Abstract
We lack a comprehensive understanding of evolutionary pattern and process because short-term and long-term data have rarely been combined into a single analytical framework. Here we test alternative models of phenotypic evolution using a dataset of unprecedented size and temporal span (over 8,000 data points). The data are body-size measurements taken from historical studies, the fossil record, and among-species comparative data representing mammals, squamates, and birds. By analyzing this large dataset, we identify stochastic models that can explain evolutionary patterns on both short and long timescales and reveal a remarkably consistent pattern in the timing of divergence across taxonomic groups. Even though rapid, short-term evolution often occurs in intervals shorter than 1 Myr, the changes are constrained and do not accumulate over time. Over longer intervals (1–360 Myr), this pattern of bounded evolution yields to a pattern of increasing divergence with time. The best-fitting model to explain this pattern is a model that combines rare but substantial bursts of phenotypic change with bounded fluctuations on shorter timescales. We suggest that these rare bursts reflect permanent changes in adaptive zones, whereas the short-term fluctuations represent local variations in niche optima due to restricted environmental variation within a stable adaptive zone.
Keywords: microevolution, stasis, macroevolution, evolutionary rate, phylogenetic signal
Evolutionary biologists working at different timescales often adopt dramatically different perspectives on the pace and process of phenotypic evolution. For example, the norm for microevolutionary studies is to observe high levels of heritable genetic variation (1, 2), strong selective pressures (3, 4), and the frequent occurrence of substantial phenotypic change on a timescale of a few to a few dozen generations (5–8). However, paleontologists working on much longer timescales have recognized an overwhelming prevalence of evolutionary stasis (9–11), although other patterns are known (12–14). On even longer timescales, comparative studies of extant species routinely record substantial divergence (15, 16). These different perspectives have generated controversy about the ability of microevolutionary process to explain macroevolutionary patterns (4, 9–10, 17–21). To resolve this debate we need models that can simultaneously account for evolutionary change across a range of timescales. Here, we make a step toward that goal by assembling and analyzing a dataset of body-size evolution across an unprecedented temporal span.
Gingerich (7) compiled a large dataset representing a collection of evolutionary rates measured over 100–107 generations in the fossil record. Analyzing this dataset, Estes and Arnold (21) found a striking pattern of bounded fluctuations in phenotype, which implies that the expected magnitude of phenotypic change is about the same regardless of whether two samples are separated by 10 generations or 1 million generations (see also ref. 22). In other words, short-term, fluctuating evolution occurs, but the changes fail to accumulate with time. This pattern predicts that closely related species should be as different as less related species, a conclusion seriously at odds with comparative studies, which often detect strong phylogenetic signal for traits related to organism size (23–26). This incongruence suggests that we need a more extensive compilation of data to get a full picture of how evolutionary patterns scale up over time.
Many studies have examined large-scale patterns of body-size evolution using either fossil (7, 21, 27, 28) or comparative data (15, 16, 29). Such studies have yielded many important insights, but no studies have yet combined paleontological and comparative data in the same modeling framework, and the puzzling inconsistencies that seem to exist between these types of studies remain. Resolving these inconsistencies is made more challenging by the different timescales, traits, and taxa analyzed in different studies. To achieve a much needed synthesis, we combine microevolutionary and fossil time series data with phylogeny-based comparative data and analyze these data with a set of evolutionary models that extends the analysis of Estes and Arnold (21). We base our analysis on a dataset that spans the broadest range of timescales yet examined (0.2 y to 357 Myr) by combining historical and contemporary field studies, fossil time series, and comparative data.
Results and Discussion
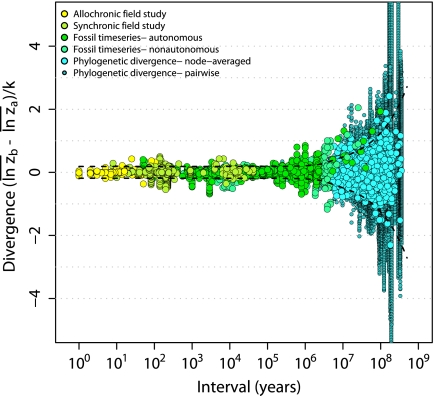
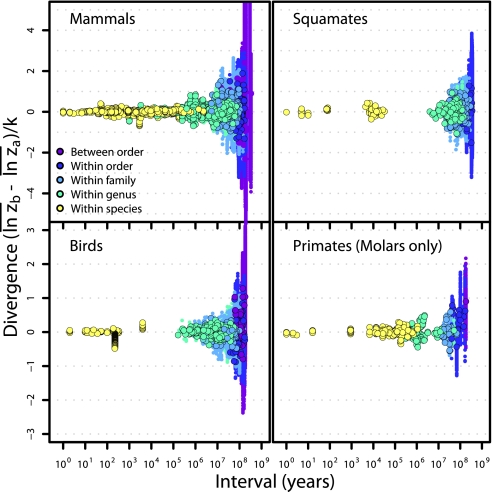
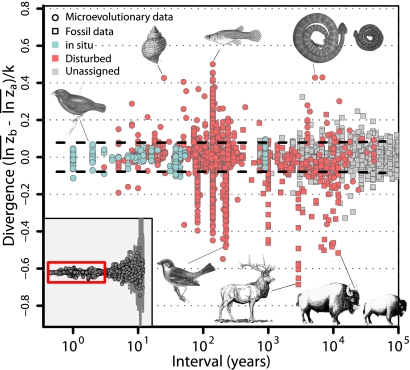
Consistent with previous studies (21, 22) we find a complete absence of a time-span effect up to ∼1 Myr, but this pattern then yields to a pattern of increasing divergence on timescales above 1–10 Myr (Fig. 1). Whereas the dominant pattern of bounded, but fluctuating changes on shorter timescales is consistent with a common paleobiological concept of stasis (9, 14), the long-term evolutionary pattern is consistent with observations of phylogenetic correlation and cumulative evolutionary change. The union of these seemingly contradictory patterns generates a remarkably continuous visual pattern reminiscent of the flared barrel of a blunderbuss firearm. We show that this “blunderbuss pattern” is consistent regardless of the method of trait standardization and whether timescale is expressed in generations or years (SI Text and Figs. S1 and S2). Because biases introduced by including different taxa and traits at different timescales may cloud our interpretation of the pattern, we examined subsets of data on divergence in the body size of mammals, squamates, and birds, as well as molar dimensions in primates (Fig. 2). The general pattern and timing are strikingly consistent across traits and taxa. Although the continuity of pattern is clear, it is less obvious what evolutionary processes can account for divergence across all timescales.
Fig. 1.
The “blunderbuss pattern”, showing the relationship between evolutionary divergence and elapsed time. Divergence is measured as the difference between the means of log-transformed size in two populations (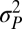 and
and 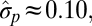 ) standardized by the dimensionality, k. Intervals represent the total elapsed evolutionary time between samples. Microevolutionary data include longitudinal (allochronic) and cross-sectional (synchronic) field studies from extant populations. Paleontological divergence is measured from time series, including both stratigraphically adjacent (autonomous) populations and averaged longer-term trends (nonautonomous). We supplement these data with node-averaged divergence between species with intervals obtained from time-calibrated phylogenies. Pairwise comparisons between species (small points) are also presented to give a visual sense of the range of divergence values across taxonomic groups. Dotted lines indicate the expected 95% confidence interval for the multiple-burst model fitted to the microevolutionary, fossil, and node-averaged phylogenetic data.
) standardized by the dimensionality, k. Intervals represent the total elapsed evolutionary time between samples. Microevolutionary data include longitudinal (allochronic) and cross-sectional (synchronic) field studies from extant populations. Paleontological divergence is measured from time series, including both stratigraphically adjacent (autonomous) populations and averaged longer-term trends (nonautonomous). We supplement these data with node-averaged divergence between species with intervals obtained from time-calibrated phylogenies. Pairwise comparisons between species (small points) are also presented to give a visual sense of the range of divergence values across taxonomic groups. Dotted lines indicate the expected 95% confidence interval for the multiple-burst model fitted to the microevolutionary, fossil, and node-averaged phylogenetic data.
Fig. 2.
Divergence patterns are similar across major groups of vertebrates. Taxonomic levels are denoted by color. For comparative data, smaller points indicate pairwise divergence measures and larger points are node-averaged divergence. Data for primates are first molar lengths and widths only.
To describe these patterns more precisely, we fitted four stochastic models to the data. The first model describes bounded evolution (BE) and was designed to fit the pattern of short-term fluctuations observed in the data. This model assumes that the amount of trait divergence over any time interval is an independent draw from a normal distribution with mean zero and variance, 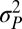 . Because this model does not allow for increasing divergence on longer timescales, we combined it with three other time-dependent stochastic models that capture this aspect of divergent evolution. The first of these unbounded model components is the well-known Brownian-motion (BM) model, which is often used to describe phylogenetic correlations. In this model the variance in trait means among replicate lineages increases linearly with time. In the single-burst (SB) model, we assume that the optimum can undergo a single large normally distributed change after a random (exponentially distributed) time interval. A special case of this model, in which the change was immediate, had the best fit to the data in Estes and Arnold (21). Finally, in the multiple-burst (MB) model, we allow multiple normally distributed changes in the optimum to occur over time according to a Poisson process.
. Because this model does not allow for increasing divergence on longer timescales, we combined it with three other time-dependent stochastic models that capture this aspect of divergent evolution. The first of these unbounded model components is the well-known Brownian-motion (BM) model, which is often used to describe phylogenetic correlations. In this model the variance in trait means among replicate lineages increases linearly with time. In the single-burst (SB) model, we assume that the optimum can undergo a single large normally distributed change after a random (exponentially distributed) time interval. A special case of this model, in which the change was immediate, had the best fit to the data in Estes and Arnold (21). Finally, in the multiple-burst (MB) model, we allow multiple normally distributed changes in the optimum to occur over time according to a Poisson process.
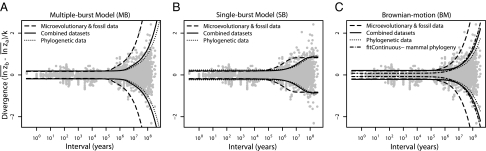
The multiple-burst model was the best-fitting model, although all unbounded models capture the expansion of variance that occurs after 1 Myr of stationary fluctuations (Figs. 1 and 3 and Table 1). The multiple-burst model was preferred over the Brownian-motion model primarily because it better explained the overdispersion of divergence values found in the data at intermediate time intervals (106–108 y) in both the fossil data and the phylogenetic data. The three models including unbounded components (BM, SB, and MB) all estimate 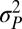 which means that 1 SD in the central band of data corresponds to an ∼10% difference in linear size traits. Some of this change reflects measurement error, but much of it probably reflects evolutionary change (SI Text). The time-dependent component of the models leads to noticeable departures from the central band only after ∼1 Myr of evolution (Table 1 and Fig. 3). For example, the expected waiting time to a displacement of the optimum in the multiple-burst model is >10 Myr (107.3976). This result implies that the distribution of divergence changes very little over the first million years, with only a modest 10% increase in the width of the 95% prediction interval for the central band occurring over this interval. By contrast, the prediction interval doubles after 5 Myr. For each displacement of the optimum, the estimated burst-size distribution predicts a size ratio between ancestor and descendant population means of 1.28.
which means that 1 SD in the central band of data corresponds to an ∼10% difference in linear size traits. Some of this change reflects measurement error, but much of it probably reflects evolutionary change (SI Text). The time-dependent component of the models leads to noticeable departures from the central band only after ∼1 Myr of evolution (Table 1 and Fig. 3). For example, the expected waiting time to a displacement of the optimum in the multiple-burst model is >10 Myr (107.3976). This result implies that the distribution of divergence changes very little over the first million years, with only a modest 10% increase in the width of the 95% prediction interval for the central band occurring over this interval. By contrast, the prediction interval doubles after 5 Myr. For each displacement of the optimum, the estimated burst-size distribution predicts a size ratio between ancestor and descendant population means of 1.28.
Fig. 3.
Best-fitting time-dependent stochastic models determined by fitting by maximum likelihood. Each model was fitted to the complete dataset, as well as to a subset including only microevolutionary and fossil data and a subset including only node-averaged phylogenetic data. Lines show the 95% confidence intervals for the model fits using the estimated parameters presented in Table 1. (A) Multiple-burst (MB) model. (B) Single-burst (SB) model. (C) Brownian-motion (BM) model. Also included is the expected 95% confidence interval for a Brownian-motion model of body mass evolution fitted to the mammalian phylogenetic data, accounting for simulated measurement error (SI Text). Although the confidence intervals for both the MB and the BM model appear very similar, the MB is a better fit to the overdispersed distribution of divergence values that occurs at intervals >1 Myr (SI Text).
Table 1.
Parameter estimates and AIC scores for four model fits to the complete dataset and subsets
| All datasets: microevolutionary, fossil, and phylogenetic | ||||
| Model | Parameter estimates |
Inverse of rate parameters | AIC | |
| Bounded evolution | 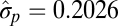 |
−2940.53 | ||
| Brownian motion | 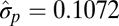 |
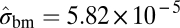 |
−7877.97 | |
| Single-burst | 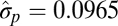 |
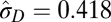 |
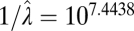 |
−9018.03 |
| Multiple-burst | 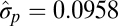 |
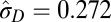 |
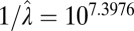 |
−9142.54 |
| Multiple-burst model fit to subsets of the data | ||||
| Dataset | Parameter estimates |
|||
| Microevolutionary and fossil | 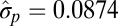 |
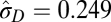 |
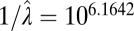 |
|
| Phylogenetic | 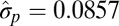 |
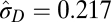 |
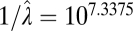 |
|
In all models, SDs are in units of the natural log size difference. The inverse of the rate parameters (1/λ) for the exponential distribution and Poisson distribution in the single-burst and multiple-burst models, respectively, can be interpreted as the average number of years until a displacement. The lowest AIC score was for the multiple-burst model.
The patterns we observe are not simple consequences of sampling bias introduced by using different data sources. Whereas fossil data could be biased by the hesitancy of paleontologists to assign ancestor–descendant relationships to highly divergent samples, we observe the opposite pattern in the dataset. When we fitted models to microevolutionary and fossil data alone and compare these with models fitted to phylogenetic data, we find that the fossil series begin to diverge more rapidly than the phylogenetic data, with otherwise remarkably similar parameter estimates (Table 1, Table S1, and Fig. 3). This pattern is not expected if fossil series are biased against evolutionary change, but may be explained, for example, by increasing probability of extinction in more rapidly evolving lineages. Alternatively, this pattern could be a consequence of the known tendency of molecular phylogenies to estimate older divergence times than the fossil record (30, 31).
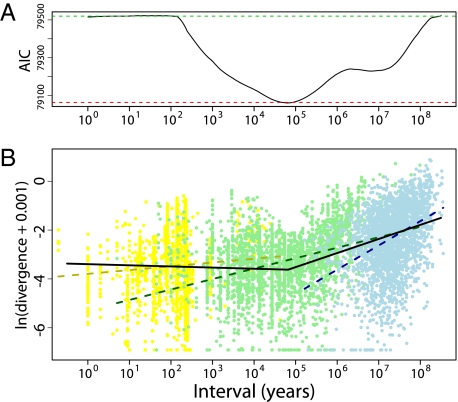
The transition from bounded evolution to steadily increasing divergence is illuminated by using linear regressions to model the relationship between absolute divergence and time. We compared a segmented regression with a single breakpoint to a model with separate regressions for each of the three subsets of data (Fig. 4 and SI Text). A segmented regression with a breakpoint at 66,000 y has a lower Akaike's information criterion (AIC) than a model in which independent regressions are fitted to each of the three major subsets of the data. This success indicates that the change in slope is not an artifact arising from differences in data sources, but instead indicates a pattern common to both phylogenetic and paleontological data (Fig. 4, Fig. S3, and SI Text).
Fig. 4.
Linear regressions of interval on the logarithm of absolute divergence, ln(|divergence| + 0.001). A positive linear relationship is predicted from Brownian-motion models of evolution. (A) AIC scores for the fit of the microevolutionary, fossil, and node-averaged phylogenetic data to linear regressions. The fit of a single linear regression to the entire dataset is the most poorly fitting model (green dashed line, AIC = 79,520). We then performed a segmented regression to find the optimal breakpoint by fitting the model iteratively while increasing the breakpoint from 0 to 8.5 log10 y with a step size of 0.01 (solid black line). The AIC for this model reaches a minimum with a breakpoint of 104.82 years (AIC = 79,059). Subsetting by datasets and fitting a regression independently to each did not improve the fit relative to the segmented regression (red dashed line, AIC = 79,062). (B) Best-fit segmented regression model provides a better fit to the data (solid line) than a model where independent linear regressions are fitted to each dataset (dashed lines). Datasets and the corresponding regression lines are indicated by color (microevolutionary, yellow; fossil, green; and phylogenetic, blue).
One potential explanation for a central, bounded band of divergence that lasts 1 Myr is that the tendency to study rapid evolution on microevolutionary timescales might mask a pattern of ever-increasing divergence in the data. In particular, evolutionary responses to disturbances such as introductions, anthropogenic disturbance, or island isolation may be especially rapid and bias microevolutionary data toward higher divergence values. To test for such bias, we examined the evolution of body size according to whether a particular study represented disturbance-mediated evolution or in situ divergence (defined in ref. 8). Disturbance-mediated evolution is clearly more prevalent in field and historical studies (Fig. 5) and also appears elevated in two fossil time series that are arguably influenced by novel selective factors: the transition of Bison antiquus to Bison bison, which is hypothesized to have been driven by community change and anthropogenic selection (31), and the evolution of a dwarf form of Cervus elaphus on the Isle of Jersey (32). Clearly, we can often identify the many causal processes that generate divergence in body size, and these are likely to be overrepresented in our microevolutionary dataset. Although these types of causal phenomena may be overrepresented in intervals spanning 1–100 y, these same phenomena are likely to occur naturally over timescales of 1,000–100,000 y, and yet we see little accumulation of divergence across these timescales.
Fig. 5.
Divergence identifiable as natural in situ variation vs. disturbance-mediated community change demonstrates that the majority of cases of rapid evolution over microevolutionary timescales are from identifiable causes such as introductions, anthropogenic disturbances, and island isolation. Highlighted examples include (clockwise, starting from left) in situ evolution of Geospiza fortis in response to natural climate variation, divergence in Nucella lapillus in response to an introduced predator, introduction of Gambusia affinis to Nevada and Hawaii, island–mainland divergence in Crotalus mitchelli, Holocene dwarfing of Bison antiquus to B. bison, dwarfing of Cervus elaphus on Jersey Island, and the introduction of Passer domesticus to North America and New Zealand. Most fossil time series cannot be assigned to either group, but are expected to record primarily in situ evolution of wide-ranging species. Dotted lines indicate the 95% confidence interval for simulated measurement error given equal means, average within-population variance, and a distribution of sample sizes matching those found in the data (SI Text).
We have taken a phenomenological approach to modeling the stable, central band of data apparent in Fig. 1 and have not attempted to account for that band with alternative models. Nevertheless, because that band is similar to a stable band analyzed by Estes and Arnold (21), we can conclude that none of the several alternative models considered by Estes and Arnold (21) can fully account for the band in Fig. 1. Those models include Brownian motion of the trait mean, Brownian and white-noise motion of an intermediate optimum (including a stationary optimum as a special case), steady directional movement of an intermediate optimum, and shifts of the trait mean between alternative adaptive peaks. All of these models fail either because they require unrealistic values for microevolutionary processes or because bounded evolution is difficult to achieve under any set of parameter values. We conclude that some unspecified process of bounded evolution is responsible for the band we observe in our data (Fig. 1). This process may include short-term movements of adaptive peaks corresponding to more or less regular, recurring fluctuations in the local environment (cf. Fig. 5 and ref. 33) as well as bounded displacements of adaptive peaks within adaptive zones. The process resembles Simpson's depiction of the bounded evolution of lineages as they radiate within an adaptive zone (34, 35). Under this interpretation, the slower time-dependent component of divergence commonly estimated from phylogenetic comparative data (14, 15) could then be due to the accumulation of rare, dramatic changes in the niche (or “primary”) optimum (18, 34, 36, 37). However, this interpretation does not address the issue of what controls the macroevolutionary dynamics of niches and results in departures from bounded evolution over longer timescales (19, 21, 37, 38).
It is tempting to turn to climate for an explanation of evolutionary bursts and suppose that the 1-Myr boundary on bounded evolution reflects limits on long-term rates of climatic and environmental change. However, substantial changes in global climate over timescales of <<1 Myr appear to result in bounded divergence characteristic of microevolutionary timescales, rather than dramatically hastening phenotypic evolution (39). Although habitat tracking by migration may mitigate the effects of such climate shifts, the data nonetheless do not strongly support a primary role for climate in driving phenotypic change.
What then, allows divergence to accumulate above 1 Myr, but not below? A particularly elegant explanation is provided by Futuyma's ephemeral-divergence model. Futuyma argues that although continuous, rapid evolution often occurs in local populations, the mosaic of niches and diverse adaptive optima of wide-ranging species prevents local evolutionary changes from spreading across the entire range (9, 40–44). Consequently, the variance of the stationary fluctuations, 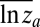 , that we estimate in our models could be interpreted as measuring the among-population geographic variation that results from ephemeral divergence in phenotypes responding to local selective pressures. Selective pressures that cause displacement of the optimum at the species level could reflect the same kind of disturbances that we observe occurring at the population level, but spread across a species’ entire range. Such significant, range-wide changes in selective optima may be sufficiently rare to explain the observed pattern of bounded evolution on timescales <1 Myr. Note that range-wide changes in adaptive optima can be accomplished by two means: either by the global spread of a selective factor across a species’ range or by the contraction of the species’ range itself (9, 41). Consequently, any process that reduces the range of a species, including speciation and taxon cycles, can potentially result in a displacement in phenotype by subsampling from the set of previously occupied adaptive niches (e.g., sampling from the distribution estimated by
, that we estimate in our models could be interpreted as measuring the among-population geographic variation that results from ephemeral divergence in phenotypes responding to local selective pressures. Selective pressures that cause displacement of the optimum at the species level could reflect the same kind of disturbances that we observe occurring at the population level, but spread across a species’ entire range. Such significant, range-wide changes in selective optima may be sufficiently rare to explain the observed pattern of bounded evolution on timescales <1 Myr. Note that range-wide changes in adaptive optima can be accomplished by two means: either by the global spread of a selective factor across a species’ range or by the contraction of the species’ range itself (9, 41). Consequently, any process that reduces the range of a species, including speciation and taxon cycles, can potentially result in a displacement in phenotype by subsampling from the set of previously occupied adaptive niches (e.g., sampling from the distribution estimated by 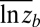 ). Species life spans are typically identified as spanning 1–5 Myr (28, 45–47) and taxon cycles have been described as spanning from 10,000 y to 10 Myr (48, 49), providing a suggestive correspondence to the rate of accumulation in phenotypic evolution that we observe in the data.
). Species life spans are typically identified as spanning 1–5 Myr (28, 45–47) and taxon cycles have been described as spanning from 10,000 y to 10 Myr (48, 49), providing a suggestive correspondence to the rate of accumulation in phenotypic evolution that we observe in the data.
Our results are qualitatively consistent with recent analyses of body-size evolution that have fitted phenomenological models to data on a more limited timescale and with more taxonomic constraint than the data that we examine here. Gillman (27) identified the striking regularity with which body-size range increases with time on macroevolutionary timescales, but did not observe this phenomenon's relationship to bounded microevolutionary divergence due to limited sampling on short timescales. Other studies used various large comparative datasets to compare the fit of Brownian motion, an Ornstein–Uhlenbeck process, and an early-burst model in which a Brownian motion rate parameter declined exponentially with time (15, 16). The early-burst model fitted best on large groups such as mammals or birds taken as a whole (16). This success indicates that the rate of evolution may be faster early in a radiation, but within subclades the early-burst model was usually outcompeted by the other models. Although Ornstein–Uhlenbeck models often are the best-fitting models in these studies in some clades and subclades, the restraining force is generally very weak (15, 16). Consequently, the qualitative picture of linearly increasing variance we observe over most macroevolutionary timescales is consistent with these results (15, 16, 27). Our analysis and treatment of the data do not allow us to make inferences about the possible role of speciation and extinction events. Those processes may or may not be important components in generating the patterns we observe. In other words, our models and conclusions allow for the possible existence of a correlation between evolutionary change and speciation and extinction events (29).
We believe the path forward will involve explicit microevolutionary interpretations of the phenomenological stochastic models used in this and similar studies (15, 16, 29). In addition, combining data across micro- and macroevolutionary timescales should facilitate interpretation of model results in terms of adaptive landscapes and ecological processes. Finally, understanding how landscape dynamics scale across species ranges is a neglected step that needs to be taken. We may need more sophisticated models that bridge between population- and range-wide evolution to understand the striking patterns of divergence that we have documented.
Materials and Methods
Measurement of Divergence and Interval.
We log-transformed all trait measurements so that divergence between any two samples is a unitless measure of the proportional change in a phenotypic trait in factors of e. Divergence between the means of two samples a and b is then measured as
where za and zb are untransformed observations in each sample. Although the difference in mean log-scaled trait values is unitless, it is not dimensionless. Consequently, we divided by k, where k is the dimensionality of the data (e.g., k = 3 for mass, k = 2 for area, and k = 1 for linear measurements). We calculated divergence from time series data, tree-based comparative data, and longitudinal and cross-sectional microevolutionary studies and calculated the total time for evolution as the total duration of independent evolutionary change separating population samples (SI Text). To minimize potential biases in trait representation across different datasets, we examine only a single trait class: morphometric traits correlated with body size on the log-linear scale. Therefore, the entire dataset approximates the evolution of a single trait: body size. Alternative standardizations are analyzed in SI Text (Fig. S1).
For each measure of divergence, we calculated a corresponding time interval. A generation timescale for these intervals is natural in the sense that many evolutionary models predict evolutionary change in generations. On the other hand, long-term trends in divergence may reflect responses to natural events that occur on timescales that cut across differences in generation time (e.g., tectonic processes, climate change). Consequently, divergence is of interest on both timescales. Preliminary analysis indicated that divergence patterns are more coincident on the absolute timescale and that estimation of generation times apparently adds some systematic bias to the overall pattern (Fig. S2). Consequently, we primarily present figures using the absolute timescale and provide generation scaling in SI Text.
Database.
We compiled datasets that measure evolutionary divergence in size-related traits from three types of data: (i) contemporary field and historical studies, (ii) fossil time series, and (iii) phylogenetic comparative data. For the first two categories, we have drawn from the original databases of Gingerich (7) and Hendry et al. (8), which also include the entirety of the size-related data used in Estes and Arnold (21). We also added 29 additional microevolutionary and paleontological studies to the dataset (SI Text and Table S2). We have supplemented these with comparative data by using databases of bird, mammal, and squamate body sizes and obtained time intervals from time-calibrated phylogenies by summing the branch lengths separating species (SI Text and Table S3). The final database includes 6,053 morphometric divergence measurements from 169 microevolutionary and paleontological studies and 2,627 node-averaged divergence estimates obtained from 37 time-calibrated phylogenies. The total time span of the complete dataset ranges from 0.2 y to 357 Myr.
Because the datasets combine many types of data across many different taxa, we risk comparing traits and taxa for which the evolutionary process is not comparable. Consequently, we examined patterns in the dataset for specific taxonomic groups (mammals, birds, squamates, and primates). For squamates, we used data on body-size divergence between mainland and land-bridge island populations, using the age of last connection between populations to estimate the time interval. For primates, we examined a single trait, first-molar size. By measuring a homologous trait in a single clade across the range of timescales included in the full dataset, we reduce the complications resulting from standardization. The first-molar size is ideal for this purpose as it is among the most abundant fossilized remains in primates and has one of the lowest sampling variances among dental traits (50).
To visualize the effect of time on evolutionary divergence in each dataset, we plotted divergence against time interval. We scaled the interval on the log10 scale to obtain resolution from microevolutionary to macroevolutionary timescales.
Stochastic Model Fitting.
We fitted four stochastic evolutionary models to the combined microevolutionary, fossil, and node-averaged phylogenetic data by maximum likelihood. These models are (i) BE, (ii) BM, (iii) SB, and (iv) MB (Fig. S4). In the bounded-evolution model, divergence is modeled as independent draws from a stationary normal distribution, N(0, 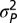 ), so that there is no time-span effect on the size of evolutionary changes. All three of the remaining models include this time-independent component of divergence, as well as a process that causes cumulative evolutionary change, resulting in increasing variance in divergence among lineages with time. In the Brownian-motion model, the distribution of replicate trait means is a normally distributed random deviate, N(0,
), so that there is no time-span effect on the size of evolutionary changes. All three of the remaining models include this time-independent component of divergence, as well as a process that causes cumulative evolutionary change, resulting in increasing variance in divergence among lineages with time. In the Brownian-motion model, the distribution of replicate trait means is a normally distributed random deviate, N(0, 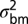 t), where t is the length of the interval. In the single-burst model, evolution is episodic. The optimum is displaced from its original position just once with a displacement magnitude that is a normally distributed deviate, N(0,
t), where t is the length of the interval. In the single-burst model, evolution is episodic. The optimum is displaced from its original position just once with a displacement magnitude that is a normally distributed deviate, N(0, 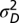 ). The timing of the displacement is not constrained as in ref. 21, but instead is modeled with an exponential distribution with rate parameter λ. In the multiple-burst model, we allowed multiple displacements to occur according to a Poisson process with rate parameter λt (as in ref. 17). As in the single-burst model, we modeled the magnitude of individual displacements as a normally distributed random deviate, N(0,
). The timing of the displacement is not constrained as in ref. 21, but instead is modeled with an exponential distribution with rate parameter λ. In the multiple-burst model, we allowed multiple displacements to occur according to a Poisson process with rate parameter λt (as in ref. 17). As in the single-burst model, we modeled the magnitude of individual displacements as a normally distributed random deviate, N(0, 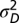 ). The distribution of phenotypes for a given number of displacements, m, is then N(0,
). The distribution of phenotypes for a given number of displacements, m, is then N(0, 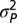 + m
+ m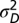 ). Note that all models include a randomly distributed normal deviate that is time independent,
). Note that all models include a randomly distributed normal deviate that is time independent,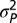 . For the best-fitting model, we bootstrapped over studies (2,000 replicates) to evaluate the sensitivity of the parameter estimates to overrepresented studies (SI Text and Fig. S5). More detail on models is provided in SI Text.
. For the best-fitting model, we bootstrapped over studies (2,000 replicates) to evaluate the sensitivity of the parameter estimates to overrepresented studies (SI Text and Fig. S5). More detail on models is provided in SI Text.
For each model, we derived a likelihood equation and estimated parameters using the functions nlm and nlminb in R (SI Text) (51). In data analysis, we assumed independence of the data, common parameters, and no autocorrelation. Developing models to deal with the complex covariance structure of this diverse dataset is beyond the scope of this paper. However, although these assumptions are clearly violated in our dataset, the exceptional size of the database should allow us to determine what types of models can explain the pattern and the associated parameter values, even if the exact differences in AIC between models are inaccurate (SI Text). We compared parameter estimates and AIC scores across models fitted to the full dataset with models fitted to subsets of data including (i) microevolutionary and fossil data only and (ii) phylogenetic data only to determine to what extent the different datasets differ in pattern. To compare our results to phylogeny-based model fitting, we fitted Brownian-motion models to the largest comparative dataset in our database (mammals) using the fitContinuous function in the Geiger package (52).
To determine how much of the estimated time-independent variance could be due to sampling error, we simulated comparisons between populations with equal means, a within-population SD of 0.055, and sample sizes drawn from a geometric distribution to match the sampling distribution of typical studies found in the database (SI Text).
Linear Regressions.
We calculated linear regressions of divergence on time. We log-transformed the absolute value of divergence, d, and regressed this value against the log10 time interval using the combined dataset of microevolutionary, fossil, and node-averaged phylogenetic data. Each of the following regressions was fitted to these data: (i) a single linear regression to the entire dataset, (ii) separate linear regressions for each dataset (microevolutionary, fossil, and phylogenetic), and (iii) a segmented regression with a single breakpoint. We compared the fits of these regressions using AIC to determine the best fitting model and whether different patterns exist between datasets (SI Text).
Supplementary Material
Acknowledgments
We thank Phil Gingerich for access to and help with his evolutionary-rate database and two anonymous reviewers for comments. We thank Lee Hsiang Liow, Trond Reitan, and Tore Schweder for helpful discussions. J.C.U. thanks the Centre for Ecological and Evolutionary Synthesis at the University of Oslo for hosting his visit. We also thank Sonya Clegg for providing unpublished data. We thank Hal and Dee Ann Story for permission to use Mr. Story's artwork and Jesse Meik, A. Michelle Lawing, and Andre Pires-da Silva for use of their photograph. This research was supported by a joint research grant from the National Science Foundation (DGE-0802268) and the Research Council of Norway (194945/V11) (to J.C.U.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.A.M. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Dryad repository http://datadryad.org/ (doi:10.5061/dryad.7d580).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014503108/-/DCSupplemental.
References
- 1.Houle D. Comparing evolvability and variability of quantitative traits. Genetics. 1992;130:195–204. doi: 10.1093/genetics/130.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen TF, Pélabon C, Houle D. Heritability is not evolvability. Evol Biol. 2011;38:258–277. [Google Scholar]
- 3.Hereford J, Hansen TF, Houle D. Comparing strengths of directional selection: How strong is strong? Evolution. 2004;58:2133–2143. doi: 10.1111/j.0014-3820.2004.tb01592.x. [DOI] [PubMed] [Google Scholar]
- 4.Bell G. Fluctuating selection: The perpetual renewal of adaptation in variable environments. Philos Trans Soc B. 2010;365:87–97. doi: 10.1098/rstb.2009.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendry AP, Kinnison MT. Perspective: The pace of modern life: Measuring rates of contemporary microevolution. Evolution. 1999;53:1637–1653. doi: 10.1111/j.1558-5646.1999.tb04550.x. [DOI] [PubMed] [Google Scholar]
- 6.Kinnison MT, Hendry AP. The pace of modern life II: From rates of contemporary microevolution to pattern and process. Genetica. 2001;112–113:145–164. [PubMed] [Google Scholar]
- 7.Gingerich PD. Rates of evolution on the time scale of the evolutionary process. Genetica. 2001;112–113:127–144. [PubMed] [Google Scholar]
- 8.Hendry AP, Farrugia TJ, Kinnison MT. Human influences on rates of phenotypic change in wild animal populations. Mol Ecol. 2008;17:20–29. doi: 10.1111/j.1365-294X.2007.03428.x. [DOI] [PubMed] [Google Scholar]
- 9.Gould SJ. The Structure of Evolutionary Theory. Cambridge, MA: Belknap Press of Harvard Univ Press; 2002. [Google Scholar]
- 10.Eldredge N, et al. The dynamics of evolutionary stasis. Paleobiology. 2005;31:133–145. [Google Scholar]
- 11.Lynch M. The rate of morphological evolution in mammals from the standpoint of the neutral expectation. Am Nat. 1990;136:727–741. [Google Scholar]
- 12.Hunt G. Fitting and comparing models of phyletic evolution: Random walks and beyond. Paleobiology. 2006;32:578–601. [Google Scholar]
- 13.Hunt G. Gradual or pulsed evolution: When should punctuational explanations be preferred? Paleobiology. 2008;34:360–377. [Google Scholar]
- 14.Hunt G. The relative importance of directional change, random walks, and stasis in the evolution of fossil lineages. Proc Natl Acad Sci USA. 2007;104:18404–18408. doi: 10.1073/pnas.0704088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmon LJ, et al. Early bursts of body size and shape evolution are rare in comparative data. Evolution. 2010;64:2385–2396. doi: 10.1111/j.1558-5646.2010.01025.x. [DOI] [PubMed] [Google Scholar]
- 16.Cooper N, Purvis A. Body size evolution in mammals: Complexity in tempo and mode. Am Nat. 2010;175:727–738. doi: 10.1086/652466. [DOI] [PubMed] [Google Scholar]
- 17.Hansen TF, Martins EP. Translating between microevolutionary process and macroevolutionary patterns: The correlation structure of interspecific data. Evolution. 1996;50:1404–1417. doi: 10.1111/j.1558-5646.1996.tb03914.x. [DOI] [PubMed] [Google Scholar]
- 18.Hansen TF, Houle D. In: Phenotypic Integration. Piggliucci M, Preston K, editors. Oxford: Oxford Univ Press; 2004. pp. 130–150. [Google Scholar]
- 19.Arnold SJ, Pfrender ME, Jones AG. The adaptive landscape as a conceptual bridge between micro- and macroevolution. Genetica. 2001;112–113:9–32. [PubMed] [Google Scholar]
- 20.Schluter D. The Ecology of Adaptive Radiation. Oxford: Oxford Univ Press; 2000. [Google Scholar]
- 21.Estes S, Arnold SJ. Resolving the paradox of stasis: Models with stabilizing selection explain evolutionary divergence on all timescales. Am Nat. 2007;169:227–244. doi: 10.1086/510633. [DOI] [PubMed] [Google Scholar]
- 22.Gingerich PD. Rates of evolution: Effects of time and temporal scaling. Science. 1983;222:159–161. doi: 10.1126/science.222.4620.159. [DOI] [PubMed] [Google Scholar]
- 23.Smith FA, et al. Similarity of mammalian body size across the taxonomic hierarchy and across space and time. Am Nat. 2004;163:672–691. doi: 10.1086/382898. [DOI] [PubMed] [Google Scholar]
- 24.Freckleton RP, Harvey PH, Pagel M. Phylogenetic analysis and comparative data: A test and review of evidence. Am Nat. 2002;160:712–726. doi: 10.1086/343873. [DOI] [PubMed] [Google Scholar]
- 25.Ashton KG. Comparing phylogenetic signal in intraspecific and interspecific body size datasets. J Evol Biol. 2004;17:1157–1161. doi: 10.1111/j.1420-9101.2004.00764.x. [DOI] [PubMed] [Google Scholar]
- 26.Blomberg SP, Garland T, Ives AR. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 27.Gillman MP. Evolutionary dynamics of vertebrate body mass range. Evolution. 2007;61:685–693. doi: 10.1111/j.1558-5646.2007.00060.x. [DOI] [PubMed] [Google Scholar]
- 28.Liow LH, et al. Higher origination and extinction rates in larger mammals. Proc Natl Acad Sci USA. 2008;105:6097–6102. doi: 10.1073/pnas.0709763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattila TM, Bokma F. Extant mammal body masses suggest punctuated equilibrium. Proc Biol Sci. 2008;275:2195–2199. doi: 10.1098/rspb.2008.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benton MJ. Early origins of modern birds and mammals: Molecules vs. morphology. BioEssays. 1999;21:1043–1051. doi: 10.1002/(SICI)1521-1878(199912)22:1<1043::AID-BIES8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 31.McDonald JN. North American Bison: Their Classification and Evolution. Berkeley: Univ of California Press; 1981. [Google Scholar]
- 32.Lister A. Critical reappraisal of the Middle Pleistocene deer species. Quaternaire. 1990;1:175–192. [Google Scholar]
- 33.Siepielski AM, DiBattista JD, Carlson SM. It's about time: The temporal dynamics of phenotypic selection in the wild. Ecol Lett. 2009;12:1261–1276. doi: 10.1111/j.1461-0248.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- 34.Simpson GG. Tempo and Mode in Evolution. New York: Columbia Univ Press; 1944. [DOI] [PubMed] [Google Scholar]
- 35.Simpson GG. The Major Features of Evolution. New York: Columbia Univ Press; 1953. [Google Scholar]
- 36.Hansen TF. Stabilizing selection and the comparative analysis of adaptation. Evolution. 1997;51:1341–1351. doi: 10.1111/j.1558-5646.1997.tb01457.x. [DOI] [PubMed] [Google Scholar]
- 37.Labra A, Pienaar J, Hansen TF. Evolution of thermal physiology in Liolaemus lizards: Adaptation, phylogenetic inertia, and niche tracking. Am Nat. 2009;174:204–220. doi: 10.1086/600088. [DOI] [PubMed] [Google Scholar]
- 38.Hansen TF, Pienaar J, Orzack SH. A comparative method for studying adaptation to a randomly evolving environment. Evolution. 2008;62:1965–1977. doi: 10.1111/j.1558-5646.2008.00412.x. [DOI] [PubMed] [Google Scholar]
- 39.Lister AM. The impact of Quaternary Ice Ages on mammalian evolution. Proc Biol Sci. 2004;359:221–241. doi: 10.1098/rstb.2003.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Futuyma DJ. On the role of species in anagenesis. Am Nat. 1987;130:465–473. [Google Scholar]
- 41.Futuyma DJ. Evolutionary constraint and ecological consequences. Evolution. 2010;64:1865–1884. doi: 10.1111/j.1558-5646.2010.00960.x. [DOI] [PubMed] [Google Scholar]
- 42.Williams GC. Natural Selection: Domains, Levels, and Challenges. New York: Oxford Univ Press; 1992. [Google Scholar]
- 43.Thompson JN. The Geographic Mosaic of Coevolution. Chicago: Univ of Chicago Press; 2005. [Google Scholar]
- 44.Lieberman BS, Dudgeon S. An evaluation of stabilizing selection as a mechanism for stasis. Palaeogeogr Palaeoclimatol Palaeoecol. 1996;127:229–238. [Google Scholar]
- 45.Liow LH, Stenseth NC. The rise and fall of species: Implications for macroevolutionary and macroecological studies. Proc Biol Sci. 2007;274:2745–2752. doi: 10.1098/rspb.2007.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alroy J, Koch PL, Zachos JC. Global climate change and North American mammalian evolution. Paleobiology. 2000;26:259–288. [Google Scholar]
- 47.Foote M. Symmetric waxing and waning of marine invertebrate genera. Paleobiology. 2007;33:517–529. [Google Scholar]
- 48.Ricklefs RE, Cox GW. Taxon cycles in the West Indian avifauna. Am Nat. 1972;106:195–219. [Google Scholar]
- 49.Ricklefs RE, Bermingham E. The concept of the taxon cycle in biogeography. Glob Ecol Biogeogr. 2002;11:353–361. [Google Scholar]
- 50.Gingerich PD. Correlation of tooth size and body size in living hominoid primates, with a note on relative brain size in Aegyptopithecus and Proconsul. Am J Phys Anthropol. 1977;47:395–398. doi: 10.1002/ajpa.1330470308. [DOI] [PubMed] [Google Scholar]
- 51.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2009. Available at http://www.R-project.org.Accessed September, 2009. [Google Scholar]
- 52.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: Investigating evolutionary radiations. Bioinformatics. 2008;24:129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.