi M, Goldstein JA. The nuclear receptors constitutive androstane receptor and pregnane X receptor cross-talk with hepatic nuclear factor 4alpha to synergistically activate the human CYP2C9 promoter. J Pharmacol Exp Ther. 2005;314:1125–33. doi: 10.1124/jpet.105.087072. [DOI] [PubMed] [Google Scholar]
- 15.Surapureddi S, Rana R, Reddy JK, Goldstein JA. Nuclear receptor coactivator 6 mediates the synergistic activation of human cytochrome P-450 2C9 by the constitutive androstane receptor and hepatic nuclear factor-4alpha. Mol Pharmacol. 2008;74:913–23. doi: 10.1124/mol.108.048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson SS, Chen Y, LeCluyse EL, Negishi M, Goldstein JA. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4alpha. Mol Pharmacol. 2005;68:747–57. doi: 10.1124/mol.105.013169. [DOI] [PubMed] [Google Scholar]
- 17.Hewetson A, Chilton BS. Progesterone-dependent deoxyribonucleic acid looping between RUSH/SMARCA3 and Egr-1 mediates repression by c-Rel. Mol Endocrinol. 2008;22:813–22. doi: 10.1210/me.2007-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.