Abstract
Genetic variation is thought to contribute to variability in platelet function; however, the specific variants and mechanisms that contribute to altered platelet function are poorly defined. With the use of a combination of fine mapping and sequencing of the platelet endothelial aggregation receptor 1 (PEAR1) gene we identified a common variant (rs12041331) in intron 1 that accounts for ≤ 15% of total phenotypic variation in platelet function. Association findings were robust in 1241 persons of European ancestry (P = 2.22 × 10−8) and were replicated down to the variant and nucleotide level in 835 persons of African ancestry (P = 2.31 × 10−27) and in an independent sample of 2755 persons of European descent (P = 1.64 × 10−5). Sequencing confirmed that variation at rs12041331 accounted most strongly (P = 2.07 × 10−6) for the relation between the PEAR1 gene and platelet function phenotype. A dose-response relation between the number of G alleles at rs12041331 and expression of PEAR1 protein in human platelets was confirmed by Western blotting and ELISA. Similarly, the G allele was associated with greater protein expression in a luciferase reporter assay. These experiments identify the precise genetic variant in PEAR1 associated with altered platelet function and provide a plausible biologic mechanism to explain the association between variation in the PEAR1 gene and platelet function phenotype.
Introduction
Genetic variation is thought to contribute to variability in platelet function under native conditions1 and after treatment with aspirin2; however, the specific variants and mechanisms that contribute to altered platelet function are poorly defined. With the use of a candidate gene approach, we previously identified a single nucleotide polymorphism (SNP; rs2768759, genetic location chromosome 1:155119087, National Center for Biotechnology Information [NCBI] build 36.3) in the platelet endothelial aggregation receptor (PEAR1, chromosome 1q23.1) gene that was associated with greater agonist-induced platelet aggregability in the presence and absence of aspirin.3 A recent genomewide association study also identified a SNP in PEAR1 (rs12566888, genetic location chromosome 1:155135671, NCBI build 36.3) associated with greater platelet aggregability in native platelet function but did not examine platelet function after aspirin treatment.4 Neither of the 2 previously identified PEAR1 SNPs is located in a functional domain of the gene, making it difficult to determine the mechanistic basis or causality for the observed gene-association findings.
With the use of fine mapping of the PEAR1 locus and sequencing of the PEAR1 gene, we have now identified a common variant (rs12041331, genetic location chromosome 1: 155136338, NCBI build 36.3) in intron 1 of PEAR1 that accounts most strongly for the association signal between the PEAR1 locus and platelet function phenotypes. Association findings were robust in our cohort; findings in persons of European ancestry (EA) were replicated down to the variant and nucleotide level in persons of African ancestry, in both the fine-mapping and sequencing approaches, and were similar across multiple platelet function phenotypes in the presence and absence of aspirin. Findings were also replicated in an independent sample of EA from the Framingham Heart Study (FHS) Offspring cohort. The major allele (G) at rs12041331 was associated with greater platelet aggregation in all assays in all ethnic groups and with an apparent allelic dose-response effect. These findings were corroborated by experiments that showed greater platelet expression of PEAR1 protein in relation to the G allele at rs12041331 of the PEAR1 gene, as well as greater luciferase expression in cells transfected with a G allele–containing reporter plasmid.
Methods
Participants and study protocol
The participants and protocol for GeneSTAR (Genetic Study of Aspirin Responsiveness) have been described in detail previously.2,5 The study was approved by the Johns Hopkins Medicine Institutional Review Board, and all participants provided written informed consent in accordance with the Declaration of Helsinki. Briefly, participants were recruited from EA and African American families with a history of premature coronary artery disease (onset < 60 years of age). Healthy family members of affected probands were eligible if they were free of clinically apparent atherosclerotic disease or other serious comorbidity. Exclusion criteria included aspirin allergy or intolerance, anemia (hematocrit < 30%), thrombocytopenia (platelet count < 100 000/μL), and leukocytosis (white blood cell count > 20 000/μL). Participants were given a supply of 81-mg chewable aspirin tablets and instructed to take 1 pill each day for 14 days. Other than study drug, participants were prohibited from taking aspirin and other nonsteroidal anti-inflammatory drugs in the 10 days before baseline measurements and throughout the 14-day treatment period.
A medical history, physical examination, and laboratory measurements were performed at baseline. Height and weight were measured, and body mass index was calculated (kg/m2). Venous blood was drawn after a 12-hour overnight fast and assayed for glucose, cholesterol, fibrinogen, and platelet function studies. Plasma levels of glucose, total cholesterol, and triglycerides were measured with standard methods (Cholestech Corporation). Plasma fibrinogen was measured with an automated optical clot detection device (Behring Coagulation System; Dade-Behring). Hypertension, cigarette smoking, diabetes, and hypercholesterolemia were defined by standard methods as previously described.2,5
The participant population and protocol in the FHS have been described in detail elsewhere.6 Briefly, FHS participants in the Offspring cohort (second generation and spouses) were eligible for participation if free of aspirin and other antiplatelet medications. Medical histories were obtained as previously described (examination cycle 5). Venous blood was drawn after an overnight fast and assayed for cholesterol and platelet function studies.
Platelet function
Platelet function in GeneSTAR participants was assessed at baseline and after 2 weeks of aspirin therapy. Platelet-rich plasma (200 000 platelets/μL) was prepared by differential centrifugation of citrated (3.2%) blood samples. Optical aggregation was measured in a PAP-4 Aggregometer (Bio/Data Corp) after stimulating samples with collagen (2 and 5 μg/mL), ADP (2 and 10μM), or epinephrine (2 and 10μM). Peak aggregation within 5 minutes of agonist stimulation was recorded as a percentage (0%-100%).
Platelet function in FHS participants was assessed in the absence of aspirin only. Platelet-rich plasma was prepared by differential centrifugation of citrated (3.8%) blood samples. Optical aggregation was measured in a 4-channel aggregometer (Bio/Data Corp) after stimulating samples with ADP (3 and 5μM) or epinephrine (1 and 3μM). Percentage of maximum aggregation was recorded 4 (for ADP) or 5 (for epinephrine) minutes after agonist stimulation.
Genotyping and imputation
A 105-kb region from 2 kb upstream of NTRK1 to 2 kb downstream of PEAR1 (genetic coordinates chromosome1:155050166 to 155154850, NCBI build 36.3) was examined. SNPs from the designated region were selected from the Illumina Human 1Mv1_C array (GeneSTAR participants), and genotyping was performed by deCODE Genetics. MaCH7 was used to impute additional SNPs in the region based on the HapMap CEU- and YRI-phased haplotypes (release 22). A total of 87 SNPs with minor allele frequency > 2% and MaCH quality scores > 0.3 were evaluated in GeneSTAR participants of EA; 78 of these 87 participants met the same criteria and were evaluated in GeneSTAR African American participants. Genotyping in FHS participants was conducted with the Affymetrix 500K array and an additional gene-focused 50K array as part of the SNP Health Association Resource project. MaCH was used to impute additional SNPs in FHS. SNPs replicated in EA and African ancestry cohorts within GeneSTAR were evaluated for replication in the FHS cohort.
Sequencing
One hundred four GeneSTAR participants, 50 with high aggregation (26 EA, 24 African American) and 54 with low aggregation (27 EA, 27 African American), were selected for sequencing of PEAR1. Criteria for high aggregation were upper quartile for aggregation after aspirin to collagen 2 μg/mL, epi 2μM, and ADP 10μM. Criteria for low aggregation were lower quartile for aggregation after aspirin to collagen 2 μg/mL, epi 2μM, and ADP 10μM. The entire PEAR1 gene, from chromosomal 1 coordinates 155129893 to 155158850 (NCBI build 36.3) were sequenced by deCODE Genetics as previously described.8 Briefly, PCR amplifications and sequencing reactions were set up on Zymark SciClone ALH300 robotic workstations and amplified on MJR Tetrads. PCR products were verified for correct length by agarose gel electrophoresis and were purified with AMPure (Agencourt Bioscience). Purified products were sequenced with an ABI PRISM Fluorescent Dye Terminator system, repurified with CleanSEQ (Agencourt), and resolved on Applied Biosystems 3730 capillary sequencers. SNP calling from primary sequence data were performed with deCODE Genetics Sequence Miner software. PEAR1 variants were confirmed by manual inspection of automated primary signal traces.
Platelet expression of PEAR1 protein
Twenty-six randomly selected participants who varied by genotype at the PEAR1 rs12041331 locus were selected to examine platelet expression of PEAR1 protein. Platelets were isolated from whole blood by differential centrifugation and lysed in 0.5% NP40, 20mM Tris, 100mM NaCl, and 1mM EDTA plus commercially available protease inhibitor cocktail (Sigma Chemical Co). Platelet proteins were reduced, boiled, electrophoresed on a 3.8% Tris-Acetate gel in Tris-Acetate SDS buffer (Invitrogen; 30 μg protein/lane), and then transferred to a nylon membrane. Blots were probed with goat anti–human PEAR1 polyclonal antibody (R&D Systems Inc) as primary antibody and donkey anti–goat IgG-HRP (Santa Cruz Biotechnology Inc) as secondary antibody. Proteins were visualized by enhanced chemiluminescence (GE Healthcare), and quantification was standardized to a control sample consisting of a pooled mixture from persons with AG genotype. Western blotting results were confirmed by quantitative ELISA in an additional 24 persons. Goat anti–human PEAR1 polyclonal antibody, goat anti–human PEAR1 biotinylated antibody, recombinant human PEAR1, and streptavidin-HRP were purchased from R&D System Inc. TMB (3,3′,5,5′-tetramethylbenzidine) substrate was purchased from Cell Signaling Technology, Inc. Ninety-six–well immunoassay plates were incubated with 0.8 μg/mL PEAR1 polyclonal antibody at 4°C overnight. Plates were washed and blocked with 1% BSA in Tris-buffered saline before adding recombinant PEAR1 protein standard or platelet lysate sample. After 2 hours at room temperature, wells were washed (0.05% Tween-20 in 1X Tris-buffered saline), incubated with biotinylated anti-PEAR1 antibody (100 μL of 50 ng/mL), and developed with streptavidin-HRP (1:200 dilution). The reaction was quenched with 50 μL of 2M H2SO4, and spectrophotometric emissions were read at 450 nm. The PEAR1 standard curve was linear between 0 and 160 ng/mL (R2 = 0.9986) and the detection limit was 5 ng/mL. The significance of differences in platelet protein expression across genotypes was determined by ANOVA.
Luciferase gene reporter assay
PGL3 basic and PGL3 promoter vectors, β-galactosidase expression vector, and assay reagents were bought from Promega Corporation. A 186-bp DNA fragment containing rs12041331 was amplified by PCR with DNA from a subject heterozygous for the rs1204331 A/G polymorphism. The primers were 5′TTCACACCCAGGCTTGAACTCGAGCAG and 5′GGACTCGAGTTCCTGGTGGACAAGA and included an XhoI site (indicated by the italicized sequence). The PGL3 promoter vector and amplified fragments were digested with XhoI and purified by gel electrophoresis, and the digested fragment was inserted into the PGL3 vector upstream of the SV40 promoter. Positive clones with the 186-bp A or G insert (designated PGL3-P-A and PGL3-P-G) were confirmed by sequencing. The PGL3 plasmids and β-galactosidase vector were cotransfected into the megakaryocytic cell line, MEG-01, and HUVECs with the use of Effectene Transfection Reagent (QIAGEN). Cells were harvested and lysed 36-48 hours after transfection. Promoter activity was expressed by relative light units of luciferase normalized to absorption of β-galactosidase activity. One-way ANOVA was used to determine the significance of differences in luciferase expression among PGL3 promoters.
Statistical analyses
Variable normality was confirmed by the Wilk-Shapiro test, and log-transformation or empirical normal quantile transformation was applied when necessary.
Genotype-phenotype associations were determined separately for EA and African American participants in the GeneSTAR cohort. Genotyped SNPs were assessed with a Likelihood Ratio Test implemented in the option ASSOC of the Merlin Genetic Analysis software; imputed SNPs were assessed with linear mixed effects models implemented in SAS (PROC MIXED Version 9.2; SAS Institute). All GeneSTAR analyses were adjusted for cardiac risk factors (age, sex, hypertension, current smoking, body mass index, diabetes, low-density lipoprotein cholesterol, and fibrinogen levels). In each case, an additive genetic model was used. A total of 87 SNPs were evaluated across 18 phenotypes. The primary phenotypes were platelet aggregation measures after exposure to aspirin. Associations were considered significant at the Bonferroni corrected level of P < 3.19 × 10−5 to account for multiple comparisons [0.05/(87 SNPs × 18 phenotypes)]. Independence of association signals from SNPs in PEAR1 and NTRK1 was determined by GEE regression models. Genotype-phenotype association signals significant at the Bonferroni threshold in both EA and African American groups were evaluated for replication in FHS participants.
Genotype-phenotype associations were determined in FHS with the use of imputed SNP dosages and a linear-mixed effects model (additive genetic model) which accounted for familial correlation with the use of the kinship coefficient matrix.9 Adjustments were made for age, sex, and principal components 1-8 on the basis of EIGENSTRAT.10
Means and variance (SD) for platelet aggregation phenotypes by genotype were calculated. The percentage of total phenotypic variance explained by gene variants for each phenotype was calculated for each ethnic group by maximum likelihood methods (ASSOC in MERLIN).
Chi-square tests were used to test for association between the allele frequencies of each genetic variant between participants in the high (case) and low (control) aggregation groups, and we combined test statistics, taking the direction of the effect (ie, the risk allele) into account. Under the null hypothesis of no association, the test statistic can be written as independent draws from a Normal (0,1); thus, their sum divided by the square root of 2 is itself a draw from a Normal (0,1). This allows for a simple and valid calculation of a combined meta-analysis P value.
Results
Characteristics of the study sample
The GeneSTAR study sample consisted of 2076 apparently healthy persons from 559 families (332 EA and 227 African American) with a family history of premature coronary artery disease (onset ≤ 60 years of age). Study participants consisted of siblings of affected probands (n = 751), adult offspring of siblings or affected probands (n = 1120), and co-parents of the offspring (n = 205). Mean age in the GeneSTAR sample was ∼ 44 years, and ∼ 40% were male. Cardiac risk factors were common in both ethnic groups (see supplemental Table 1 for full sample characteristics of the GeneSTAR cohort, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Genotype-phenotype associations
A total of 87 SNPs in a 105-kb region from 2 kb upstream of NTRK1 to 2 kb downstream of PEAR1 (genetic coordinates chromosome1:155050166 to 155154850, NCBI build 36.3) were evaluated for associations with platelet function phenotypes. The primary phenotype was aggregation after aspirin treatment, and several agonists and concentrations were evaluated because of their biologic relevance to platelet function and their reported relation to clinical disease.11–13 Secondary phenotypes were aggregation before aspirin treatment (ie, baseline) and after aspirin treatment adjusted for baseline aggregation (a measure of change in platelet function in response to aspirin). Pairwise correlations (r) among aggregation phenotypes ranged between 0.0019 and 0.8889 in EA and between 0.0012 and 0.9188 in African Americans; correlations between phenotypes involving different agonists were generally < 0.5.
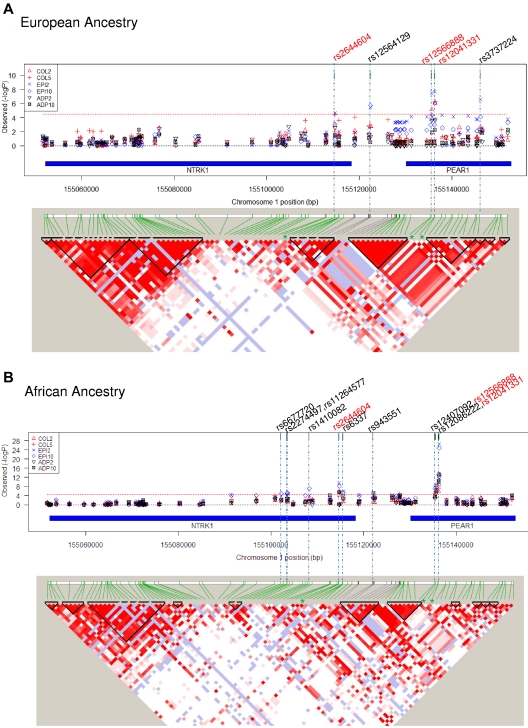
Tests for association were performed for the 87 SNPs against 18 phenotypes in the EA participants, and significance was assessed with stringent Bonferroni criteria [Bonferroni-corrected P < 3.19 × 10−5; 0.05/(87 SNPs × 18 phenotypes)]. Five SNPs in this region met these criteria with ≥ 1 of the phenotypes after aspirin therapy in EA participants: 1 SNP in NTRK1, 1 SNP in the intergenic region between NTRK1 and PEAR1, and 3 SNPs in PEAR1 (Figure 1A; Table 1). The strongest and most consistent signal across phenotypes was from rs12041331 in intron 1 of PEAR1. This SNP was in very tight linkage disequilibrium (LD; r2 = 0.97) with rs12566888, ∼ 600 bp upstream and also in intron 1. Although the previously reported intergenic SNP in the distal promoter region of PEAR1 (rs2768759) was strongly associated with collagen aggregation (P = 8.70 × 10−5),3 it failed the significance threshold for this study (see supplemental Table 2A for full list of SNPs and genotype-phenotype association βs and P values in EA participants).
Figure 1.
Gene variants in NTRK1 and PEAR1 associated with platelet function after aspirin treatment and linkage disequilibrium at the locus. (A) EA ancestry and (B) African ancestry. In both figures, the top panel shows the strength of association between individual SNPs along the locus (genetic coordinates chromosome 1:155050166 to 155154850, NCBI build 36.3) and platelet function phenotypes. Col2 and 5 indicate aggregation to collagen 2 and 5 μg/mL, respectively; ADP2 and 10, aggregation to ADP 2 and 10μM, respectively; Epi2 and 10, aggregation to epinephrine 2 and 10μM, respectively. Red dotted line signifies Bonferroni corrected significance threshold (3.19 × 10−5); rs numbers across the top signify SNPs reaching the significance threshold (red font denotes significance for both ancestries, black font denotes significance for one ancestry). Chromosomal coordinates for the NTRK1 and PEAR1 genes within the locus are shown by blue bars. In both figures, the bottom panel shows linkage equilibrium among SNPs in the region. The pairwise strength of association between SNPs (D') is signified by depth of color of shaded areas (white = 0, dark red = 1.0). Areas within triangles signify LD blocks.
Table 1.
Significant associations (P < 3.1 × 10 × 10−5) of PEAR1 and NTRK1 gene variants with platelet function phenotypes after aspirin treatment
| Chrom1 position | SNP rs no. | Gene | Role | MA(F) | Col2 | Col5 | ADP2 | ADP10 | Epi2 | Epi10 |
|---|---|---|---|---|---|---|---|---|---|---|
| European ancestry | ||||||||||
| 155114730 | rs2644604 | NTRK1 | Intron | A (0.112) | .0011 | 3.59 × 10−5 | .0026 | .0334 | 2.17 × 10−5 | .0008 |
| 155122419 | rs12564129 | Intergenic | C (0.047) | .0015 | .0025 | .0505 | .2934 | 3.36 × 10−6 | 1.77 × 10−6 | |
| 155135671 | rs12566888 | PEAR1 | Intron | T (0.092) | 6.99 × 10−6 | 5.43 × 10−5 | .0002 | .0125 | 3.48 × 10−8 | 4.50 × 10−5 |
| 155136338 | rs12041331 | PEAR1 | Intron | A (0.091) | 8.76 × 10−7 | 4.57 × 10−7 | .0002 | .0060 | 2.22 × 10−8 | 7.55 × 10−7 |
| 155146204 | rs3737224 | PEAR1 | Coding-syn | T (0.117) | .0022 | .0041 | .1729 | .0099 | 2.69 × 10−7 | 7.93 × 10−5 |
| African ancestry* | ||||||||||
| 155114730 | rs2644604 | NTRK1 | Intron | A (0.284) | 4.76 × 10−5 | 1.60 × 10−6 | 3.33 × 10−6 | 3.48 × 10−6 | 3.01 × 10−10 | 7.95 × 10−9 |
| 155135671 | rs12566888 | PEAR1 | Intron | G (0.463) | 7.67 × 10−7 | 1.20 × 10−8 | 2.19 × 10−5 | 3.68 × 10−7 | 3.30 × 10−9 | 3.49 × 10−9 |
| 155136338 | rs12041331 | PEAR1 | Intron | A (0.371) | 1.15 × 10−9 | 7.83 × 10−14 | 8.53 × 10−13 | 4.73 × 10−14 | 2.31 × 10−27 | 1.58 × 10−25 |
P values are shown for each genotype-phenotype association signal.
MA(F) indicates minor allele (frequency); Col2 and 5, aggregation to collagen 2 and 5 μg/mL, respectively; ADP2 and 10, aggregation to ADP 2 and 10μM, respectively; and Epi2 and 10, aggregation to epinephrine 2 and 10μM, respectively.
Significant SNPs in common with participants of EA.
Eleven SNPs showed a significant (P < 3.19 × 10−5) association with ≥ 1 of the phenotypes after aspirin therapy in African Americans: 6 SNPs in NTRK1, 1 SNP in the intergenic region, and 4 SNPs in PEAR1 (Figure 1B). Among these 11 SNPs, 3 replicated results seen in participants of EA: rs2644604 in NTRK1 and rs12041331 and rs12566888 in PEAR1. Similar to EA, the strongest and most consistent genotype-phenotype signals in this independent group of African Americans came from the PEAR1 intron 1 SNP, rs12041331 (Table 1; see supplementary Table 2B for full list of SNPs and genotype-phenotype association βs and P values in African ancestry participants).
Examination of the secondary phenotypes (ie, baseline platelet aggregation and after aspirin aggregation adjusted for baseline) for association signals with the 3 replicated gene variants (rs2644604, rs12566888, rs12041331) showed similar results. Strong association signals were observed for each of the 3 SNPs with ≥ 1 of the secondary phenotypes, and the strongest and most consistent signals across phenotypes and ancestry groups were seen for the PEAR1 intron 1 variant, rs12041331 (Table 2).
Table 2.
Genotype-phenotype associations for secondary platelet aggregation phenotypes in GeneSTAR (for SNPs common to participants of EA and African ancestry)
| SNP | Baseline aggregation phenotypes |
After aspirin aggregation phenotypes adjusted for baseline |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Col2 | Col5 | ADP2 | ADP10 | Epi2 | Epi10 | Col2 | Col5 | ADP2 | ADP10 | Epi2 | Epi10 | |
| European ancestry | ||||||||||||
| rs2644604 | .0798 | .0161 | .0002 | .0002 | .0012 | .0060 | .0011 | .0001 | .2318 | .0587 | .0003 | .0097 |
| rs12566888 | .0229 | .1252 | 2.60 × 10−6 | .0371 | 7.08 × 10−5 | .0005 | 1.12 × 10−5 | .0001 | .1327 | .0288 | 9.72 × 10−6 | .0026 |
| rs12041331 | .3077 | .0166 | .0001 | .0045 | 3.08 × 10−5 | .0010 | 8.98 × 10−6 | 1.60 × 10−5 | .0730 | .0276 | .0003 | .0015 |
| African ancestry | ||||||||||||
| rs2644604 | 5.23 × 10−7 | .0157 | .0002 | .0020 | 3.58 × 10−7 | 2.83 × 10−6 | 0.0003 | 5.78 × 10−5 | .0034 | .0013 | .0001 | 1.70 × 10−5 |
| rs12566888 | .0002 | .1867 | 3.12 × 10−5 | 6.83 × 10−7 | 1.39 × 10−8 | 7.59 × 10−8 | 3.47 × 10−6 | 1.12 × 10−8 | .0143 | .0029 | .0024 | .0016 |
| rs12041331 | .0002 | .0020 | 2.28 × 10−9 | 6.29 × 10−8 | 9.97 × 10−17 | 1.81 × 10−12 | 1.40 × 10−7 | 4.89 × 10−12 | 3.55 × 10−5 | 1.56 × 10−7 | 6.31 × 10−6 | 9.10 × 10−7 |
P values are shown for each genotype-phenotype association signal.
Col2 and 5 indicates aggregation to collagen 2 and 5 μg/mL, respectively; ADP2 and 10, aggregation to adenosine diphosphate 2 and 10μM, respectively; and Epi 2 and 10, aggregation to epinephrine 2 and 10μM, respectively.
The association of baseline platelet phenotypes with the 3 gene variants was further replicated among persons of European ancestry in the FHS Offspring cohort. Platelet aggregation phenotypes were harmonized to allow comparisons across studies. Both SNPs in intron 1 of PEAR1 were strongly associated with platelet aggregation to ADP and epinephrine in the FHS participants (Table 3). LD between rs12041331 and rs12566888 was very high (r2 = 0.85). Association signals of the NTRK1 gene variant with ADP and epinephrine aggregation were present but less robust than were observed for the PEAR1 SNPs, similar to findings in GeneSTAR (Table 3).
Table 3.
Genotype-phenotype associations for secondary platelet aggregation phenotypes in FHS participants with EA
| SNP | Baseline aggregation phenotypes |
|||
|---|---|---|---|---|
| ADP3 | ADP5 | Epi1 | Epi3 | |
| rs2644604 | 2.96 × 10−5 | .0212 | .0112 | .1224 |
| rs12566888 | 6.72 × 10−8 | .0012 | 9.38 × 10−6 | .1910 |
| rs1241331 | 1.51 × 10−7 | 8.14 × 10−04 | 1.64 × 10−05 | .1562 |
P values are shown for each genotype-phenotype association signal.
ADP3 and 5 indicates aggregation to ADP 3 and 10μM, respectively; and Epi1 and 3, aggregation to epinephrine 1 and 3μM, respectively.
We sought to determine the extent to which the associations between platelet phenotype and the rs12041331 gene variant in PEAR1 were independent of the rs2644604 gene variant in NTRK1. Pairwise LD (r2) between these 2 variants in participants of EA and African ancestry in GeneSTAR were 0.315 and 0.068, respectively. Inclusion of both SNPs in a single model to test for association with each phenotype reduced the effect sizes (β coefficients) and P values for both; however, rs12041331 of PEAR1 remained more strongly associated with all platelet function phenotypes than rs2644604 of NTRK1 both in the presence and absence of aspirin (see supplemental Table 3A-B). For example, P values for 15 of 18 genotype-phenotype associations in African Americans remained < 3.19 × 10−5 for rs12041331 in combined models, but none retained this significance level for the NTRK1 SNP. LD between the 2 SNPs in intron 1 of PEAR1 was too high (r2 = 0.97 in EA) to examine independence.
The G allele (major allele) of rs12041331 was associated with greater platelet aggregation in the presence and absence of aspirin treatment (Table 4). This relation was present in both EA and African American samples despite differences in allele frequency. Furthermore, the magnitude of effect of the G allele on platelet aggregation was substantive and similar in both races, with aggregation for the GG genotype being 60%-100% greater than for the AA genotype in most of the phenotypes examined. The proportion of total variance in platelet phenotype attributable to the rs12041331 variant ranged between 0.1% and 15% and was generally higher for aspirin-treated phenotypes than phenotypes in the absence of aspirin (average = 6% vs 3%, respectively) and for participants of African ancestry than EA (average = 7% vs 2%, respectively; Table 4). The G allele of rs12041331 was also associated with greater platelet aggregation in the FHS cohort (supplemental Table 4), consistent with findings in GeneSTAR.
Table 4.
Mean values for platelet aggregation phenotypes by PEAR1 rs12041331 genotype (AA, AG, GG) in participants of EA and African ancestry
| EA |
African ancestry |
|||||||
|---|---|---|---|---|---|---|---|---|
| AA, mean ± SD (N) | AG, mean ± SD (N) | GG, mean ± SD (N) | PVL, % | AA, mean ± SD (N) | AG, mean ± SD (N) | GG, mean ± SD (N) | PVL, % | |
| Phenotype after aspirin | ||||||||
| Col2 | 4.9 ± 7.8 (10) | 10 ± 12 (205) | 15 ± 13 (1026) | 2.92 | 4.9 ± 7.7 (113) | 14 ± 16 (373) | 20 ± 19 (349) | 6.57 |
| Col5 | 11 ± 11 (10) | 24 ± 19 (204) | 32 ± 21 (1023) | 2.58 | 14 ± 16 (112) | 29 ± 23 (368) | 37 ± 24 (342) | 8.08 |
| ADP2 | 15 ± 13 (10) | 32 ± 17 (187) | 36 ± 17 (912) | 1.35 | 25 ± 16 (107) | 31 ± 17 (359) | 38 ± 18 (326) | 7.09 |
| ADP10 | 57 ± 19 (10) | 66 ± 13 (202) | 69 ± 13 (992) | 0.712 | 58 ± 16 (112) | 65 ± 14 (368) | 71 ± 12 (339) | 7.14 |
| Epi2 | 13 ± 7.7 (10) | 18 ± 11 (205) | 23 ± 12 (1025) | 2.95 | 10 ± 8 (113) | 17 ± 11 (373) | 25 ± 17 (349) | 14.6 |
| Epi10 | 17 ± 8 (10) | 25 ± 14 (204) | 30 ± 14 (1021) | 2.28 | 14 ± 10 (112) | 22 ± 13 (369) | 31 ± 18 (341) | 13.6 |
| Phenotype at baseline | ||||||||
| Col2 | 43 ± 36 (10) | 61 ± 32 (205) | 66 ± 28 (1016) | 0.110 | 43 ± 35 (112) | 62 ± 31 (374) | 69 ± 26 (354) | 1.88 |
| Col5 | 69 ± 33 (10) | 81 ± 19 (205) | 83 ± 17 (1015) | 0.616 | 70 ± 29 (110) | 81 ± 21 (373) | 85 ± 15 (354) | 1.43 |
| ADP2 | 23 ± 20 (10) | 37 ± 25 (188) | 46 ± 26 (901) | 1.69 | 30 ± 26 (106) | 37 ± 28 (352) | 47 ± 28 (330) | 5.50 |
| ADP10 | 73 ± 25 (10) | 76 ± 16 (202) | 80 ± 14 (1004) | 0.854 | 69 ± 19 (112) | 74 ± 19 (373) | 80 ± 15 (351) | 4.28 |
| Epi2 | 32 ± 39 (10) | 47 ± 36 (205) | 57 ± 33 (1012) | 1.85 | 27 ± 33 (111) | 46 ± 35 (373) | 62 ± 34 (353) | 9.92 |
| Epi10 | 45 ± 38 (10) | 66 ± 31 (205) | 72 ± 27 (1012) | 1.15 | 39 ± 37 (112) | 61 ± 34 (374) | 72 ± 30 (354) | 7.22 |
PVL indicates proportion of total phenotypic variance attributable to the locus; Col2 and 5, aggregation to collagen 2 and 5 μg/mL, respectively; ADP2 and 10, aggregation to ADP 2 and 10μM, respectively; and Epi2 and 10, aggregation to epinephrine 2 and 10 μM, respectively
PEAR1 sequencing and relation of gene variants to platelet function
The SNP association analyses in GeneSTAR and FHS participants identified a strong relation between platelet function phenotype and 2 highly correlated gene variants in intron 1 of PEAR1. To further characterize genetic variants in PEAR1 we sequenced the entire PEAR1 gene (chromosome 1 coordinates: 155129813 to 155159018, NCBI build 36.3) in 104 GeneSTAR participants, for example, in 50 who demonstrated high platelet aggregability and in 54 who demonstrated low aggregability after aspirin. A total of 226 SNPs and 11 indels were identified by sequencing, which included 205 SNPs not included in the original set of genotyped or imputed SNPs. Allele frequencies of each genetic variant between participants in the high (case) and low (control) aggregation groups were compared within groups of EA and African ancestry. (The frequencies of indels were too rare to perform meaningful statistical analyses.) A meta-analysis was then performed across EA and African ancestry groups, accounting for direction of allelic effect. RS12041331 in intron 1 was the PEAR1 gene variant in the sequence data that was most strongly associated with platelet function (P = 2.07 × 10−6; see Table 5 for top 10 variants; see also supplemental Table 5 for all variants), and its association was the strongest of 31 variants identified in a 5-kb region centered on rs12041331.
Table 5.
Top 10 sequence variants in PEAR1 related to after aspirin platelet function phenotype in meta-analysis of data from participants of EA and African ancestry
| Chromosome 1 position | SNP rs no. | Role | P for EA | P for African ancestry | P for meta-analysis |
|---|---|---|---|---|---|
| 155134608 | Intron 1 | .0047 | .0377 | .0005 | |
| 155135472 | rs12407092 | Intron 1 | .0017 | .0778 | .0005 |
| 155135671 | rs12566888 | Intron 1 | .0111 | .0044 | .0001 |
| 155136056 | Intron 1 | .0047 | .0543 | .0008 | |
| 155136338 | rs12041331 | Intron 1 | .0210 | 1.06 × 10−5 | 2.07 × 10−6 |
| 155150241 | .0451 | .0222 | .0024 | ||
| 155151208 | .0042 | .0978 | .0014 | ||
| 155153725 | utr-3 | .0039 | .0475 | .0006 | |
| 155152115 | rs4661012 | utr-3 | .0124 | .0146 | .0005 |
| 155154204 | utr-3 | .0059 | .0463 | .0008 |
PEAR1 intron 1 gene variant rs12041331 and expression of PEAR1 protein in platelets
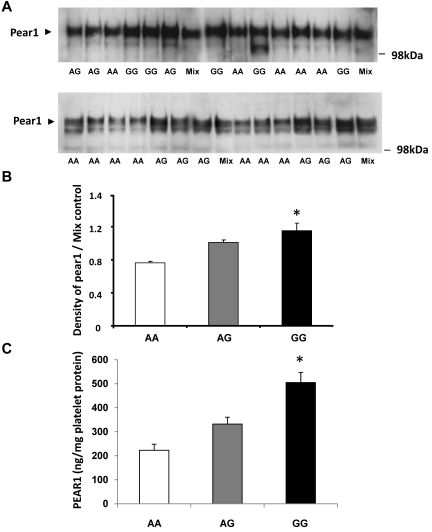
Platelet expression of PEAR1 was quantified by Western blots with the use of protein lysates obtained from 26 persons whose genotype differed at the rs12041331 locus (Figure 2A). PEAR1 protein expression was greatest for GG homozyotes, intermediate for GA heterozygotes, and least for AA homozygotes, consistent with the relation of the G allele to greater platelet aggregation (Figure 2B). Western blot findings were confirmed in an additional 24 subjects (8 of each genotype) with the use of ELISA to quantify platelet expression of PEAR1 (Figure 2C).
Figure 2.
Platelet expression of PEAR1 protein is related to genetic variation in PEAR1 at rs12041331. Western blotting analysis of platelet lysates (30 μg/lane) from randomly selected persons who varied by genotype (AA, AG, GG) at rs12041331. (A) Representative Western blots. (B) Summary results for platelet expression by genotype normalized to control (AG pooled mixture). (C) Platelet expression of PEAR1 protein assessed by quantitative ELISA. Results are displayed as mean ± SD; *P < .05 for difference among groups by ANOVA.
PEAR1 intron 1 gene variant rs12041331 and luciferase expression in a reporter assay
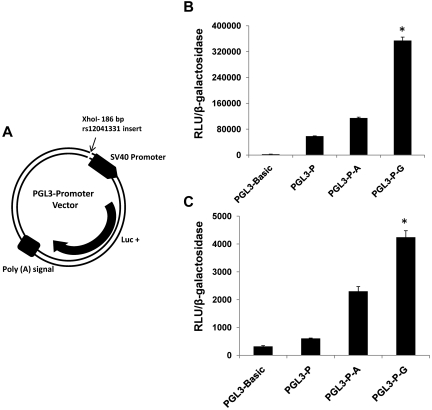
The ability for DNA sequence variation at rs12041331 to modify gene expression was examined with a luciferase reporter assay. Oligonucleotides containing the G allele or A allele were inserted into a PGL3 expression plasmid and transfected into MEG-01 cells and HUVECs. Cultured cells transfected with the G allele–containing plasmid showed greater expression of luciferase than cells with the A allele–containing vector (Figure 3).
Figure 3.
PEAR1 rs12041331 gene variant modifies expression of luciferase in a gene reporter assay. (A) PGL3 promoter vector construct showing the insertion site for a 186-bp sequence of intron1 of PEAR1 spanning the A allele or G allele of rs12041331. Luc+ indicates luciferase gene. (B) Expression of luciferase in MEG-01 cells cotransfected with plasmids containing PGL3 and β-galactosidase. (C) Expression of luciferase in HUVECs cotransfected with plasmids containing PGL3 and β-galactosidase. Luciferase expression is shown by relative light units of luciferase (RLUs) normalized to absorption (OD) of β-galactosidase activity. PGL3 basic indicates luciferase gene plasmid without promoter; PGL3-P, luciferase gene plasmid with SV40 promoter; PGL3-P-A, luciferase gene plasmid with SV40 promoter and A allele–containing insert cloned from PEAR1; PGL3-P-G, luciferase gene plasmid with SV40 promoter and G allele–containing insert cloned from PEAR1. Results are displayed as mean ± SD; *P < .001 for difference among groups by ANOVA; n = 3.
Discussion
Variability in platelet response to aspirin is highly heritable2 and is strongly associated with risk of myocardial infarction and stroke.14,15 We have now identified a common genetic variant in intron 1 of the PEAR1 gene, rs12041331, that accounts for ≤ 15% of the total phenotypic variance in platelet function after aspirin exposure and ≤ 10% of native platelet function (ie, before aspirin treatment). Findings were strongest for persons of African ancestry but were robust in persons of EA as well, being replicated in an independent sample of EA participants from the FHS, and were demonstrable across multiple platelet function phenotypes. Genetic variation at the rs12041331 locus showed a dose-response relation with platelet expression of PEAR1 protein and with luciferase expression in in vitro cell systems, providing a plausible biologic connection between the intron 1 variant and platelet function phenotype.
With the use of a candidate gene approach, we previously identified a relation between rs2768759, located in the intergenic region between PEAR1 and NTRK1, and platelet aggregation phenotype.3 With the use of fine mapping, this study confirms the important contribution of the PEAR1 gene to variability in human platelet function and identifies a variant within intron 1 that is most closely associated with platelet response to aspirin treatment. The previously documented association between rs2768759 and platelet function3 failed to meet the Bonferroni significance threshold for this study. LD between rs2768759 and rs12041331 was modest (r2 = 0.163 in EA and r2 = 0.076 in African Americans), and, as expected, inclusion of both SNPs in regression models had no effect on the relation between rs12041331 and platelet function phenotypes (data not shown). Results from this investigation are consistent with those from a recently reported genomewide association study in which an intronic PEAR1 variant (rs12566888) was associated with native platelet function.4 Furthermore, our experiments identify the intron1 variant rs12041331, which is in tight LD with rs12566888, as the most probable variant to explain genotype-phenotype association findings and extend findings to platelet aspirin response phenotypes, which are related to clinical outcome in human studies.14,15
Jones et al16 reported an association between genetic variation in PEAR1 and native platelet function with the use of a candidate SNP approach in 500 persons of Northern European descent. A block of 4 SNPs in strong LD (rs3737224, rs41273215, rs41299597, and rs822442) were reported to be associated with baseline platelet activation by collagen-related peptide, and a fifth independent SNP (rs11264579) was associated with baseline platelet activation by ADP; rs3737224 and rs41299597 were also reported to be associated with surface expression of PEAR1 protein after platelet activation. In the present study, rs3737224 was strongly associated with platelet activation by epinephrine in persons of EA (and less strongly to collagen), but no association was found in persons of African descent before or after aspirin treatment. We also found no significant relation between rs11264579 and any platelet activation phenotype in either ethnic group. Similarly, sequencing studies did not show a relation between any of the SNPs identified by Jones et al16 and platelet function in either ethnic group or in meta-analysis. In addition, association signals of rs3737224 and rs11264579 with baseline platelet aggregation in FHS participants were either weak (strongest signal for aggregation to 3μM epinephrine, P = .02) or not present. In contrast, we report that the locus defined by rs12041331 in intron 1 of PEAR1 is strongly associated with platelet activation by multiple agonists, both in the presence and absence of aspirin, in 3 separate cohorts of different ethnicity, and with platelet expression of PEAR1 protein. Although we cannot preclude the possibility that other PEAR1 SNPs, including those reported by Jones et al,16 may contribute to variability in baseline platelet activation among persons of EA, our results suggest that rs12041331 is a much greater contributor to variation in human platelet function and response to aspirin treatment and is a more probable causative locus.
Association findings in 2 different population ancestry groups (European and African) and 2 different cohorts (GeneSTAR and FHS) suggested that genetic variation in a 600-bp region (between rs12041331 and rs12566888) in intron 1 of PEAR1 was the most prominent source of the observed phenotypic variations in platelet function. Association approaches such as the unbiased and comprehensive genomewide association study model have been productive in discovering genes that control the risk of complex diseases and phenotypes. However, it has been difficult for gene-association studies to obtain adequate coverage of the genome for African-origin populations,17considering the shortcoming of relying only on Yoruban genomes (ie, YRI) to represent variation in African Americans.18 The reliance on tests for association with a set of SNPs that may inadequately tag the true causal variant in African admixed populations, given the higher breakdown in LD in this population, renders replication between ancestry groups at a SNP-for-SNP level difficult. Nonetheless, we demonstrate replication of the association signal at rs12041331 as being the strongest in both ancestry groups and document with sequence data that there are no additional variants within PEAR1 that approach its signal strength.
We found a dose-response relation between the number of G alleles at rs12041331 and the level of PEAR1 protein expressed in platelets, providing a potential biologic explanation for the genotype-phenotype association findings. PEAR1 is an integral membrane protein that is highly expressed on the surface of platelets and endothelial cells.19 It contains 15 extracellular growth factor-type domains and an intracellular tail that becomes phosphorylated on tyrosine and serine residues after agonist stimulation and/or close platelet-platelet apposition or both.19 Although the precise role of PEAR1 in platelet function has not been defined, our findings suggest an important physiologic role for PEAR1 in human platelet aggregation that is worthy of further investigation.
Results from PEAR1 expression studies in platelets and luciferase expression studies in a megakaryocytic cell line and HUVECs consistently showed a relation between the G allele variant and greater protein expression. The molecular basis for this effect is not known; however, data are consistent with the hypothesis that the rs12041331 variant in intron 1 defines a transcription factor binding site capable of regulating gene expression. A bioinformatics search with the Transcription Element Search System tool (available at http://www.cbil.upenn.edu/tess) identified several potential transcription factor binding sites for the PEAR1 intronic sequence flanking the A/G dimorphism, and predictions differed for the A and G alleles. Intronic sequences are well recognized to regulate gene expression through the carriage of enhancer elements and alternative splice sites and to provide a substrate pool for reshuffling of genetic sequences20; however, the importance of intronic variants to expression of clinically important complex phenotypes is unknown. Additional experiments beyond the scope of this work are required to provide a more complete description of the mechanistic link between intronic variation in the PEAR1 gene and differences in platelet structure and function.
The clinical applicability of our findings is not defined at this time. Although platelet aggregability after aspirin treatment is associated with increased risk of myocardial infarction and stroke,14,15 clinical outcome was not assessed in our cohort of asymptomatic persons. Additional studies will be needed to determine the importance of genetic variation in PEAR1 to cardiovascular disease expression and clinical response to antiplatelet therapy.
Supplementary Material
Acknowledgments
This work was supported by the National Heart, Lung, and Blood Institute (NHLBI) through the PROGENI (U01 HL72518) and STAMPEED (R01 HL087698-01) consortia and by grant R01-HL-48157. A portion of this work was supported by resources of the Johns Hopkins General Clinical Research Center, funded through the National Center for Research Resources (grant M01-RR000052). This work was also supported by the NHLBI's Framingham Heart Study (contract no. N01-HC-25195) and its contract with Affymetrix Inc for genotyping services (contract no. N02-HL-6-4278). A portion of this research used the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center.
This research was conducted in part with the use of data and resources from the NHLBI's Framingham Heart Study of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: N.F., C.J.O., D.M.B., and L.C.B. led the study; N.F. took primary responsibility for drafting the manuscript; L.R.Y., R.M., A.D.J., R.Q., M-H.C., C.J.O., A.D.F., P.F.B., D.M.B., and L.C.B. contributed to and edited the manuscript; N.F., L.R.Y., R.M., J.E.H.-G., D.M.B., and L.C.B. were involved in the guidance, collection, and analysis of GeneStar phenotype data; G.H.T. and C.J.O. were involved in the guidance, collection, and analysis of Framingham phenotype data; L.R.Y., R.M., J.E.H.-G., B.S., A.D.J., M.-H.C., and I.R. conducted SNP association analyses and determined the percentage of variance explained; L.R.Y;, R.M., B.S., R.Q., and I.R. conducted regression analyses and meta-analyses on sequencing data; A.G. and U.T. conducted the sequencing experiments; and X.P.Y. and A.D.F. conducted the Western blotting, luciferase reporter, and ELISA experiments. All authors read and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nauder Faraday, 298 Meyer Bldg, Johns Hopkins Hospital, 600 N Wolfe St, Baltimore, MD 21287; e-mail: nfaraday@jhmi.edu.
References
- 1.Bray PF, Mathias RA, Faraday N, et al. Heritability of platelet function in families with premature coronary artery disease. J Thromb Haemost. 2007;5(8):1617–1623. doi: 10.1111/j.1538-7836.2007.02618.x. [DOI] [PubMed] [Google Scholar]
- 2.Faraday N, Yanek LR, Mathias R, et al. Heritability of platelet responsiveness to aspirin in activation pathways directly and indirectly related to cyclooxygenase-1. Circulation. 2007;115(19):2490–2496. doi: 10.1161/CIRCULATIONAHA.106.667584. [DOI] [PubMed] [Google Scholar]
- 3.Herrera-Galeano JE, Becker DM, Wilson AF, et al. A novel variant in the platelet endothelial aggregation receptor-1 gene is associated with increased platelet aggregability. Arterioscler Thromb Vasc Biol. 2008;28(8):1484–1490. doi: 10.1161/ATVBAHA.108.168971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson AD, Yanek LR, Chen MH, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet. 2010;42(7):608–613. doi: 10.1038/ng.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker DM, Segal J, Vaidya D, et al. Sex differences in platelet reactivity and response to low-dose aspirin therapy. JAMA. 2006;295(12):1420–1427. doi: 10.1001/jama.295.12.1420. [DOI] [PubMed] [Google Scholar]
- 6.O'Donnell CJ, Larson MG, Feng D, et al. Genetic and environmental contributions to platelet aggregation. The Framingham Heart Study. Circulation. 2001;103(25):3051–3056. doi: 10.1161/01.cir.103.25.3051. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 9.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26(4):580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 11.Gum PA, Kottke-Marchant K, Welsh PA, White J, Topol EJ. A prospective, blinded determination of the natural history of aspirin resistance among stable patients with cardiovascular disease. J Am Coll Cardiol. 2003;41(6):961–965. doi: 10.1016/s0735-1097(02)03014-0. [DOI] [PubMed] [Google Scholar]
- 12.Ohmori T, Yatomi Y, Nonaka T, et al. Aspirin resistance detected with aggregometry cannot be explained by cyclooxygenase activity: involvement of other signaling pathway(s) in cardiovascular events of aspirin-treated patients. J Thromb Haemost. 2006;4(6):1271–1278. doi: 10.1111/j.1538-7836.2006.01958.x. [DOI] [PubMed] [Google Scholar]
- 13.Gori AM, Marcucci R, Paniccia R, et al. Thrombotic events in high risk patients are predicted by evaluating different pathways of platelet function. Thromb Haemost. 2008;100(6):962–964. doi: 10.1160/th08-06-0374. [DOI] [PubMed] [Google Scholar]
- 14.Snoep JD, Hovens MMC, Eikenboom JCJ, van der Bom JG, Huisman MV. Association of laboratory-defined aspirin resistance with a higher risk of recurrent cardiovascular events: a systematic review and meta-analysis. Arch Intern Med. 2007;167(15):1593–1599. doi: 10.1001/archinte.167.15.1593. [DOI] [PubMed] [Google Scholar]
- 15.Krasopoulos G, Brister SJ, Beattie WS, Buchanan MR. Aspirin “resistance” and risk of cardiovascular morbidity: systematic review and meta-analysis. BMJ. 2008;336(7637):195–198. doi: 10.1136/bmj.39430.529549.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones CI, Bray S, Garner SF, et al. A functional genomics approach reveals novel quantitative trait loci associated with platelet signaling pathways. Blood. 2009;114(7):1405–1416. doi: 10.1182/blood-2009-02-202614. [DOI] [PubMed] [Google Scholar]
- 17.Mathias RA, Grant AV, Rafaels N, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125(2):336–346. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tishkoff SA, Reed FA, Friedlaender FR, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324(5930):1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nanda N, Bao M, Lin H, et al. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. J Biol Chem. 2005;280(26):24680–24689. doi: 10.1074/jbc.M413411200. [DOI] [PubMed] [Google Scholar]
- 20.Fedorova L, Fedorov A. Introns in gene evolution. Genetica. 2003;118(2–3):123–131. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.