For both naked and enveloped viruses, viral entry, genome replication, and egress involve specific interactions with the membranes of a susceptible host cell. Their characterization has led to the discovery of lipid-active compounds as potential antiviral drugs.
Abstract
Viruses intricately interact with and modulate cellular membranes at several stages of their replication, but much less is known about the role of viral lipids compared to proteins and nucleic acids. All animal viruses have to cross membranes for cell entry and exit, which occurs by membrane fusion (in enveloped viruses), by transient local disruption of membrane integrity, or by cell lysis. Furthermore, many viruses interact with cellular membrane compartments during their replication and often induce cytoplasmic membrane structures, in which genome replication and assembly occurs. Recent studies revealed details of membrane interaction, membrane bending, fission, and fusion for a number of viruses and unraveled the lipid composition of raft-dependent and -independent viruses. Alterations of membrane lipid composition can block viral release and entry, and certain lipids act as fusion inhibitors, suggesting a potential as antiviral drugs. Here, we review viral interactions with cellular membranes important for virus entry, cytoplasmic genome replication, and virus egress.
Viruses are obligatory intracellular parasites that are simple in structure and composition, but engage in multiple and complex interactions with their host. All viruses contain a nucleic acid genome encased in a protein shell, the capsid. Although the capsid represents the outermost structure of naked viruses, it is surrounded by a host cell-derived membrane, in the case of enveloped viruses. Virus replication occurs exclusively inside the respective host cell. Accordingly, viruses have to cross the host cell boundary at least twice during their replication cycle, for entry and exit. In enveloped viruses, this occurs by fusion of the incoming virus with, and budding of the nascent virus through a cellular membrane. Entry of naked viruses requires transient disturbance of a cellular (mostly endosomal) membrane to transfer the viral genome into the cytoplasm, but this disturbance must not compromise cell viability to ensure for successful viral replication (reviewed in Tsai 2007). The entry mechanisms of naked viruses are not well understood at present, but were shown to involve membrane insertion of amphipathic capsid domains (picornaviruses) (Fricks and Hogle 1990; Hogle 2002), a phospholipase activity of the viral capsid (parvoviruses) (Zadori et al. 2001; Farr et al. 2005), retrograde transfer through the translocon at the endoplasmic reticulum (ER) (polyomaviruses) (Tsai et al. 2003; Magnuson et al. 2005; Schelhaas et al. 2007; Tsai 2007; Qian et al. 2009; Tsai and Qian 2010), lytic activity of a capsid protein inducing positive membrane curvature (adenoviruses) (Maier and Wiethoff 2010; Maier et al. 2010), or membrane pore formation by a myristoylated protein (reoviruses) (Liemann et al. 2002; Zhang et al. 2009). Naked viruses generally exit the infected cell by membrane disruption through cell lysis, but transient envelopment followed by exit through the secretory route has been described for rotaviruses (Cuadras et al. 2006).
The genomes of most DNA and of some RNA viruses have to enter the nucleus for replication. Nuclear entry may be facilitated by disruption of the nuclear envelope during mitosis (in the case of most retroviruses with the exception of lentiviruses), but mostly depends on karyophilic properties of viral structural proteins or on the association of viral components with nuclear import factors and subsequent transport through the nuclear pore complex. Nuclear exit of viral components (proteins and nucleic acids) generally also occurs through the nuclear pore, with the notable exception of herpesvirus capsids, which assemble in the nucleus and exit by consecutive envelopment and deenvelopment at the inner and outer nuclear membrane, followed by secondary envelopment in the cytoplasm to yield the complete virion (Mettenleiter et al. 2009).
The finding that lipid interactions, including membrane envelopment, membrane fusion, and membrane remodeling are crucial for successful replication of many viruses triggered studies on compounds that either affect lipid biosynthesis or bind and extract specific lipids regarding their effects on viral replication. Compounds affecting cholesterol (e.g., β-cyclodextrin, amphotericin B methyl ester, statins) or sphingolipids (e.g., L-cycloserine, Lysenin) (reviewed in Chan et al. 2010; Waheed and Freed 2010), as well as plant-derived compounds (Verma et al. 2009) and synthetic compounds which emulate natural lipids (M Lorizate and H-G Kräusslich, unpub. data), were shown to interfere with viral infectivity at different stages of virus replication. Their applicability as antiviral compounds will depend on achieving specificity for the viral membrane or pathway, however, which may be difficult for abundant cellular lipids.
In the following, we discuss viral interactions with cellular membranes important for virus entry, cytoplasmic genome replication, and virus egress.
VIRUS ENTRY
Viral infection of animal cells requires transfer of the viral genome into the host cell cytoplasm, either at the plasma membrane or at the endomembrane system. For enveloped viruses, transfer is achieved by fusion of the viral and cellular membranes, whereas naked viruses need to transiently destabilize the target membrane without compromising its overall integrity. Virus entry is a stochastic event, but not a passive process. Productive entry and downstream events rely on normal cellular processes, including endocytosis (clathrin-mediated, clathrin-independent pathway, raft-dependent pathways, and macropinocytosis), vesicular trafficking, and membrane fusion (Fig. 1). Virus entry and post entry stages have been the subject of several excellent recent reviews (Smith and Helenius 2004; Sieczkarski and Whittaker 2005; Marsh and Helenius 2006; Tsai 2007; Weissenhorn et al. 2007; Harrison 2008; Haywood 2010; Kielian et al. 2010; Mercer et al. 2010; Schelhaas 2010) and will therefore only be briefly summarized here.
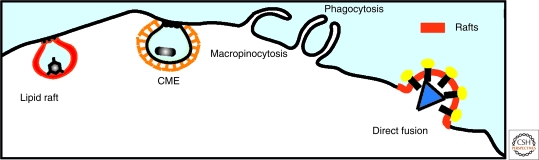
Figure 1.
Pathways of viral entry. Viruses achieve host cell entry in two principal ways: by direct fusion at the plasma membrane or following an endocytic pathway. The major endocytic pathways operating in mammalian cells that are exploited by viruses are clathrin-mediated endocytosis (CME), lipid raft pathway, clathrin-independent pathways, macropinocytosis, and phagocytosis.
Virus entry is specific for susceptible host cells and depends on viral surface proteins and host cell receptors. Most cellular receptors are surface proteins of various functions, but sugars (i.e., sialic acid for influenza virus) and lipids can also function as receptors. Examples for the latter are gangliosides which serve as receptors for members of the polyomavirus family (Smith et al. 2003; Tsai et al. 2003; Low et al. 2006; Sapp and Day 2009; Ewers et al. 2010; Tsai and Qian 2010) and phosphatidylserine which can be used by vesicular stomatitis virus (VSV) (Carneiro et al. 2002, 2006). Virus entry may also be enhanced by nonspecific binding, thus increasing viral residence times at the cell surface. This is commonly achieved by glycosaminoglycans (e.g., heparan sulfate), which promote cell attachment of many different viruses by ionic interactions (Spillmann 2001; Liu and Thorp 2002). Several glycosphingolipids may have a similar function for human immunodeficiency virus (HIV) attachment in certain cell types. Fusion commonly does not occur at the site of initial attachment, but after further trafficking either on the cell surface or through the endosomal pathway. Virus particles attached to cell filopodia can reach the cell body by an actomyosin-dependent “surfing” process, initially discovered for retroviruses, but later also observed for other virus groups (reviewed in Sattentau 2008; Mothes et al. 2010). In the case of coxsackievirus entry into epithelial cells, the virus first attaches to the GPI-anchored protein decay-accelerating factor with subsequent Abl- and Rac-dependent actin rearrangements leading to virus movement on the cell surface to tight junctions, where it meets its actual entry receptor (coxsackie-adenovirus receptor [CAR]) (Coyne and Bergelson 2006). The tight junction complex also seems to be important for hepatitus C virus (HCV) entry which requires four different cell surface molecules: CD81, scavenger receptor B, and the tight junction proteins claudin and occludin (reviewed in Ploss and Rice 2009).
Intuitively, direct fusion of the cell surface attached virus with the plasma membrane would appear to be the most likely pathway for entry. On the contrary, however, most viruses enter cells through the endosomal route. This has several advantages: (1) all virion components are completely removed from the cell surface, hiding them from the immune system; (2) endosomal transport carries the virus across the cortical actin, thus overcoming a barrier and delivering the genome deeply into the cytosol; (3) endosomal acidification can serve as a cue for conformational changes of viral surface proteins thus triggering the fusion process at the desired stage. All viruses requiring low pH for fusion traffic through the endosomal route, whereas pH-independent viruses can fuse either at the cell surface or from the endosome. Accordingly, these viruses induce syncytia between infected and noninfected cells confirming their capacity for plasma membrane fusion. Nevertheless, exclusive plasma membrane fusion appears to occur rarely—if at all—and even those viruses that can fuse at the plasma membrane appear to commonly take an endosomal route (Permanyer et al. 2010). This can be host cell dependent as shown for herpesviruses (reviewed in Heldwein and Krummenacher 2008; Akhtar and Shukla 2009), and it is conceivable that the density and motility of cell surface receptors and the kinetics of endocytosis determine where the virus fuses in such cases. Furthermore, virus attachment to cell surface receptors commonly activates signaling pathways that may trigger surface trafficking of cell-bound particles, virion endocytosis, and downstream processes (Greber 2002; Coyne and Bergelson 2006; Coyne et al. 2007).
Many viruses rely on lipid rafts for entry. For instance, several nonenveloped viruses exploit raft-dependent entry pathways requiring cholesterol. These viruses associate with detergent-resistant membranes at the plasma membrane and with liquid ordered (lo) phases in giant unilamellar vesicles (Ewers et al. 2010). Lipid rafts also play a role in enveloped virus entry as can be inferred from the usage of raft-associated viral receptors (i.e., GPI-anchored or raft-associated trans-membrane receptors) and the dependency of entry on raft integrity and cholesterol (Chan et al. 2010). Ceramides inhibit virus entry, probably by self-segregating into ceramide-rich microdomains (Cremesti et al. 2002) that laterally segregate cholesterol from raft domains (Megha and London 2004). This membrane rearrangement could lead to the dispersion of surface receptors, reducing the possibility of effective virus–host interaction (Cremesti et al. 2002; Finnegan et al. 2004; Megha and London 2004).
Membrane Fusion
Viral membrane fusion is mechanistically similar to SNARE-mediated cellular vesicle fusion processes (Frolov et al. 2011), except that the entire fusion machinery is provided by the virus and thus resides on a single membrane. Furthermore, viral envelopes have not been shown to redistribute lipids between leaflets and are metabolically inert using only the energy released by conformational changes of virion proteins. Viral fusion proteins are classified into three distinct classes (class I, II, and III), which differ in oligomeric state and structure, but follow a mechanistically similar reaction cascade (Weissenhorn et al. 2007; Harrison 2008; Backovic and Jardetzky 2009; Kielian et al. 2010). Fusion is activated by specific triggers like low pH or the engagement of a cellular coreceptor, leading to large structural rearrangements in the viral fusion protein and exposure of a hydrophobic peptide, loop, or patch (the so-called fusion peptide).
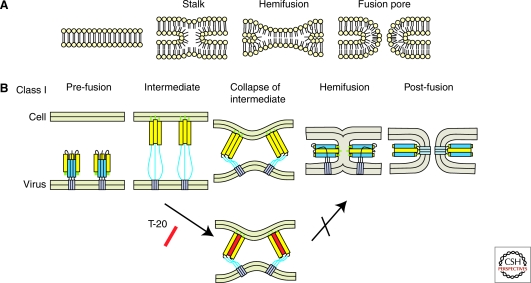
Virus membranes have a strong positive curvature relative to the host cell membrane. After attachment and triggering, the fusion peptide inserts into the target membrane, and virus-cell fusion proceeds through formation of a transient lipid stalk (Fig. 2), where the outer leaflets of the viral and cellular membranes are already mixed, while the inner leaflets are still separated. This so-called hemifusion intermediate is characterized by the transition from strong positive to strong negative curvature. The energy for this unfavorable conversion is provided by the conformational rearrangements of the viral glycoproteins with membrane distortion by the two membrane-inserted segments (trans-membrane domain and fusion peptide) lowering the energy barrier. Subsequently, the inner leaflets merge, and the hemifusion stalk opens to form the fusion pore. Finally, the pore becomes stable and expands leading to complete fusion, which requires overcoming an additional energetic hurdle (Chernomordik and Kozlov 2005). This is mediated by refolding of the fusion protein into a rigid rod-like conformation comprising a six-helix bundle with the two membrane-interacting regions of the protein located at the same end of the rod. The energy barrier to overcome the hemifusion stalk is ∼40–50 Kcal/mol, corresponding to the free energy released by the collapse of one or two fusion protein trimers (Danieli et al. 1996; Kuzmin et al. 2001; Chernomordik et al. 2006; Yang et al. 2006; Harrison 2008). It appears likely, however, that multiple trimers are required for productive viral membrane fusion.
Figure 2.
Membrane fusion. (A) Stalk mechanisms of lipid bilayer fusion. (B) Fusion models promoted by class I fusion proteins. The lower panel depicts the T-20 mode of action inhibiting transition from the prehairpin structure to six-helix bundle formation by direct binding to the intermediate.
According to the described mechanism, viral fusion proteins generally carry a hydrophobic fusion peptide and two amphipathic helical regions. Besides these, a membrane proximal external region (MPER) (Salzwedel et al. 1999) of the fusion protein can also be involved in the fusion process, possibly destabilizing the viral membrane (Saez-Cirion et al. 2002; Buzon et al. 2010) by partitioning into the membrane interface (reviewed in Shai 2001; Nieva and Agirre 2003; Lorizate et al. 2008).
Fusion Inhibitors and Role of Lipids
Entry of many viruses involves lipid rafts, where viral receptors and/or coreceptors are often localized, but lipids also play more direct roles in viral entry. Cholesterol and sphingolipids are both required for alphavirus fusion, whereas lipid rafts appear dispensable (reviewed in Kielian et al. 2010). The molecular shape of membrane lipids strongly influences virus–cell fusion and may even be exploited for the design of antiviral compounds. Cylindrical lipids, with hydrophobic tails and hydrophilic heads of similar cross section, are likely to form lamellar bilayers lacking curvature. Insertion of cone-shaped lipids with larger hydrophobic tails favors negative curvature, whereas insertion of inverted cone-shaped lipids with larger hydrophilic heads favors positive curvature. Accordingly, the outer leaflets of viral envelopes are often enriched in glycolipids with relatively large hydrophilic heads. Viral membrane fusion and infectivity can be modulated by inserting curvature-affecting lipids into the viral or cellular membrane. The presence of cone-shaped lipids like phosphatidylethanolamine (PE) or diacylglycerol (DAG) in the outer leaflet promotes hemifusion, whereas inducing positive curvature inhibits stalk formation and consequently membrane fusion. This has been shown for lysophospholipids and lysophosphoglycans with hydrophilic heads of much larger diameter than their hydrophobic tails, which act as near-universal fusion inhibitors (Chernomordik et al. 1995). However, these molecules are often cytotoxic or rapidly degraded, preventing their application as antivirals. Recently, synthetic and nontoxic rigid amphipathic fusion inhibitors (RAFIs) were designed on the basic principle of inverted cone-shaped lipids (St Vincent et al. 2010). These compounds insert into the viral membrane and promote positive curvature, thus increasing the energy barrier for fusion. RAFIs were shown to inhibit fusion of several unrelated enveloped viruses at nanomolar concentration, while being nontoxic and inactive against nonenveloped viruses. A similar principle may also apply to another recently reported, but chemically unrelated compound, LJ001, which inhibits fusion of several unrelated enveloped viruses as well (Wolf et al. 2010). Such molecules are attractive candidates for broad-spectrum antivirals against a wide variety of enveloped viruses.
Viral membrane fusion may also be inhibited by targeting the fusion machinery. T-20, a peptide derived from the carboxy-terminal heptad repeat region of the HIV fusion protein, is clinically used to treat HIV/AIDS patients, and is the first approved entry inhibitor (Kilby and Eron 2003). It functions by competitively binding the amino-terminal heptad repeat and thus preventing six-helix-bundle formation (Fig. 2B). This principle also applies to other viruses, and related fusion inhibitors have been designed from their respective sequences. Small molecules dominantly competing with the fusogenic gp41 helical sequences for binding to hydrophobic grooves transiently exposed at the ectodomain surface have also been described as HIV fusion inhibitors (Doms and Moore 2000; Eckert and Kim 2001; Frey et al. 2006). A naturally produced blood plasma oligopeptide called VIRIP (virus-inhibitory peptide) is capable of blocking HIV-1 infection by targeting the fusion protein as well (Münch et al. 2007), and oligopeptides that target or are derived from the fusion protein sequence have been shown to inhibit cell–cell syncytia formation (Owens et al. 1990; Kliger et al. 1997; Gomara et al. 2006).
ROLE OF MEMBRANES IN VIRUS REPLICATION
Interaction with and alteration of cellular membranes plays an important role in genome replication of many viruses (Miller and Krijnse-Locker 2008). Poxviruses (e.g., Vaccinia virus) generate a transient membrane-shielded replication factory by rearranging ER-derived vesicles around viral replication sites to form a structure resembling a cytoplasmic “mini-nucleus” (Tolonen et al. 2001). Positive-strand RNA viruses induce formation of a large number of cytoplasmic vesicles (Salonen et al. 2005), specialized sites for genome replication. The vesicular membranes are in most cases ER-derived (flaviviruses, picornaviruses, SARS-coronavirus), but may also originate from endosomes/lysosomes (togaviruses) or mitochondria (nodaviruses) (Miller and Krijnse-Locker 2008), and can be embedded in extended membrane networks or membranous webs (Egger et al. 2002; Knoops et al. 2008; Welsch et al. 2009; Hsu et al. 2010). Recent analyses indicated that replication vesicles in Semliki Forest virus (SFV)-infected cells originate from the plasma membrane and undergo long distance endosomal transport on microtubules which is dependent on actomyosin and phosphatidyl inositol-3-kinase (PI3K) (Spuul et al. 2010). The induction of such novel cytoplasmic membrane compartments by virus infection has been suggested to (1) increase the local concentration of components required for replication; (2) provide a scaffold for anchoring the replication complex; (3) confine the process of RNA replication to a specific cytoplasmic location; (4) prevent the activation of host defense mechanisms by sequestering the replication complexes; and (5) link virus replication and assembly.
Induction of the vesicular networks involves rearrangement of existing cellular membranes by viral proteins but probably also de novo synthesis of lipids. HCV infection has been reported to cause a protein kinase B dependent inactivation of the cellular AMP-activated protein kinase (AMPK), which is required for formation of the HCV-induced membranous web. Accordingly, HCV replication could be blocked by restoring AMPK-activity (Mankouri et al. 2010). Recent analysis of host factors required for Dengue virus (DENV) replication showed that fatty acid synthetase is recruited to viral replication sites through the viral NS3 protein. This led to an increase in the rate of fatty acid biosynthesis in DENV-infected cells with de novo synthesized lipids preferentially cofractionating with the viral genome (Heaton et al. 2010). It appears likely that a similar increase in lipid biosynthesis will be found for other viruses employing membranous replication factories. Three-dimensional analysis of the architecture of DENV replication sites revealed a membrane network with replication vesicles that have cytosolic pores for genome exit in close proximity with virus assembly and budding sites (Welsch et al. 2009). These results suggest an intricate coupling of plus-strand RNA virus genome replication and virion morphogenesis at virus-induced membrane networks, and similar structures have been observed or are predicted for other replication sites.
Viral factors inducing membrane networks required for RNA replication are sometimes membrane-spanning proteins (e.g., NS4B of HCV or NS4A of DENV), although many are soluble proteins, whose mechanism of action is currently unknown. A recent study revealed that picornavirus 3A, a small tail-anchored membrane protein, remodels the host cell secretory pathway to generate a replication compartment with unique lipid and protein composition (Hsu et al. 2010). This is achieved by 3A interacting with the small Ras family GTPase Arf1 and its guanine nucleotide exchange factor GBF1, which specifically promotes phosphatidyl inositol-4-kinase IIIβ (PI4KIIIβ) localization to the compartment, while the closely related isoform PI4KIIIα is not recruited. The 3A containing membranes are also reduced in COP-I binding, causing a defect in their anterograde membrane trafficking. Recruitment of PI4KIIIβ increased the concentration of the phosphoinositide PI4P in the membrane of the virus-induced compartment fivefold thus facilitating membrane binding of the soluble viral RNA polymerase 3D, which specifically interacts with PI4P (Hsu et al. 2010). The viral 3D enzyme does not contain a known PI4P-binding motif and elucidating its binding mode may lead to discovery of similar interactions in other viruses. Conceivably, 3D enzymatic activity may even be stimulated by PI4P binding, thus coupling the formation of the replication compartment not only with recruitment of the replication machinery, but also with induction of its activity.
PI4KIIIβ has been suggested to play a similar role in HCV replication, which also appears to make use of a PI4P-rich lipid environment for its replication (Hsu et al. 2010). A role of PI4KIIIβ was not confirmed in several recent siRNA screens for host factors of HCV replication, however, and this difference has been attributed to HCV strain variation (Reiss et al. 2011). A more robust phenotype was observed for PI4KIIIα in these screens (Tai et al. 2009; Vaillancourt et al. 2009), which is recruited to HCV replication sites and activated by the viral NS5A protein (Reiss et al. 2011). This activation is required for the functional integrity of the viral replication organelle (the membranous web) and PI4P levels are increased in HCV-infected cells in tissue culture and in infected livers in vivo (Hsu et al. 2010; Reiss et al. 2011). Although PI4KIIIα is not required for replication of the related DENV, PI4KIII isoforms appear to be key cellular proteins exploited by several RNA viruses for their replication, and may be candidates for the development of broad-spectrum antivirals targeting essential host cell factors and active against several virus families.
VIRUS MORPHOGENESIS AND RELEASE
Most enveloped viruses acquire their envelope by budding through a cellular membrane. All viral membranes are thus derived from cellular membranes, but may differ from these in protein and lipid content. Budding and release of an infectious virion requires trafficking and assembly of its essential components (genome, inner structural proteins, surface glycoproteins, and accessory components) to the budding site, induction of membrane curvature by the nascent virion and scission of the viral from the cellular membrane. These processes mechanistically resemble the formation of cellular vesicles and make use of cellular components, but are generally orchestrated by viral machinery (Welsch et al. 2007).
Virus envelopment may occur at the plasma membrane or at internal membranes depending on the virus (Fig. 3A). Most retro-, paramyxo-, and orthomyxoviruses (including HIV, measles virus, and influenza virus) bud at the plasma membrane, whereas flaviviruses such as DENV bud at the ER membrane, and secondary envelopment of herpesvirus capsids exported from the nucleus occurs at membranes of the TGN or endosomes (Mettenleiter et al. 2009). Viruses that bud into the cellular endomembrane system are generally released via the secretory pathway.
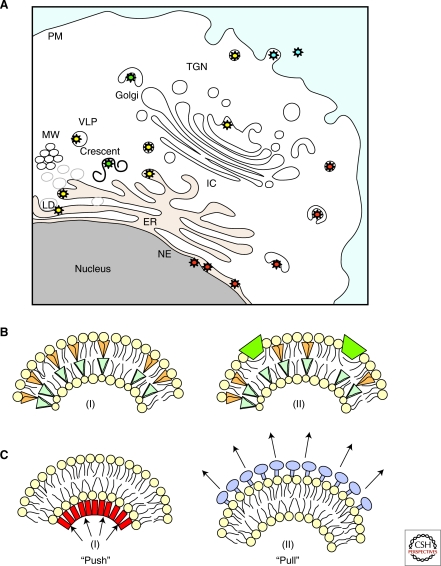
Figure 3.
Membranes implicated in enveloped virus budding and mechanisms of curvature induction. (A) Virus envelopment can occur at the plasma membrane (particles depicted in blue) or into the lumen of organelles (i.e., ER, LD, Golgi, and TGN) along the secretory pathway (particles depicted in yellow and green). Herpesviruses undergo sequential envelopment, de-envelopment and re-envelopment that take place at the nucleus and TGN (particles depicted in red). (MW, membranous web; LVP, lipo-viro-particles; LD, lipid droplets.) (B) Factors producing membrane curvature include (I) lipid molecules with different shapes, (II) shallow insertions of hydrophobic or amphipathic protein domains into one of the membrane monolayers. (C) Membrane scaffolding driven by inner structural proteins of the virion (“Push”; i.e., viral core proteins) (I) or by surface proteins on the outer membrane (“Pull”; i.e., viral envelope proteins) (II).
Membrane envelopment of viral cores is different for Vaccinia virus, in which the first stage of virion assembly comprises a crescent-shaped membrane precursor whose identity and origin has been the subject of long-standing discussion. Recent analyses using deep-etch microscopy as well as cryo electron microscopy and tomography indicated that these crescents are uniformly curved open cytoplasmic membrane sheets consisting of a single bilayer and supported by a hexagonal viral protein lattice (Heuser 2005; Chlanda et al. 2009). Open membrane sheets are generally not observed in the cytoplasm and have been proposed to be derived from virus-induced rupture of small vesicles connected to the crescent (Chlanda et al. 2009). The viral factor that induces membrane rupture is unknown, but the major protein of viral crescent, A17, has been recently shown not to be responsible (P Chlanda, unpubl.). The open crescent subsequently closes into a spherical structure once the viral genome and associated factors are enwrapped leading to formation of the infectious mature virion, which may undergo secondary envelopment at the TGN (Sodeik and Krijnse-Locker 2002).
HCV also shows an unusual pathway of virion morphogenesis that is tightly linked to lipid biosynthesis. The virus presumably buds at the ER membrane, but makes use of a specialized export route. Infectious HCV particles from patient plasma have an unexpectedly low density because of their association with host-derived apolipoproteins and lipids (André et al. 2002). Treatment with lipoprotein lipase inhibits HCV infectivity confirming that lipoproteins are important for infectivity (Andreo et al. 2007), and infectious HCV is therefore considered a “lipo-viro-particle” (LVP) (Fig. 3A) (André et al. 2005). The association with cellular lipoproteins appears to occur during HCV morphogenesis, which depends on components of the very low-density lipoprotein (VLDL) pathway (Huang et al. 2007; Syed et al. 2010; Bartenschlager et al. 2011). This conclusion is supported by the recent analysis of the HCV lipid composition (see below). Importantly, HCV core protein is specifically recruited to and associates with cytosolic lipid droplets (LD) that are thought to serve as assembly platform (Miyanari et al. 2007; Shavinskaya et al. 2007). Transfer of core to the lipid droplet has recently been shown to require diacylglycerol acyltransferase I, which serves as essential host factor for HCV morphogenesis (Herker et al. 2010). As in other flaviviruses, HCV genome replication at the membranous web, virus assembly and budding into the ER are tightly coupled processes, where the replicated viral genomes must associate with LD-associated core proteins for production of virus progeny. It is currently not clear, however, whether this involves transfer of replicated viral genomes to the core protein or—vice versa—transfer of HCV core from LD to the site of genome replication (Bartenschlager et al. 2011). It is assumed that the assembled nucleocapsid subsequently buds at the ER membrane, where it may associate with lumenal lipid droplets, which are precursors of VLDL, thus yielding HCV release via the VLDL pathway.
Induction of Membrane Curvature in Virus Budding
All budding processes require the generation of membrane curvature, which may be achieved by different mechanisms. These include the insertion of wedge-shaped lipid molecules or of shallow hydrophobic protein regions into the membrane and the formation of membrane scaffolds on the outer or inner membrane leaflet (Fig. 3B) (Kozlov et al. 2010). In most cellular vesicles, curvature is generated by assembling a protein coat on the cytoplasmic face of the membrane (see Frolov et al. 2011) or by assembling a protein lattice at the neck of the nascent bud. Viruses cannot make use of cellular coats because the topology of their budding (away from the cytoplasm) is opposite to that of most cellular vesiculation processes. A notable exception is vesicle formation at the multivesicular body (MVB), and it is thus not surprising that many enveloped viruses have hijacked part of the ESCRT (endosomal sorting complex required for transport) machinery involved in this process (see below).
Depending on the topology of the assembly machinery viruses can be grouped into different classes (Fig. 3C). In one group, budding is driven by assembly of the membrane glycoproteins on the surface of the nascent bud, similar to the action of cellular coat proteins, but on the other side of the membrane (“pulling force”). Examples are the HA protein of influenza virus (Chen et al. 2007; Chen and Lamb 2008) and the surface glycoproteins of the flavivirus tick borne encephalitis virus and of hepatitis B virus (HBV) (Allison et al. 1995; Vennema et al. 1996). Expression of these glycoproteins leads to release of subviral particles even in the absence of other viral components. In a second group of viruses, budding is driven by the inner structural proteins exerting a “pushing force” to deform the membrane. The most prominent example are retroviruses including HIV, in which expression of only the Gag protein is sufficient for assembly and release of virus-like particles (VLP) resembling the immature virion (Gheysen et al. 1989; Bieniasz 2009). It is likely that Gag lattice formation at the membrane and lipid interactions provide the energy for membrane bending, thus wrapping the plasma membrane around the nascent virion. VLP release was also observed on expression of matrix proteins of several other viruses suggesting that curvature induction is also facilitated by the inner structural proteins in these cases (reviewed in Welsch et al. 2007; Kozlov et al. 2010). Alphaviruses including SFV provide an example, in which both the surface glycoproteins and the nucleocapsid form a regular protein lattice contributing to virus budding, and the induction of curvature occurs by their concerted action on both sides of the membrane (Garoff et al. 2004).
Association of Viral Components with Membranes and Membrane Microdomains
Although viral membrane glycoproteins are, as a rule, cotranslationally inserted into the ER membrane (Doms et al. 1993), the inner structural proteins involved in viral nucleocapsid formation generally lack membrane-spanning domains. Their recruitment to the budding membrane may be facilitated by specific membrane targeting and retention signals and/or by interaction with trans-membrane components of the virus through recognition signals in the cytoplasmic domain of the viral glycoproteins. Congregation of viral components at the budding membrane may be facilitated by their cosorting into the same membrane microdomain. A specific interaction of trans-membrane proteins and viral capsid proteins or preassembled nucleocapsids leading to their recruitment to the respective budding regions has been observed, for example, herpesviruses, HBV, and some retroviruses (Wilk et al. 2001; Sfakianos and Hunter 2003; Mettenleiter et al. 2009; Patient et al. 2009). In these cases, virus envelopment and release are completely dependent on the viral glycoproteins, whereas assembly of the viral nucleocapsid is not.
Many viruses have been suggested to bud from raft-like membrane microdomains based on their lipid composition (see below) and the incorporation of raft-associated proteins into the virion (reviewed in Ono 2010; Waheed and Freed 2010). Association with membrane microdomains serves to concentrate and partition viral components, while reducing or excluding host cell membrane proteins. Lipid rafts are small, short-lived sterol- and sphingolipid-rich domains with a lo membrane structure and an estimated diameter on the order of 10–50 nm (see Simons and Sampaio 2011). They are thus much smaller than viral envelopes. Accordingly, virus formation is unlikely to occur from a single raft, and raft-dependent viruses have to organize their membrane either by recruitment and coalescence of pre-existing small rafts into larger microdomains or by de novo assembly of a microdomain.
The Gag protein of HIV is targeted to the host cell plasma membrane with subsequent sorting into detergent-resistant microdomains; budding is cholesterol- and sphingolipid-dependent and the virus is enriched in raft-associated proteins and lipids. These data and the copatching of HIV proteins with GM1 on incubation with labeled cholera toxin suggest that acquisition of the viral envelope at the plasma membrane involves the clustering of rafts. Plasma membrane targeting of HIV Gag requires myristoylation and a basic patch in its amino-terminal MA (matrix) domain. Mutation of the myristoylation signal led to a loss of membrane targeting and virus release, whereas mutation or deletion of basic residues caused retargeting of the budding process to ER (Fäcke et al. 1993) or endosomal membranes (Ono et al. 2000), suggesting that the latter signal is responsible for plasma membrane specificity. Correct HIV Gag targeting further depends on the plasma membrane specific phosphoinositide phosphatidyl inositol (4,5) bisphosphate (PI(4,5)P2), with depletion or delocalization of PI(4,5)P2 causing a severe reduction of HIV budding and loss or redirection of Gag membrane targeting, respectively (Ono et al. 2004). Gag plasma membrane binding thus is mediated by the combined effects of (1) membrane insertion of its amino-terminal myristic acid (initially sequestered in a hydrophobic pocket and only exposed on membrane binding); (2) electrostatic interactions with acidic phosphoplipids (which are strongly enriched in the HIV lipidome, see below); and (3) specific recognition of PI(4,5)P2 (Fig. 4). Acylation of viral structural proteins has been found for many other viral matrix proteins (lining the inner leaflet of the virion membrane). Given that the binding energy of a myristoyl group is insufficient to stably link a protein to a lipid bilayer, acylation is likely to be necessary but not sufficient in these cases as well. PI(4,5)P2 is also important for the release of the retrovirus Mason-Pfizer monkey virus, and it will be important to determine its role in membrane targeting and budding of other plasma-membrane-associated viruses.
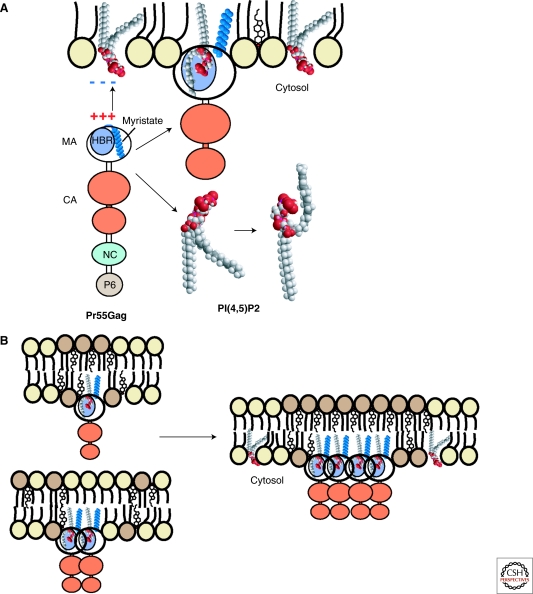
Figure 4.
Membrane association and targeting of HIV Gag and its association with lipid rafts. (A) The HIV-1 Gag polyprotein contains a highly basic region (HBR) in its amino-terminal MA domain that binds to negatively charged phospholipids and specifically interacts with PI(4,5)P2. This binding induces exposure of the sequestered myristate moiety of Gag and concomitantly sequesters the unsaturated PI(4,5)P2 acyl chain. (B) Gag-membrane binding creates a more saturated lipid environment that may promote membrane-raft coalescence depending on Gag multimerization.
Structural analysis of the HIV MA domain in complex with a truncated derivative of PI(4,5)P2 unraveled the mode of the protein-lipid interaction, and also suggested a model how HIV acquires its specific lipid composition (Saad et al. 2006). PI(4,5)P2 adopts an unusual conformation, in which the inositol head group and 2′-fatty acid bind to a hydrophobic cleft of MA, and the 1′-fatty acid and exposed myristoyl group bracket a conserved basic surface patch previously implicated in membrane binding (Saad et al. 2006). These findings indicate that PI(4,5)P2 acts as both a trigger of the myristyl switch and as a membrane anchor. Exposure and membrane insertion of the saturated myristate group is predicted to occur concomitant with removal of the unsaturated 2′ acyl group of PI(4,5)P2 from the membrane (Fig. 4). These alterations would lead to the presence of two saturated acyl chains instead of the saturated 1′ and the unsaturated 2′ acyl group of PI(4,5)P2. Given that there are ∼30 lipid molecules per Gag in the inner leaflet of the virion membrane (Brügger et al. 2006), this cannot account for the unique lipid composition of HIV, but formation of the immature Gag protein lattice with consequent increase in local concentration of saturated lipids may change the environment to be more conducive for lipids of the lo phase. This suggests that membrane binding, Gag assembly, and lipid sorting may be interdependent and mutually stimulating processes. It appears likely that similar mechanisms also operate in other viruses, but are currently less well understood.
Lipid Composition of Different Viruses
Differences in lipid composition between viral membranes and the cell membranes they are derived from had already been suggested in early studies, indicating lipid sorting in virus release (reviewed in Waheed and Freed 2010). Aloia et al. (1988, 1993) showed that the HIV membrane is enriched in SM, PE, PS, PC, and cholesterol with a decreased fluidity when compared with the plasma membrane of the producer cell. Similar increases in membrane order were suggested for influenza virus, whereas the membranes of, for example, VSV and SFV, are suggested to be less ordered (Scheiffele et al. 1999; Blom et al. 2001). Recent advances in lipid mass spectrometry allow for the first time a comprehensive analysis of the entire lipid composition (the lipidome) of purified viruses including determination of side chains. Several such studies have considerably advanced our knowledge about viral lipid composition, while deficiencies in purification of cellular membranes, and in particular of the plasma membrane, still obscure our view of the process of lipid sorting.
Quantitative analysis of the lipid constituents of HIV revealed a strong enrichment of the raft lipids SM, cholesterol, and plasmalogen-PE with an increase in saturated fatty acids compared to the producer cell. The inner leaflet of the viral membrane was enriched in PS, and the overall lipid composition of HIV strongly resembled that of detergent-resistant membranes isolated from producer cells (Brügger et al. 2006). This and the observation that the native HIV membrane shows a lo structure (Lorizate et al. 2009) provided direct evidence for its raft-like nature. Comparing the HIV membrane with the plasma membrane of the producer cell further revealed an enrichment of PI(4,5)P2 in the virus, consistent with the suggested model of Gag membrane binding and organization (Chan et al. 2008, see above). These investigators further reported an enrichment of cholesterol, ceramide, and glycosphingolipids for both HIV and murine leukemia virus when compared with the host cell plasma membrane, although no difference in sphingolipids was observed.
Quantitative lipidomic analyses were also performed for VSV, SFV, and more recently HCV. Consistent with the finding that SFV and VSV were not raft-associated, their lipidomes showed a close resemblance with each other and with the plasma membrane they were derived from (Kalvodova et al. 2009). Blom et al. (2001) analyzed the lipid composition of VSV and influenza virus budded from fibroblasts and observed an enrichment of glycosphingolipids in influenza virus in comparison to VSV, also consistent with the assignment of influenza virus as being raft-associated. A very different picture was observed for HCV, whose lipidome closely resembled that of LDL and VLDL with cholesteryl esters accounting for almost half of the total HCV lipids (Merz et al. 2010). This is consistent with the described tight link of HCV assembly and release with VLDL synthesis and secretion.
Scission of Viral and Cellular Membranes
Enveloped virus release by scission of the viral and cellular membranes has long been thought to be a spontaneous event. Early experiments with HIV-1 already identified peptide motifs in the structural Gag protein, whose mutation led to arrested late budding structures with ready-made virions remaining stuck at the cell surface through a thin membrane tether (Göttlinger et al. 1991). Such “late domains” were subsequently identified in many enveloped viruses and shown to interact with the ESCRT machinery. ESCRT has been implicated in topologically similar processes including intralumenal vesicle (ILV) formation at the MVB and midbody formation in cytokinesis. Cellular receptor sorting into MVBs requires the regulated assembly of ESCRT-0 followed by ESCRTs I, II, and III and the vacuolar protein sorting (Vps) 4 complex, whereas enveloped virus release generally requires only a subset of components always including ESCRT-III and Vps4. These two complexes thus appear to constitute the core fission machinery with upstream factors mainly involved in protein sorting and membrane bending. In the case of HIV-1, cryo electron-tomography revealed that the immature extracellular virus consists of a truncated sphere of Gag, whereas variants carrying late domain mutations showed a complete Gag sphere (Carlson et al. 2008). These results suggested an active role of ESCRT in virion release causing ESCRT dependent virus egress before Gag assembly is completed, possibly acting when the bud neck has achieved a suitable diameter.
ESCRT-III and Vps4 are critical for the release of all ESCRT-dependent viruses. Prior to recruitment, ESCRT-III proteins are autoinhibited in the cytosol and activation is required for membrane targeting and lattice polymerization. Polymerization is a general feature of all ESCRT-III proteins, but appears to be restricted to the bud neck, consistent with their exclusion from the vesicle. Several models for ESCRT-III mediated membrane scission have been proposed based on available structural information and in vitro vesiculation experiments. All models suggest ESCRT-III polymer-dependent membrane constriction on the inside of the bud neck leading to membrane scission; in one model this is mediated by coiling of polymeric filaments and in the other by formation of a cone-shaped polymer scaffold or “dome” (Fig. 5). Given that CHMP4 can form filamentous polymers that induce membrane deformation and tubulation (Hanson et al. 2008) the first model suggests that such filaments may grow into a circular structure at the membrane with their sequential constriction mediating scission. This may occur by sliding of one end of the filament over the other with consecutive sliding steps leading to narrowing of the bud radius provided that lipid diffusion across the filament barrier is inhibited (“spiral constriction”). A variant of the sliding model suggests sequential Vps4-mediated removal of CHMP4 subunits from the polymeric filament with consequent bud constriction; the ATPase activity of Vps4 would provide the required energy in this case (“purse-string”). However, Vps4 is not required for vesicle abscission in vitro (Wollert and Hurley 2010). The second model is based on the observation that polymers of CHMP2A and CHMP3 form cone-shaped structures binding to lipids on their convex surface, and suggests that such structures can form a scaffold for narrowing the bud neck (Lata et al. 2008). Modeling studies indicated two quasi-equilibrium states for the prefission state of such structures showing either a wide (∼25 nm) or a narrow (∼3 nm) neck (Fabrikant et al. 2009). For a certain CHMP-membrane affinity, the narrow neck becomes energetically favorable and would undergo spontaneous fission. The membrane neck would thus be held under stress, which is relieved on scission with sudden release of membrane stress by Vps4-mediated disassembly of the ESCRT-III lattice possibly accelerating fission. This is consistent with Vps4 not being essential for membrane scission in vitro, but suggests that it may be functionally important for the scission process in vivo. Interestingly, a recent fluorescence microscopy study revealed recruitment of Vps4 complexes to nascent HIV-1 budding sites prior to virion release (Baumgärtel et al., submitted), indicating an active contribution of Vps4 to virus membrane scission beyond recycling of ESCRT components.
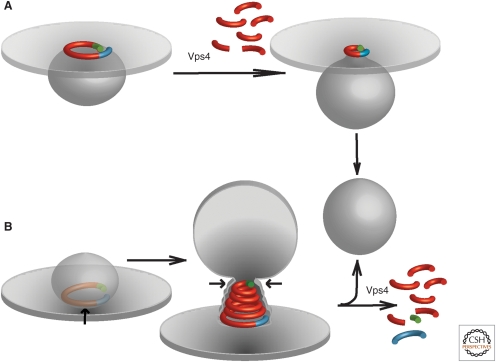
Figure 5.
Models for virus membrane fission. (A) “Purse-string” model based on data from Saksena et al. (2009). A single ESCRT-III filament (red) with asymmetric ends (blue/green) is used to delineate and later constrict the neck of an evolving vesicle. Vps4 is proposed to disassemble the filament from one end to constrict the string, but Vps4-independent sliding may also achieve this constriction. (B) “Spiral constriction” model based on data from Lata et al. (2008). A growing ESCRT-III spiral surrounds and eventually constricts a cargo-containing membrane domain, forcing cargo at the center into an evolving vesicle. Membrane scission has been suggested to be mediated by membrane adhesion on a dome-like protein scaffold formed by the ESCRT-III complex (Fabrikant et al. 2009).
ESCRT-dependence of enveloped virus release is generally assumed if late domains are present in viral structural proteins and if virus release is inhibited by dominant negative Vps4, although the latter may also act indirectly. ESCRT components clearly have an important role in the release of many, but certainly not all enveloped viruses. Important exceptions are the herpesvirus human cytomegalovirus (Fraile-Ramos et al. 2007), human influenza virus (Chen and Lamb 2008), and respiratory syncytial virus (Utley et al. 2008). These viruses may recruit alternative cellular machinery or employ viral proteins mediating membrane scission. A recent study revealed that the influenza virus M2 protein contains an amphipathic helix that is necessary and sufficient for vesiculation in vitro and for influenza virus budding in tissue culture (Rossman et al. 2010a,b). M2 is a trans-membrane protein that forms a homotetramer with proton-selective ion channel activitiy in the virion membrane. Rossman et al. (2010b) showed that M2 binds to low cholesterol (ld) membrane regions inducing positive curvature, and preferentially sorts to the phase boundary of phase-separated vesicles causing extrusion of the lo domain, dependent on the presence of the amphipathic helix. Furthermore, M2 localizes to the neck of influenza virus buds in virus-producing cells and mutation of its amphipathic helix leads to late budding arrest similar to late domain mutations in other enveloped viruses. These results suggest that M2 serves an analogous function as the ESCRT-III/Vps4 complex in other viruses, but by a completely different mechanism. The influenza virus membrane is enriched in cholesterol and is likely to be more lo than the surrounding plasma membrane, creating line tension at the phase boundary demarcating the viral bud (Kozlov 2010). M2 appears to specifically sort to this phase boundary and may modulate line tension by membrane interaction of its amphipathic helices. This suggests the following model: the influenza surface protein HA induces membrane bending (Chen and Lamb 2008) and recruits the matrix protein M1, which in turn recruits the M2 tetramer. M2 preferentially sorts to the phase boundary of phase-separated membranes, which leads to its concentration at the bud neck and promotes membrane scission and virus release.
CONCLUDING REMARKS
Lipids have long been known as structural elements of viral and cellular membranes, but recent studies revealed their involvement in the intricate virus-cell interaction in many more ways. Thus, lipids have been shown to play a role at various stages in viral replication, including entry, uncoating, genome replication, assembly, and release. Technical advances in lipid identification and quantitation, in lipid imaging and concerning the knockdown of factors involved in lipid metabolism made this progress possible and paved the ground for future detailed analyses of lipid involvement in viral replication. Given the high number of very recent advances published in 2009 and 2010, it is easy to predict that this area of research is only in its infancy and many more exciting discoveries lie ahead. Understanding the manifold roles of lipids in viral replication also led to the discovery of lipid-active compounds as potential antivirals, but current compounds largely lack specificity and are thus unacceptably toxic. Exploiting specific lipid requirements of individual pathogens or whole virus groups and delineating the intricate interactions of these pathogens with cellular lipids and the modification of their respective metabolism may, however, provide new approaches for antiviral therapies in the future.
ACKNOWLEDGMENTS
We thank R. Bartenschlager and S. Welsch for critical review of the manuscript. We apologize to those colleagues whose work could not be cited due to space limitation.
Research in the authors’ laboratory is supported by a grant from the Deutsche Forschungsgemeinschaft within TRR83.
REFERENCES
- Akhtar J, Shukla D 2009. Viral entry mechanisms: Cellular and viral mediators of herpes simplex virus entry. FEBS J 276: 7228–7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SL, Stadler K, Mandl CW, Kunz C, Heinz FX 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J Virol 69: 5816–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia RC, Tian H, Jensen FC 1993. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci 90: 5181–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia RC, Jensen FC, Curtain CC, Mobley PW, Gordon LM 1988. Lipid composition and fluidity of the human immunodeficiency virus. Proc Natl Acad Sci 85: 900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Bréchot C, Paranhos-Baccalà G, Lotteau V 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol 76: 6919–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- André P, Perlemuter G, Budkowska A, Brechot C, Lotteau V 2005. Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis 25: 93–104 [DOI] [PubMed] [Google Scholar]
- Andreo U, Maillard P, Kalinina O, Walic M, Meurs E, Martinot M, Marcellin P, Budkowska A 2007. Lipoprotein lipase mediates hepatitis C virus (HCV) cell entry and inhibits HCV infection. Cell Microbiol 9: 2445–2456 [DOI] [PubMed] [Google Scholar]
- Backovic M, Jardetzky TS 2009. Class III viral membrane fusion proteins. Curr Opin Struct Biol 19: 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Penin F, Lohmann V, André P 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol 19: 95–103 [DOI] [PubMed] [Google Scholar]
- Bieniasz PD 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5: 550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom TS, Koivusalo M, Kuismanen E, Kostiainen R, Somerharju P, Ikonen E 2001. Mass spectrometric analysis reveals an increase in plasma membrane polyunsaturated phospholipid species upon cellular cholesterol loading. Biochemistry 40: 14635–14644 [DOI] [PubMed] [Google Scholar]
- Brügger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Kräusslich HG 2006. The HIV lipidome: A raft with an unusual composition. Proc Natl Acad Sci 103: 2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W 2010. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog 6: e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, Briggs JA, Glass B, Riches JD, Simon MN, Johnson MC, Müller B, Grünewald K, Kräusslich HG 2008. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe 4: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro FA, Bianconi ML, Weissmuller G, Stauffer F, Da Poian AT 2002. Membrane recognition by vesicular stomatitis virus involves enthalpy-driven protein–lipid interactions. J Virol 76: 3756–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro FA, Lapido-Loureiro PA, Cordo SM, Stauffer F, Weissmuller G, Bianconi ML, Juliano MA, Juliano L, Bisch PM, Da Poian AT 2006. Probing the interaction between vesicular stomatitis virus and phosphatidylserine. Eur Biophys J 35: 145–154 [DOI] [PubMed] [Google Scholar]
- Chan RB, Tanner L, Wenk MR 2010. Implications for lipids during replication of enveloped viruses. Chem Phys Lipids 163: 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R, Uchil PD, Jin J, Shui G, Ott DE, Mothes W, Wenk MR 2008. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J Virol 82: 11228–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372: 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Morita E, Lamb RA 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol 81: 7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM 2005. Membrane hemifusion: Crossing a chasm in two leaps. Cell 123: 375–382 [DOI] [PubMed] [Google Scholar]
- Chernomordik LV, Zimmerberg J, Kozlov MM 2006. Membranes of the world unite! J Cell Biol 175: 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L, Chanturiya A, Green J, Zimmerberg J 1995. The hemifusion intermediate and its conversion to complete fusion: Regulation by membrane composition. Biophys J 69: 922–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlanda P, Carbajal MA, Cyrklaff M, Griffiths G, Krijnse-Locker J 2009. Membrane rupture generates single open membrane sheets during vaccinia virus assembly. Cell Host Microbe 6: 81–90 [DOI] [PubMed] [Google Scholar]
- Coyne CB, Bergelson JM 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124: 119–131 [DOI] [PubMed] [Google Scholar]
- Coyne CB, Kim KS, Bergelson JM 2007. Poliovirus entry into human brain microvascular cells requires receptor-induced activation of SHP-2. EMBO J 26: 4016–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremesti AE, Goni FM, Kolesnick R 2002. Role of sphingomyelinase and ceramide in modulating rafts: Do biophysical properties determine biologic outcome? FEBS Lett 531: 47–53 [DOI] [PubMed] [Google Scholar]
- Cuadras MA, Bordier BB, Zambrano JL, Ludert JE, Greenberg HB 2006. Dissecting rotavirus particle-raft interaction with small interfering RNAs: Insights into rotavirus transit through the secretory pathway. J Virol 80: 3935–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danieli T, Pelletier SL, Henis YI, White JM 1996. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol 133: 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms RW, Moore JP 2000. HIV-1 membrane fusion: Targets of opportunity. J Cell Biol 151: F9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms RW, Lamb RA, Rose JK, Helenius A 1993. Folding and assembly of viral membrane proteins. Virology 193: 545–562 [DOI] [PubMed] [Google Scholar]
- Eckert DM, Kim PS 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc Natl Acad Sci 98: 11187–11192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol 76: 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers H, Romer W, Smith AE, Bacia K, Dmitrieff S, Chai W, Mancini R, Kartenbeck J, Chambon V, Berland L, et al. 2010. GM1 structure determines SV40-induced membrane invagination and infection. Nat Cell Biol 12: 11–18; sup pp 11–12 [DOI] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM 2009. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol 5: e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fäcke M, Janetzko A, Shoeman RL, Kräusslich HG 1993. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol 67: 4972–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr GA, Zhang LG, Tattersall P 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc Natl Acad Sci 102: 17148–17153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan CM, Rawat SS, Puri A, Wang JM, Ruscetti FW, Blumenthal R 2004. Ceramide, a target for antiretroviral therapy. Proc Natl Acad Sci 101: 15452–15457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile-Ramos A, Pelchen-Matthews A, Risco C, Rejas MT, Emery VC, Hassan-Walker AF, Esteban M, Marsh M 2007. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol 9: 2955–2967 [DOI] [PubMed] [Google Scholar]
- Frey G, Rits-Volloch S, Zhang XQ, Schooley RT, Chen B, Harrison SC 2006. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc Natl Acad Sci 103: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricks CE, Hogle JM 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol 64: 1934–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov VA, Shnyrova AV, Zimmerberg J 2011. Lipid polymorphisms and membrane shape. Cold Spring Harb Perspect Biol 3: a004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H, Sjoberg M, Cheng RH 2004. Budding of alphaviruses. Virus Res 106: 103–116 [DOI] [PubMed] [Google Scholar]
- Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59: 103–112 [DOI] [PubMed] [Google Scholar]
- Gomara MJ, Lorizate M, Huarte N, Mingarro I, Perez-Paya E, Nieva JL 2006. Hexapeptides that interfere with HIV-1 fusion peptide activity in liposomes block GP41-mediated membrane fusion. FEBS Lett 580: 2561–2566 [DOI] [PubMed] [Google Scholar]
- Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci 88: 3195–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber UF 2002. Signalling in viral entry. Cell Mol Life Sci 59: 608–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol 180: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC 2008. Viral membrane fusion. Nat Struct Mol Biol 15: 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood AM 2010. Membrane uncoating of intact enveloped viruses. J Virol 84: 10946–10955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci 107: 17345–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EE, Krummenacher C 2008. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci 65: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV Jr, Ott M 2010. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med 16: 1295–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser J 2005. Deep-etch EM reveals that the early poxvirus envelope is a single membrane bilayer stabilized by a geodetic “honeycomb” surface coat. J Cell Biol 169: 269–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle JM 2002. Poliovirus cell entry: Common structural themes in viral cell entry pathways. Annu Rev Microbiol 56: 677–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, et al. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141: 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M Jr, Ye J 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci 104: 5848–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvodova L, Sampaio JL, Cordo S, Ejsing CS, Shevchenko A, Simons K 2009. The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J Virol 83: 7996–8003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M, Chanel-Vos C, Liao M 2010. Alphavirus entry and membrane fusion. Viruses 2: 796–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilby JM, Eron JJ 2003. Novel therapies based on mechanisms of HIV-1 cell entry. N Engl J Med 348: 2228–2238 [DOI] [PubMed] [Google Scholar]
- Kliger Y, Aharoni A, Rapaport D, Jones P, Blumenthal R, Shai Y 1997. Fusion peptides derived from the HIV type 1 glycoprotein 41 associate within phospholipid membranes and inhibit cell–cell fusion. Structure–function study. J Biol Chem 272: 13496–13505 [DOI] [PubMed] [Google Scholar]
- Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov MM 2010. Biophysics: Joint effort bends membrane. Nature 463: 439–440 [DOI] [PubMed] [Google Scholar]
- Kozlov MM, McMahon HT, Chernomordik LV 2010. Protein-driven membrane stresses in fusion and fission. Trends Biochem Sci 35: 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin PI, Zimmerberg J, Chizmadzhev YA, Cohen FS 2001. A quantitative model for membrane fusion based on low-energy intermediates. Proc Natl Acad Sci 98: 7235–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W 2008. Helical structures of ESCRT-III are disassembled by VPS4. Science 321: 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC 2002. Structure of the reovirus membrane-penetration protein, μ1, in a complex with is protector protein, σ3. Cell 108: 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Thorp SC 2002. Cell surface heparan sulfate and its roles in assisting viral infections. Med Res Rev 22: 1–25 [DOI] [PubMed] [Google Scholar]
- Lorizate M, Brügger B, Akiyama H, Glass B, Müller B, Anderluh G, Wieland FT, Kräusslich HG 2009. Probing HIV-1 membrane liquid order by Laurdan staining reveals producer cell-dependent differences. J Biol Chem 284: 22238–22247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorizate M, Huarte N, Saez-Cirion A, Nieva JL 2008. Interfacial pre-transmembrane domains in viral proteins promoting membrane fusion and fission. Biochim Biophys Acta 1778: 1624–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low JA, Magnuson B, Tsai B, Imperiale MJ 2006. Identification of gangliosides GD1b and GT1b as receptors for BK virus. J Virol 80: 1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson B, Rainey EK, Benjamin T, Baryshev M, Mkrtchian S, Tsai B 2005. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol Cell 20: 289–300 [DOI] [PubMed] [Google Scholar]
- Maier O, Galan DL, Wodrich H, Wiethoff CM 2010. An N-terminal domain of adenovirus protein VI fragments membranes by inducing positive membrane curvature. Virology 402: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier O, Wiethoff CM 2010. N-terminal α-helix-independent membrane interactions facilitate adenovirus protein VI induction of membrane tubule formation. Virology 408: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri J, Tedbury PR, Gretton S, Hughes ME, Griffin SD, Dallas ML, Green KA, Hardie DG, Peers C, Harris M 2010. Enhanced hepatitis C virus genome replication and lipid accumulation mediated by inhibition of AMP-activated protein kinase. Proc Natl Acad Sci 107: 11549–11554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh M, Helenius A 2006. Virus entry: Open sesame. Cell 124: 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megha, London E 2004. Ceramide selectively displaces cholesterol from ordered lipid domains (rafts): Implications for lipid raft structure and function. J Biol Chem 279: 9997–10004 [DOI] [PubMed] [Google Scholar]
- Mercer J, Schelhaas M, Helenius A 2010. Virus entry by endocytosis. Annu Rev Biochem 79: 803–833 [DOI] [PubMed] [Google Scholar]
- Merz A, Long G, Hiet MS, Bruegger B, Chlanda P, André P, Wieland F, Krijnse-Locker J, Bartenschlager R 2010. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem 286: 3018–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter TC, Klupp BG, Granzow H 2009. Herpesvirus assembly: An update. Virus Res 143: 222–234 [DOI] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J 2008. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6: 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9: 1089–1097 [DOI] [PubMed] [Google Scholar]
- Mothes W, Sherer NM, Jin J, Zhong P 2010. Virus cell-to-cell transmission. J Virol 84: 8360–8368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch J, Standker L, Adermann K, Schulz A, Schindler M, Chinnadurai R, Pohlmann S, Chaipan C, Biet T, Peters T, et al. 2007. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell 129: 263–275 [DOI] [PubMed] [Google Scholar]
- Nieva JL, Agirre A 2003. Are fusion peptides a good model to study viral cell fusion? Biochim Biophys Acta 1614: 104–115 [DOI] [PubMed] [Google Scholar]
- Ono A 2010. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol Cell 102: 335–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Orenstein JM, Freed EO 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J Virol 74: 2855–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci 101: 14889–14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens RJ, Tanner CC, Mulligan MJ, Srinivas RV, Compans RW 1990. Oligopeptide inhibitors of HIV-induced syncytium formation. AIDS Res Hum Retroviruses 6: 1289–1296 [DOI] [PubMed] [Google Scholar]
- Patient R, Hourioux C, Roingeard P 2009. Morphogenesis of hepatitis B virus and its subviral envelope particles. Cell Microbiol 11: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permanyer M, Ballana E, Este JA 2010. Endocytosis of HIV: Anything goes. Trends Microbiol 18: 543–551 [DOI] [PubMed] [Google Scholar]
- Ploss A, Rice CM 2009. Towards a small animal model for hepatitis C. EMBO Rep 10: 1220–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M, Cai D, Verhey KJ, Tsai B 2009. A lipid receptor sorts polyomavirus from the endolysosome to the endoplasmic reticulum to cause infection. PLoS Pathog 5: e1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, et al. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9: 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA 2010a. Influenza virus M2 ion channel protein is necessary for filamentous virion formation. J Virol 84: 5078–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Lamb RA 2010b. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142: 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci 103: 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Cirion A, Nir S, Lorizate M, Agirre A, Cruz A, Perez-Gil J, Nieva JL 2002. Sphingomyelin and cholesterol promote HIV-1 gp41 pretransmembrane sequence surface aggregation and membrane restructuring. J Biol Chem 277: 21776–21785 [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD 2009. Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Ahola T, Kaariainen L 2005. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol 285: 139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzwedel K, West JT, Hunter E 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol 73: 2469–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp M, Day PM 2009. Structure, attachment and entry of polyoma- and papillomaviruses. Virology 384: 400–409 [DOI] [PubMed] [Google Scholar]
- Sattentau Q 2008. Avoiding the void: Cell-to-cell spread of human viruses. Nat Rev Microbiol 6: 815–826 [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Rietveld A, Wilk T, Simons K 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem 274: 2038–2044 [DOI] [PubMed] [Google Scholar]
- Schelhaas M 2010. Come in and take your coat off—How host cells provide endocytosis for virus entry. Cell Microbiol 12: 1378–1388 [DOI] [PubMed] [Google Scholar]
- Schelhaas M, Malmstrom J, Pelkmans L, Haugstetter J, Ellgaard L, Grünewald K, Helenius A 2007. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131: 516–529 [DOI] [PubMed] [Google Scholar]
- Sfakianos JN, Hunter E 2003. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic 4: 671–680 [DOI] [PubMed] [Google Scholar]
- Shai Y 2001. Molecular recognition within the membrane milieu: implications for the structure and function of membrane proteins. J Membr Biol 182: 91–104 [DOI] [PubMed] [Google Scholar]
- Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem 282: 37158–37169 [DOI] [PubMed] [Google Scholar]
- Sieczkarski SB, Whittaker GR 2005. Viral entry. Curr Top Microbiol Immunol 285: 1–23 [DOI] [PubMed] [Google Scholar]
- Simons K, Sampaio JL 2011. Membrane organization and lipid rafts. Cold Spring Harb Perspect 3: a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AE, Helenius A 2004. How viruses enter animal cells. Science 304: 237–242 [DOI] [PubMed] [Google Scholar]
- Smith AE, Lilie H, Helenius A 2003. Ganglioside-dependent cell attachment and endocytosis of murine polyomavirus-like particles. FEBS Lett 555: 199–203 [DOI] [PubMed] [Google Scholar]
- Sodeik B, Krijnse-Locker J 2002. Assembly of vaccinia virus revisited: De novo membrane synthesis or acquisition from the host? Trends Microbiol 10: 15–24 [DOI] [PubMed] [Google Scholar]
- Spillmann D 2001. Heparan sulfate: anchor for viral intruders? Biochimie 83: 811–817 [DOI] [PubMed] [Google Scholar]
- Spuul P, Balistreri G, Kaariainen L, Ahola T 2010. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J Virol 84: 7543–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Vincent MR, Colpitts CC, Ustinov AV, Muqadas M, Joyce MA, Barsby NL, Epand RF, Epand RM, Khramyshev SA, Valueva OA, et al. 2010. Rigid amphipathic fusion inhibitors, small molecule antiviral compounds against enveloped viruses. Proc Natl Acad Sci 107: 17339–17344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed GH, Amako Y, Siddiqui A 2010. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab 21: 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5: 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen N, Doglio L, Schleich S, Krijnse Locker J 2001. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol Biol Cell 12: 2031–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosteson MT, Chow M 1997. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J Virol 71: 507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B 2007. Penetration of nonenveloped viruses into the cytoplasm. Annu Rev Cell Dev Biol 23: 23–43 [DOI] [PubMed] [Google Scholar]
- Tsai B, Qian M 2010. Cellular entry of polyomaviruses. Curr Top Microbiol Immunol 2100: 177–194 [DOI] [PubMed] [Google Scholar]
- Tsai B, Gilbert JM, Stehle T, Lencer W, Benjamin TL, Rapoport TA 2003. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J 22: 4346–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley TJ, Ducharme NA, Varthakavi V, Shepherd BE, Santangelo PJ, Lindquist ME, Goldenring JR, Crowe JE Jr 2008. Respiratory syncytial virus uses a Vps4-independent budding mechanism controlled by Rab11-FIP2. Proc Natl Acad Sci 105: 10209–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt FH, Pilote L, Cartier M, Lippens J, Liuzzi M, Bethell RC, Cordingley MG, Kukolj G 2009. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology 387: 5–10 [DOI] [PubMed] [Google Scholar]
- Vennema H, Godeke GJ, Rossen JW, Voorhout WF, Horzinek MC, Opstelten DJ, Rottier PJ 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J 15: 2020–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma PC, Basu V, Gupta V, Saxena G, Rahman LU 2009. Pharmacology and chemistry of a potent hepatoprotective compound Picroliv isolated from the roots and rhizomes of Picrorhiza kurroa royle ex benth. (kutki). Curr Pharm Biotechnol 10: 641–649 [DOI] [PubMed] [Google Scholar]
- Waheed AA, Freed EO 2010. The role of lipids in retrovirus replication. Viruses 2: 1146–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Hinz A, Gaudin Y 2007. Virus membrane fusion. FEBS Lett 581: 2150–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Müller B, Kräusslich HG 2007. More than one door—Budding of enveloped viruses through cellular membranes. FEBS Lett 581: 2089–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5: 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk T, Geiselhart V, Frech M, Fuller SD, Flügel RM, Löchelt M 2001. Specific interaction of a novel foamy virus Env leader protein with the N-terminal Gag domain. J Virol 75: 7995–8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MC, Freiberg AN, Zhang T, Akyol-Ataman Z, Grock A, Hong PW, Li J, Watson NF, Fang AQ, Aguilar HC, et al. 2010. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci 107: 3157–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Hurley JH 2010. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464: 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kurteva S, Ren X, Lee S, Sodroski J 2006. Subunit stoichiometry of human immunodeficiency virus type 1 envelope glycoprotein trimers during virus entry into host cells. J Virol 80: 4388–4395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadori Z, Szelei J, Lacoste MC, Li Y, Gariepy S, Raymond P, Allaire M, Nabi IR, Tijssen P 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev Cell 1: 291–302 [DOI] [PubMed] [Google Scholar]
- Zhang L, Agosto MA, Ivanovic T, King DS, Nibert ML, Harrison SC 2009. Requirements for the formation of membrane pores by the reovirus myristoylated micro1N peptide. J Virol 83: 7004–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]