Abstract
Genetic tools have greatly aided in tracing the sources and colonization history of introduced species. However, recurrent introductions and repeated shuffling of populations may have blurred some of the genetic signals left by ancient introductions. Styela plicata is a solitary ascidian distributed worldwide. Although its origin remains unclear, this species is believed to have spread worldwide by travelling on ship's hulls. The goals of this study were to infer the genetic structure and global phylogeography of S. plicata and to look for present-day and historical genetic patterns. Two genetic markers were used: a fragment of the mitochondrial gene Cytochrome Oxidase subunit I (COI) and a fragment of the nuclear gene Adenine Nucleotide Transporter/ADP-ATP Translocase (ANT). A total of 368 individuals for COI and 315 for ANT were sequenced from 17 locations worldwide. The levels of gene diversity were moderate for COI to high for ANT. The Mediterranean populations showed the least diversity and allelic richness for both markers, while the Indian, Atlantic and Pacific Oceans had the highest gene and nucleotide diversities. Network and phylogenetic analyses with COI and ANT revealed two groups of alleles separated by 15 and 4 mutational steps, respectively. The existence of different lineages suggested an ancient population split. However, the geographic distributions of these groups did not show any consistent pattern, indicating different phylogeographic histories for each gene. Genetic divergence was significant for many population-pairs irrespective of the geographic distance among them. Stochastic introduction events are reflected in the uneven distribution of COI and ANT allele frequencies and groups among many populations. Our results confirmed that S. plicata has been present in all studied oceans for a long time, and that recurrent colonization events and occasional shuffling among populations have determined the actual genetic structure of this species.
Introduction
Biological introductions have notably increased during the last century, posing a major threat to global biodiversity and altering the structure and function of many communities [1]–[7]. Despite some relatively recent attempts to buffer the ecological impact of these introductions [e.g. 8]–[10], oceans remain one of the most affected ecosystems [7], [11]–[17]. Among other transport vectors, non-native species arrive to new locations through ships' hulls and sea chests, in ballast water or with spats for mariculture. Thus, the increasing activity in maritime traffic and aquaculture has favoured the introduction of marine species all over the world [13], [18]–[19]. The establishment of new genetic variants and spread of exotic species has also been facilitated by a proliferation of harbours and other artificial structures along the coast [20]–[25].
Genetic diversity plays a crucial role on the successful establishment of an introduced species or variant in a new area [26]–[30]. The development of genetic tools and markers has widely contributed to enhance our knowledge on these species. A throughout assessment of the genetic structure of an introduced species, including its history of subdivision and gene flow, allows the identification of range expansions, colonization events, and an understanding of the invasive potential and the relative contributions of artificial and natural dispersal [e.g. 31]–[34].
The increasing pace of introductions has also fostered increased awareness. Monitoring and control programs have been established, and recent introductions are more easily detected and inventoried than in the past [e.g. 17]. However, historical invasions may still remain hidden. Some species could have arrived to a new location long before the distribution ranges of autochthonous species were assessed, and be now regarded as native [35], [36]. Cosmopolitan or broadly distributed species, particularly those thriving in harbours and artificial substrata, are likely to be “pseudoindigenous” species [36]. Lack of historical records in many regions, taxonomic flaws and cryptic speciation further complicate the issue [e.g. 37], [38]. In addition, and despite the new methods available [e.g., 33], our ability to extract information may be limited by our knowledge and access to native populations, recurrent introduction events, and shuffling of populations during a long period of time (i.e. centuries).
The paramount importance of ascidians for the study of marine introductions is well recognized, as they represent one of the most common invaders [39], [40]. Ascidians have short-lived larvae, thus anthropogenic transport can greatly increase their dispersal abilities. The rate of introduction of non-indigenous ascidians has been increasing in the last decades [40], mostly linked to ship traffic or aquaculture activities [e.g. 39], [41]–[45]. However, some species may have been translocated centuries ago and have now become ancient introductions whose origins are poorly known [46]. These ancient colonizers are often species commonly found in harbours and man-made substrates, have broad distribution ranges and, while naturalized in many areas, continue to be introduced in new regions of the globe [e.g. 47]–[50].
Styela plicata (Lesueur, 1823) (Tunicata, Ascidiacea) is a solitary ascidian commonly found inhabiting marinas and harbours of warm and temperate oceans, usually at high-densities. In spite of its broad geographical distribution, the native range of this species is not yet elucidated [46]. Evidence to date suggests that S. plicata is native to the NW Pacific Ocean [36], [51]–[54]. In fact, the description of this species was based on an individual found on a ship's hull in Philadelphia (NE USA), and no other individual was observed in the surrounding natural substrata [55]. All records of S. plicata are based on observations of man-made structures, except in Japan, where this species has been observed to grow in natural habitats [Nishikawa pers. comm., 54]. A series of unique characteristics has allowed S. plicata to thrive in these diverse environments and outcompete other benthic invertebrates. S. plicata can physiologically adapt to widely fluctuating environments, particularly to changes in temperature and salinity [56], [57]. This species can also tolerate highly polluted waters [58], grows rapidly until reaching sexual maturity [59]–[61], and is capable of self-fertilization (authors' current research).
To gain insight into the invasive potential of this species, we analyzed the genetic structure of seventeen populations covering most of S. plicata's distribution range. Using a mitochondrial (COI) and a nuclear (ANT) marker, we attempted to infer the global phylogeography of S. plicata, understand its dispersion patterns, and assess the diversity and connectivity of introduced populations.
Methods
Sampling
Samples of Styela plicata were collected in 2009 and 2010 from seventeen localities (Table 1): two from the Mediterranean Sea (Iberian Peninsula), three from the north-eastern Atlantic Ocean (Iberian Peninsula, Canary Islands), two from the north-western Atlantic Ocean (US east coast), one from the south-western Atlantic ocean (Brazil), five from the north-western Pacific Ocean (Japan and China), one from the south-western Pacific Ocean (Australia), one from the north-eastern Pacific Ocean (US west coast), and two from the south-western Indian Ocean (South Africa). These locations were chosen to cover as much of the distribution range of this widespread species as possible. All specimens were collected from artificial substrata (harbours, marinas or decks), except for one population collected from natural substratum in Sakushima Island (Japan). The shortest distance by sea between location pairs was calculated using the “measure line” tool of Google Earth (version 3.0, Google Inc., Amphitheatre Parkway, CA, USA). S. Plicata samples were obtained according to current Spanish regulations. Samples from outside Spain were collected by national researchers following their country regulations. This species is not protected by any law and all sampling was conducted outside protected areas.
Table 1. Population code, name, geographical region (including country), and GPS position for the populations of Styela plicata analyzed in this study.
| Code | Population | Geographical Region/Country | Latitude/Longitude |
| AR | Arenys de Mar | NW Mediterranean Sea/Spain | 41°34′36″N/2°33′32″E |
| JA | Javea | NW Mediterranean Sea/Spain | 38°47′52″N/0°11′06″E |
| SP | San Fernando | NE Atlantic Ocean/Spain | 36°27′36″N/6°12′13″W |
| FE | Ferrol | NE Atlantic Ocean/Spain | 43°29′00″N/8°14′00″W |
| TEN | Tenerife | NE Atlantic Ocean/Spain | 28°00′24″N/16°39′38″W |
| KNY | Knysna | SW Indian Ocean/South Africa | 34°2′28″S/23°2′38″E |
| PE | Port Elizabeth | SW Indian Ocean/South Africa | 33°57′49″S/25°38′16″E |
| NC | North Carolina | NW Atlantic Ocean/USA | 34°8′24″N/77°51′44″W |
| SC | South Carolina | NW Atlantic Ocean/USA | 32°12′57″N/80°46′49″W |
| CAL | California | NE Pacific Ocean/USA | 32°47′00″N/117°09′00″W |
| BRA | Santa Catarina | SW Atlantic Ocean/Brasil | 26°46′30″S/48°36′34″W |
| AM | Manly | SW Pacific Ocean/Australia | 33°47′43″S/151°17′38″E |
| WAK | Wakayama | NW Pacific Ocean/Japan | 34°11′17″N/135° 8′48″E |
| OKI | Okinawajima | NW Pacific Ocean/Japan | 26°19′29″N/127°50′15″E |
| MIS | Misaki | NW Pacific Ocean/Japan | 36°9′21″N/133°18′52″E |
| SKS | Sakushima Island | NW Pacific Ocean/Japan | 34°43′00″N/137°02′00″E |
| HK | Hong Kong | NW Pacific Ocean/China | 22°24′00″N/114°21′00′E |
All specimens were collected from depths that ranged between 0 and 2 m by pulling up harbour ropes, removing specimens from submersed docks and pilings, or pulling individuals from rocky assemblages (natural population). Samples were dissected in situ and a piece of muscular tissue from the mantle or the siphon was immediately preserved in absolute ethanol. Ethanol was changed after a few hours, and samples were then stored at −20°C until DNA extraction.
DNA extraction and sequencing
Total DNA was extracted using the REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich). The universal primers LCO1490 and HCO2198 described in Folmer et al. [62] were used to amplify a fragment of the mitochondrial gene Cytochrome Oxidase subunit I (COI) from 368 individuals. The primer set designed by Jarman et al. [63] was used to amplify a fragment of the single-copy nuclear Adenine Nucleotide Transporter (ANT) gene. Based on the resulting sequences, we also designed the specific primers ANTf_Splic (5′-TTG GCA GCT GAT ATT GGA AAA GG-3′) and ANTr_Splic (5′-CCA GAC TGC ATC ATC ATK CG-3′), using the software Primer 3 v.0.4.0. [64]. Amplifications were carried out for 315 individuals using Jarman et al. [63] primers or the newly designed ones.
For both genes, amplifications were performed in a final volume of 20 µL using 10 µL of REDExtract-N-amp PCR reaction mix (Sigma-Aldrich), 1 µL of each primer (10 µM) for ANT or 0.8 µL for COI, and 2 µL of template DNA. The PCR program for ANT consisted of an initial denaturing step at 94°C for 2 min, 30 amplification cycles (denaturing at 94°C for 1 min, annealing at 58°C for 30 seconds and extension at 72°C for 30 seconds), and a final extension at 72°C for 6 min, on a PCR System 9700 (Applied Biosystems). The PCR program for COI was as described above, except for the amplification cycles, which were done at 94°C for 45 seconds, 50°C for 45 seconds and 72°C for 50 seconds. PCR products were purified using MultiScreen® filter plates (Millipore), labelled using BigDye® Terminator v.3.1 (Applied Biosystems) and sequenced on an ABI 3730 Genetic Analyzer (Applied Biosystems) at the Scientific and Technical Services of the University of Barcelona (Spain). Other samples were directly sent for purification and sequencing to Macrogen Inc. (Seoul, Korea Korea). From the resulting sequences, we discarded low quality reads for ANT, hence the lower number of specimens sequenced for this marker.
Sequences were edited and aligned using BioEdit® v.7.0.5.3 [65]. Some ANT sequences showed a deletion of 22 amino acids, thus heterozygotes had unequal lengths and had to be manually reconstructed by carefully analyzing both forward and reverse chromatograms. The allelic phase for ANT genotypic data was analyzed using fastPHASE 1.1 [66] implemented in the software DnaSP v.5 [67]. We also used the Recombination Detection Program (RDP3) [68] to test for recombination in our nuclear sequences. Sequences obtained in this study have been deposited in GenBank (accession numbers HQ916425 to HQ916446 for COI, and HQ916363 to HQ916423 for ANT).
Population genetics
Number of alleles (Nh), gene diversity (Hd), and nucleotide diversity (π) were computed with DnaSP v.5 [67]. Allelic richness was calculated using the program Contrib v.1.02, which implements a rarefaction method to obtain estimates independently of sample size [106]. Genetix v.4.05.2 [69] was used to calculate inbreeding coefficients for the ANT data obtained with fastPHASE. The nearly unbiased estimation of allelic differentiation between populations was based on the adjusted Dest measure described by Jost [70], and calculated for each marker with SPADE [71]. The mean and SE values obtained with SPADE from 1,000 bootstrap replicates were used to calculate the confidence intervals and the degree of significance of the differentiation values (using a normal approximation). To correct for multiple comparisons, we set the p-value at 0.009, following the Benjamini and Yekutieli False Discovery Rate correction [72]. A value of D was deemed significant when the confidence interval around its mean did not contain 0. An analysis of molecular variance (AMOVA) was performed to examine population structure, and its significance was tested running 10,000 permutations in Arlequin v.3.1 [73]. The correlation of genetic and geographical distances was tested for all pairs of populations with a Mantel test [74] and 10,000 permutations using Arlequin.
Visual assessment of between-population differentiation was achieved by performing a discriminant analysis of principal components (DAPC) [75] on a dataset comprising information obtained from both genes. This recently developed technique extracts information from genetic datasets (multivariate in nature) by first performing a principal component analysis (PCA) on groups or populations, and then using the PCA factors as variables for a discriminant analysis (DA). The previous PCA step ensures that the variables input to DA meet the requirements of having less variables (alleles) than number of observations (individuals) and not having any correlation between variables [75]. DA seeks to maximize the inter-group component of variation. We performed DAPC analyses on both genes combined by using the adegenet package for R [76]. DAPC was performed (function dapc) using pre-defined groups corresponding to populations or groups of populations (see Results). Variables were centred but not scaled. In all analyses, 50 principal components of PCA were retained and input to DA. DA also provided estimates of the probability with which the analysis recovers the true membership of the individuals. Finally, in order to detect population growth and infer population demographic events, we computed Tajima's D [77], Fu's Fs [78], R2 [79], and the raggedness index (based on the mismatch distribution) [80], using DnaSP.
Phylogenetic and phylogeographical analyses
The complete dataset was used to construct a median-joining network for each marker using Network v.4.5.1.6 [81]. Resulting loops for the ANT network were solved using criteria derived from the coalescent theory [82], [83]. For the COI network, only one loop was observed but it could not be resolved.
Phylogenetic analyses were conducted using Styela gibbsii as an outgroup (acc. number HQ916447 for COI and HQ916424 for ANT). The best-fit model of nucleotide substitution for each marker was selected using jModeltest v.0.1.1 [84], [85], with the Akaike Information Criterion (AIC) for COI, and the corrected version for small samples (AICc) for ANT. The positions corresponding to the indel detected for ANT were not included in the analysis (see Results). For Bayesian inference (BI), MrBayes v.3.1.2 software [86] was used to infer tree topologies, implementing the corresponding likelihood model for each gene fragment. For each gene, the program was run with 1 million generations with a sample frequency of 100 (10,000 final trees). After verifying that stationarity had been reached (i.e. the average standard deviation of split frequencies between two independent chains reached less than 0.01), the first 1,000 trees were discarded in both cases as burnin. Majority-rule consensus trees were generated from the remaining 9,000 trees. Bayesian posterior probabilities were used as a measure of support for the branch nodes obtained. The obtained trees were drawn with FigTree v.1.2.2. DnaSP was used to perform the McDonald & Kreitman test [87], and check whether patterns of variation among groups of sequences were consistent with predictions for a neutral model.
Results
Mitochondrial gene
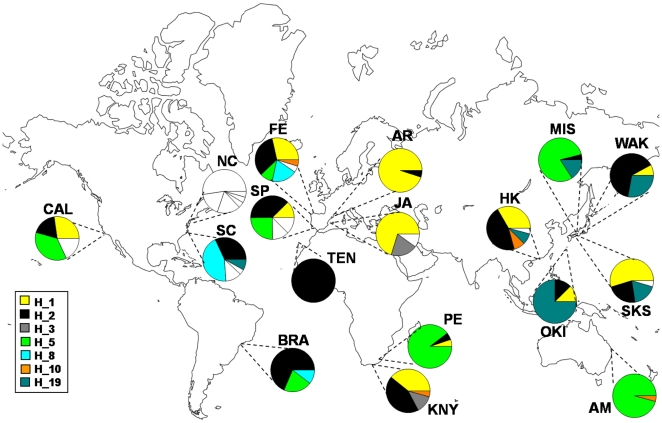
For the mitochondrial COI gene, 368 sequences with a final alignment length of 624 bp were obtained. In total, we found 22 haplotypes with 38 polymorphic sites (6%), 6 of which corresponded to non-synonymous substitutions. The majority of haplotypes obtained (68%) corresponded to private haplotypes, most of which were found in the north-western Atlantic Ocean (Fig. 1). Remarkably, the six haplotypes found for the North Carolina population (NC) were private. The number of haplotypes per location ranged between one in Tenerife and six in Ferrol and North Carolina (Table 2, Table S1). Regarding the oceanic basins, the Atlantic and Pacific Ocean had higher haplotype diversity (17 and 8 haplotypes, respectively) than the Mediterranean Sea and the Indian Ocean (4 and 5 haplotypes, respectively; Table 2). Mean and total haplotype diversity (Hd) were 0.497 (±0.266 SD) and 0.810 (±0.010 SD), respectively. Mean nucleotide diversity was 0.0055 (±0.005 SD), while total nucleotide diversity (π) was 0.0135 (±0.0006 SD). Variation in haplotype and nucleotide diversity between populations within basins was considerable. For instance, the populations of Knysna (KNY) and Port Elizabeth (PE) located in the Indian Ocean, had a haplotype diversity of 0.668 and 0.205 respectively. The California population (CAL) presented the highest haplotype and nucleotide diversity values (0.800 and 0.01684, respectively; Table 2). The higher allelic richness values (obtained after rarefaction to a common sample size of 11 and 40 genes per populations and basins) were found for the San Fernando (SP, 3.747) and Ferrol populations (FE, 3.793), while the lower values corresponded to the populations of Manly (AM, 0.458) and Arenys de Mar (AR, 0.555). When comparing between basins, the Atlantic Ocean showed the highest allelic richness, whereas the Mediterranean Sea had the lowest value (Table 2).
Figure 1. Map showing the sampling sites of Styela plicata.
Pie charts represent haplotype frequencies for the COI gene in each population analyzed. Private haplotypes are shown in white.
Table 2. Diversity measures for the studied populations of Styela plicata.
| Population | COI | ANT | |||||||||||||||
| N | r | Hd±SD | π±SD | Nh (private) | N | r | Hd±SD | π±SD | Nh (private) | Fis | Hexp | Hobs | |||||
| AR | 20 | 0.555 | 0.100 | (±0.088) | 0.00016 | (±0.00014) | 2 | 19 | 3.733 | 0.620 | (±0.072) | 0.02012 | (±0.00260) | 6 | 0.241* | 0.620 | 0.474 |
| JA | 20 | 1.785 | 0.484 | (±0.113) | 0.00388 | (±0.00095) | 3 (1) | 20 | 3.307 | 0.494 | (±0.088) | 0.01670 | (±0.00319) | 5 (1) | 0.802* | 0.494 | 0.100 |
| SP | 16 | 3.747 | 0.775 | (±0.068) | 0.01484 | (±0.00200) | 5 (2) | 17 | 8.434 | 0.791 | (±0.065) | 0.02831 | (±0.00345) | 11 (3) | 0.266* | 0.795 | 0.588 |
| FE | 21 | 3.793 | 0.795 | (±0.051) | 0.00835 | (±0.00274) | 6 (1) | 13 | 7.363 | 0.822 | (±0.059) | 0.02258 | (±0.00253) | 9 (2) | 0.259* | 0.822 | 0.615 |
| TEN | 24 | 0.000 | 0.000 | (±0.000) | 0.00000 | (±0.00000) | 1 | 29 | 5.349 | 0.743 | (±0.040) | 0.03475 | (±0.00176) | 10 (1) | −0.210* | 0.744 | 0.897 |
| KNY | 23 | 2.354 | 0.668 | (±0.057) | 0.00359 | (±0.00101) | 4 | 19 | 8.145 | 0.828 | (±0.044) | 0.03608 | (±0.00144) | 12 (4) | −0.018 | 0.828 | 0.842 |
| PE | 20 | 1.158 | 0.195 | (±0.115) | 0.00532 | (±0.00304) | 3 | 12 | 14.83 | 0.953 | (±0.029) | 0.03889 | (±0.00212) | 17 (3) | 0.040 | 0.953 | 0.917 |
| NC | 23 | 3.323 | 0.692 | (±0.085) | 0.00374 | (±0.00094) | 6 (6) | 18 | 8.927 | 0.789 | (±0.065) | 0.02859 | (±0.00429) | 13 (8) | 0.586* | 0.792 | 0.333 |
| SC | 25 | 2.976 | 0.710 | (±0.060) | 0.00491 | (±0.00046) | 5 (2) | 18 | 7.277 | 0.807 | (±0.050) | 0.02797 | (±0.00251) | 11 (1) | 0.022 | 0.807 | 0.790 |
| CAL | 11 | 3.000 | 0.800 | (±0.075) | 0.01684 | (±0.00270) | 4 (1) | 11 | 5.000 | 0.818 | (±0.049) | 0.04023 | (±0.00248) | 6 | −0.236 | 0.818 | 1.000 |
| BRA | 19 | 1.818 | 0.503 | (±0.113) | 0.01100 | (±0.00294) | 3 | 17 | 6.882 | 0.775 | (±0.052) | 0.03290 | (±0.00199) | 10 (2) | −0.301* | 0.775 | 1.000 |
| AM | 24 | 0.458 | 0.083 | (±0.005) | 0.00294 | (±0.00264) | 2 | 22 | 3.140 | 0.596 | (±0.058) | 0.01101 | (±0.00118) | 5 | 0.242 | 0.596 | 0.455 |
| WAK | 25 | 1.690 | 0.527 | (±0.084) | 0.00212 | (±0.00035) | 3 | 24 | 7.863 | 0.806 | (±0.043) | 0.03334 | (±0.00222) | 14 (3) | −0.035 | 0.806 | 0.833 |
| OKI | 24 | 1.717 | 0.424 | (±0.112) | 0.00162 | (±0.00042) | 3 | 16 | 4.972 | 0.766 | (±0.044) | 0.03892 | (±0.00176) | 7 | −0.233 | 0.766 | 0.938 |
| MIS | 25 | 1.361 | 0.347 | (±0.108) | 0.01043 | (±0.00309) | 3 | 22 | 6.178 | 0.780 | (±0.044) | 0.03019 | (±0.00208) | 10 (1) | −0.230* | 0.780 | 0.955 |
| SKS | 24 | 2.437 | 0.663 | (±0.065) | 0.00175 | (±0.00033) | 4 (1) | 24 | 4.536 | 0.714 | (±0.044) | 0.03725 | (±0.00128) | 8 (1) | −0.414* | 0.714 | 1.000 |
| HK | 24 | 2.891 | 0.692 | (±0.065) | 0.00269 | (±0.00061) | 5 (1) | 13 | 9.614 | 0.834 | (±0.044) | 0.02363 | (±0.00199) | 12 (5) | −0.177 | 0.855 | 1.000 |
| MED | 40 | 3.000 | 0.314 | (±0.091) | 0.00226 | (±0.00073) | 4 (1) | 39 | 5.377 | 0.554 | (±0.058) | 0.01833 | (±0.00176) | 7 (1) | 0.494* | 0.554 | 0.282 |
| ATL | 128 | 9.419 | 0.759 | (±0.034) | 0.01373 | (±0.00098) | 17 (12) | 124 | 17.60 | 0.852 | (±0.015) | 0.03269 | (±0.00089) | 34 (20) | 0.155* | 0.858 | 0.726 |
| PAC | 157 | 4.544 | 0.768 | (±0.011) | 0.01380 | (±0.00076) | 8 (3) | 132 | 13.55 | 0.803 | (±0.016) | 0.03200 | (±0.00078) | 27 (10) | −0.067* | 0.809 | 0.864 |
| IND | 43 | 3.930 | 0.717 | (±0.038) | 0.01566 | (±0.00085) | 5 | 31 | 21.00 | 0.883 | (±0.027) | 0.03683 | (±0.00103) | 22 (8) | 0.013 | 0.883 | 0.871 |
| Total | 368 | 8.124 | 0.810 | (±0.010) | 0.01348 | (±0.00057) | 22 | 315 | 16.32 | 0.820 | (±0.012) | 0.03214 | (±0.00059) | 61 | 0.098* | 0.824 | 0.743 |
Number of individuals analyzed per population (N). Allelic richnesstandarized across populations (r), Gene (Hd) and nucleotidic (π) diversity, and their corresponding standard deviations in brackets. Number of alleles per population (Nh), with private alleles shown in brackets. Inbreeding coefficient (Fis) for ANT. Asterisks represent significant coefficients at P<0.05. Hexp represents the expected heterozygosity and Hobs represents the observed heterozygosity.
Jost's adjusted estimator (Dest) was used to assess the allelic differentiation between populations for each marker, showing high values of differentiation (mean Dest = 0.660). The COI data revealed high differentiation between many population-pairs, as 88 comparisons out of 136 resulted in significant differences after correction for multiple comparisons (Table 3). For instance, the North Carolina population had no alleles in common with any other population (Fig. 2), and many other populations (e.g. Port Elizabeth, Manly, Misaki, Okinawajima) also differed considerably in their allele composition. No particular pattern was found for the only population collected from natural substratum (Sakushima Island, SKS), which was significantly different from half of the remaining populations.
Table 3. Jost's Dest population differentiation statistic between populations of Styela plicata for the COI (upper diagonal) and ANT (lower diagonal) markers.
| AR | JA | SP | FE | TEN | KNY | PE | NC | SC | CAL | BRA | AM | WAK | OKI | MIS | SKS | HK |
| 0.067 | 0.753 | 0.483 | 0.948 | 0.366 | 0.938 | 1 | 0.973 | 0.521 | 0.951 | 1 | 0.844 | 0.832 | 0.997 | 0.162 | 0.442 | |
| 0 | 0.76 | 0.452 | 1 | 0.299 | 0.944 | 1 | 1 | 0.481 | 1 | 1 | 0.888 | 0.841 | 1 | 0.132 | 0.439 | |
| 0.036 | 0.082 | 0.114 | 0.381 | 0.219 | 0.504 | 1 | 0.52 | 0.086 | 0.129 | 0.575 | 0.269 | 0.841 | 0.502 | 0.458 | 0.177 | |
| 0.032 | 0.129 | 0.015 | 0.45 | 0.05 | 0.767 | 1 | 0.241 | 0.184 | 0.246 | 0.835 | 0.311 | 0.804 | 0.793 | 0.185 | 0.032 | |
| 0.49 | 0.49 | 0.346 | 0.486 | 0.351 | 0.942 | 1 | 0.506 | 0.702 | 0.091 | 1 | 0.135 | 0.842 | 0.952 | 0.666 | 0.303 | |
| 0.281 | 0.289 | 0.116 | 0.258 | 0.058 | 0.923 | 1 | 0.557 | 0.325 | 0.29 | 0.997 | 0.238 | 0.774 | 0.965 | 0.101 | −0.03 | |
| 0.522 | 0.567 | 0.342 | 0.391 | 0.318 | 0.138 | 1 | 0.969 | 0.312 | 0.656 | 0.003 | 0.941 | 0.981 | 0.015 | 0.93 | 0.925 | |
| 0.978 | 0.99 | 0.992 | 0.945 | 0.897 | 0.832 | 0.715 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 0.098 | 0.157 | 0 | 0.042 | 0.213 | 0.086 | 0.231 | 0.978 | 0.771 | 0.333 | 1 | 0.409 | 0.771 | 0.946 | 0.732 | 0.493 | |
| 0.35 | 0.358 | 0.176 | 0.314 | 0 | 0 | 0.134 | 0.923 | 0.07 | 0.44 | 0.386 | 0.6 | 0.857 | 0.316 | 0.338 | 0.338 | |
| 0.269 | 0.267 | 0.125 | 0.273 | 0.018 | 0 | 0.274 | 0.97 | 0.074 | 0 | 0.716 | 0.105 | 0.842 | 0.662 | 0.639 | 0.229 | |
| 0.134 | 0.113 | 0.099 | 0.189 | 0.461 | 0.319 | 0.509 | 1 | 0.09 | 0.297 | 0.284 | 1 | 1 | 0.027 | 1 | 0.994 | |
| 0.538 | 0.543 | 0.353 | 0.51 | 0 | 0.06 | 0.3 | 0.966 | 0.212 | 0 | 0.02 | 0.482 | 0.432 | 0.876 | 0.425 | 0.128 | |
| 0.261 | 0.273 | 0.161 | 0.274 | 0.084 | 0.015 | 0.117 | 0.875 | 0.106 | 0 | 0.025 | 0.321 | 0.142 | 0.798 | 0.503 | 0.637 | |
| 0.479 | 0.499 | 0.315 | 0.457 | 0 | 0.066 | 0.281 | 0.95 | 0.157 | 0 | 0.025 | 0.427 | 0 | 0.107 | 0.925 | 0.935 | |
| 0.22 | 0.21 | 0.128 | 0.259 | 0.051 | 0.001 | 0.273 | 0.937 | 0.082 | 0 | 0 | 0.248 | 0.093 | 0 | 0.071 | 0.101 | |
| 0.525 | 0.636 | 0.585 | 0.388 | 0.826 | 0.754 | 0.789 | 0.94 | 0.604 | 0.759 | 0.758 | 0.718 | 0.822 | 0.774 | 0.777 | 0.761 |
Values in bold represent significant comparisons after FDR correction (see text).
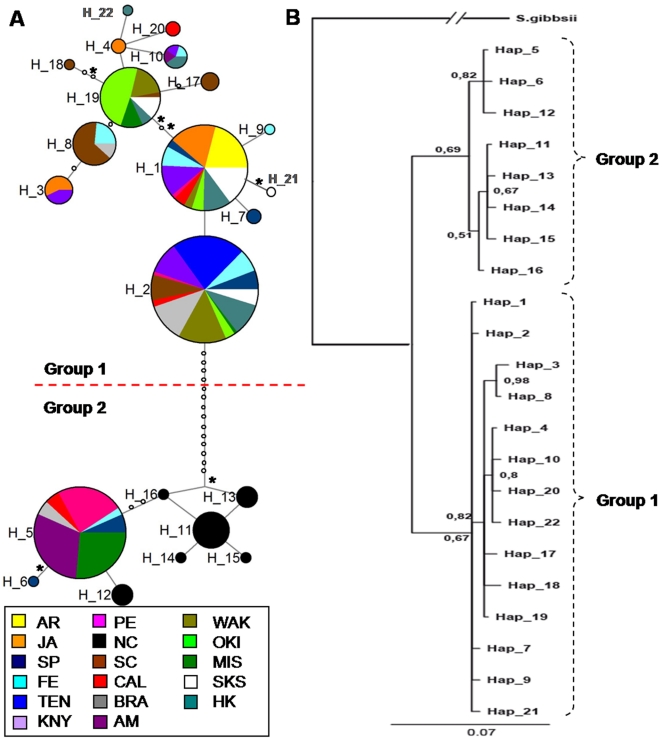
Figure 2. Network and phylogeny for COI.
a) Median-joining haplotype network for Styela plicata using COI results. Area of circles is proportional to the number of individuals found for each haplotype. Partitions inside the circles represent the proportion of each population within each haplotype. Small circles represent missing haplotypes. Lines between circles represent one mutational step and non-synonymous substitutions are indicated with an asterisk; b) Phylogeny of partial COI gene sequences using Bayesian inference. The congeneric species Styela gibbsii was used as an outgroup. Posterior probabilities are indicated when >0.5.
The results of the hierarchical AMOVA showed higher within population variability (58.41%) than the one between populations (41.59%, P<0.001, Table 4). AMOVA analyses performed by grouping populations according to their oceanic basin revealed that most of the genetic diversity was due to variability within populations (56.97%, P<0.001), and among populations within basins (34.36%, P<0.001). However, no significant differences in genetic structure were detected between basins (8.67%, P = 0.055 for COI; Table 4). Accordingly, the Mantel test showed no correlation between genetic differentiation and geographical distance between populations (r = 0.00009, P = 0.434).
Table 4. Analysis of the molecular variance (AMOVA) for the COI and ANT genetic markers.
| Source of variation | df | Sum of squares | Variance components | Variation (%) | P value | Fixation indices |
| a) COI | ||||||
| AMOVA without groups | ||||||
| Among populations without groups | 16 | 63.536 | 0.17255 Va | 41.59* | 0.000 | FST: 0.41589 |
| Within populations | 351 | 85.064 | 0.24235 Vb | 58.41 | ||
| Total | 367 | 148.601 | 0.4149 | |||
| AMOVA between basins | ||||||
| Among groups | 3 | 19.279 | 0.03690 Va | 8.67 | 0.055 | FCT : 0.08673 |
| Among populations within groups | 13 | 44.257 | 0.14618 Vb | 34.36* | 0.000 | FSC : 0.37624 |
| Within populations | 351 | 85.064 | 0.24235 Vc | 56.97* | 0.000 | FST : 0.43034 |
| Total | 367 | 148.601 | 0.42543 | |||
| b) ANT | ||||||
| AMOVA without groups | ||||||
| Among populations without groups | 16 | 28.988 | 0.03892 Va | 9.40* | 0.000 | FST: 0.09397 |
| Within populations | 613 | 230.022 | 0.37524 Vb | 90.6 | ||
| Total | 629 | 259.01 | 0.41416 | |||
| AMOVA between basins | ||||||
| Among groups | 3 | 7.806 | 0.00670 Va | 1.61 | 0.127 | FCT : 0.01610 |
| Among populations within groups | 13 | 21.182 | 0.03412 Vb | 8.20* | 0.000 | FSC : 0.08336 |
| Within populations | 613 | 230.022 | 0.37524 Vc | 90.19* | 0.000 | FST : 0.09812 |
| Total | 629 | 259.01 | 0.41606 |
Analyses are presented for the total of populations without grouping, and pooling populations from the same oceanic basin together (Mediterranean, Atlantic, Pacific and Indian). Va, Vb and Vc are the associated covariance components. FSC, FST and FCT are the F-statistics.
Overall, neutrality tests were not significant (Table 5), and hence did not support any lack of equilibrium due to selection or population size changes at any level (either partitioned by populations or oceanic basins). The only exceptions encountered were for the Australian population of Manly (AM), with significantly negative Tajima's D values, and for Sakushima and the Group 1 of haplotypes (see below), with a significant raggedness index (Table 5).
Table 5. Demographic parameters of S. plicata populations for each genetic marker (COI and ANT), calculated for each population and samples grouped by basin and by group (1 and 2 for COI, and A and B for ANT).
| COI | ANT | |||||||
| D | Fs | R2 | r | D | Fs | R2 | r | |
| AR | −1.16439 | −0.879 | 0.218 | 0.650 | 1.29064 | 2.347 | 0.169 | 0.243 |
| JA | 0.74648 | 3.941 | 0.173 | 0.462 | 0.25898 | 2.715 | 0.126 | 0.345 |
| SP | 2.15635 | 6.162 | 0.229 | 0.103 | 0.59380 | 0.232 | 0.143 | 0.077 |
| FE | −0.83585 | 3.033 | 0.104 | 0.112 | 1.04251 | −0.535 | 0.170 | 0.032 |
| TEN | 0.00000 | 0.000 | 0.000 | 0.000 | 2.32335 | 5.011 | 0.187 | 0.190 |
| KNY | −0.27356 | 2.391 | 0.123 | 0.149 | 2.15146 | 1.718 | 0.196 | 0.068 |
| PE | −1.29958 | 5.371 | 0.090 | 0.658 | 1.83362 | −4.076* | 0.197 | 0.021 |
| NC | −0.14467 | 0.419 | 0.124 | 0.127 | −0.15150 | −0.920 | 0.112 | 0.044 |
| SC | 0.52180 | 2.497 | 0.153 | 0.348 | 0.63874 | −0.198 | 0.141 | 0.140 |
| CAL | 1.81929 | 6.420 | 0.239 | 0.155 | 2.46514 | 5.670 | 0.229 | 0.119 |
| BRA | 0.55113 | 9.699 | 0.164 | 0.483 | 0.94915 | 0.814 | 0.152 | 0.101 |
| AM | −2.53406** | 5.308 | 0.200 | 0.854 | 0.83652 | 2.602 | 0.149 | 0.366 |
| WAK | 1.64264 | 2.196 | 0.220 | 0.384 | 2.48268 | 0.904 | 0.201 | 0.066 |
| OKI | 0.64968 | 1.430 | 0.169 | 0.360 | 3.02590 | 6.494 | 0.235 | 0.215 |
| MIS | 0.82576 | 10.821 | 0.163 | 0.578 | 1.06354 | 1.146 | 0.152 | 0.150 |
| SKS | 0.05885 | 0.400 | 0.136 | 0.043* | 3.17433 | 7.094 | 0.226 | 0.244 |
| HK | 0.13328 | 0.478 | 0.137 | 0.069 | 0.50405 | −0.338 | 0.141 | 0.046 |
| MED | −0.71549 | 1.657 | 0.087 | 0.482 | 1.01299 | 2.380 | 0.139 | 0.286 |
| ATL | 1.10126 | 3.816 | 0.125 | 0.109 | 1.02151 | −7.404* | 0.114 | 0.046 |
| PAC | 2.66373 | 15.635 | 0.172 | 0.103 | 1.72095 | −2.885 | 0.136 | 0.081 |
| IND | 2.31343 | −0.246 | 0.108 | 0.033 | 2.44640 | −1.956 | 0.190 | 0.029 |
| Group 1(A) | −0.84647 | −2.032 | 0.054 | 0.024* | −0.04229 | −11.460** | 0.083 | 0.066 |
| Group 2(B) | −0.53974 | −0.488 | 0.075 | 0.360 | −0.29695 | −6.598 | 0.067 | 0.140 |
Asterisks represent significant results:
*P<0.05;
**P<0.002.
Tajima's D, Fu's Fs statistic, Ramos-Onsins & Rozas's statistic (R2), and the raggedness index (r).
The network obtained for the COI gene (Fig. 2a) revealed two divergent lineages (hereafter called Group 1 and Group 2) separated by 15 mutational steps and without any intermediate haplotype in between. McDonald-Kreitman (MK) test of neutrality showed that there were no differences between proportions of silent and replacement sites within and between these two groups (P = 0.64). Sequences from both Group 1 and 2 are found in all basins and coexist in most populations; except for the absence of Group 2 in the Mediterranean. Judging by their high frequency, wide geographical distribution, and central position in the network, H_2 may be the ancestral haplotype of Group I. No clear result was obtained for group 2, as the most abundant haplotype (H_5) occupied a distal position within the group. (Fig. 2a). The BI tree reconstructed with COI haplotypes showed two moderately supported clades exhibiting 3.27% sequence divergence among them (Fig. 2b). These two clades matched exactly with Group 1 and 2 described for the COI network (Fig. 2a). Haplotype H_2 (inferred as ancestral) held a basal position within Group 1, while no evidence for a basal haplotype or group of haplotypes was found for Group 2.
Nuclear gene
For the ANT gene, we obtained 315 sequences of 220 bp. The ANT fragment targeted here includes an intron in many metazoans [63]. However, in our case, all sequences could be translated to amino acids and final sequence length was in accordance with what has been found for species without an intron in this position [63]. Our resulting dataset contained 80 homozygotes, which allowed a reliable reconstruction of the gametic phase of the heterozygotes (>95% confidence). No evidence was detected for recombination within our sequences. In total we obtained 61 alleles (Tables S2 and S3), 34 in the Atlantic (20 of which were exclusive to this basin) and 27 in the Pacific (Table 2). A deletion of 22 amino acids was found in 5 alleles (Table S2). Once more, the Mediterranean showed the lowest number of alleles (7, of which only one was private). Mean and total haplotype diversity (Hd) were 0.761 (±0.011 SD) and 0.820 (±0.012 SD), respectively. Mean nucleotide diversity was 0.0295 (±0.008 SD), while total nucleotide diversity (π) was 0.0321 (±0.0006 SD). Gene and nucleotide diversity did not differ between basins, except for the Mediterranean (Table 2). The South African populations of Knysna (KNY) and Port Elizabeth (PE) showed the highest values for genetic diversity, followed by most Pacific populations and some Atlantic ones (Table 2). Port Elizabeth (PE) was also the population showing the highest allelic richness (14.830) followed by Hong Kong (HK, 9.614), North Carolina (NC, 8.927) and Knysna (KNY, 8.145). As found for the mitochondrial gene, the lowest value of allelic richness corresponded to Manly (AM, 3.140). Low values were also retrieved for the Mediterranean populations of Javea (JA, 3.307) and Arenys de Mar (AR, 3.733). Comparisons between basins indicated that the Indian Ocean had the highest allelic richness, while the Mediterranean had the lowest (Table 2). Eight populations had less heterozygotes than expected, five of which (Arenys de Mar, Javea, San Fernando, Ferrol and North Carolina) deviated significantly from Hardy-Weinberg equilibrium (significant Fis values). Interestingly, 9 populations had an excess of heterozygotes (and negative Fis), and in 4 of them (Tenerife, Brasil, Misaki, Sakushima) these inbreeding coefficients were significant. Per basins, there was a heterozygote deficit in all populations except for the Pacific, and this deficit was most marked for the Mediterranean group of populations (0.282 Hobs vs. 0.554 Hexp).
Jost's adjusted estimator showed lower values of differentiation for the nuclear intron ANT (mean Dest = 0.324) than for the mitochondrial COI. Dest values obtained for the ANT gene revealed fewer significant differences in pair-wise comparisons (45 out of 136). As before, the North Carolina population was significantly different from all the others (Table 3). Interestingly, the Sakushima population (on natural substratum) only differed from the North Carolina and Hong Kong populations.
The hierarchical AMOVA analyses showed that most of the observed variability was found within populations (90.6%), and only a small but significant 9.4% (p<0.001) of variability was found among these populations (Table 4). When grouping populations according to their oceanic basins, AMOVA analyses' results were similar to those found for the mitochondrial marker. Most of the genetic diversity was due to variability within populations (90.19%, P<0.001), and among populations within basins (8.20%, P<0.001). No significant differences in genetic structure were detected between basins (1.61%, P = 0.127 ; Table 4). As found for COI, the Mantel test showed no correlation between genetic differentiation and geographical distance between populations (r = 0.000001, P = 0.243). Regarding the neutrality test, the same trend of COI was observed for ANT, with most tests being non-significant. However, Fu's Fs were significant for the Atlantic Ocean and the Port Elizabeth population (Table 5).
Network analyses showed a considerable amount of loops that were unambiguously resolved following coalescent rules (Fig. 3a). None of these loops affected the main structures shown in the network. However, the relationship among alleles should be considered with caution and no clear ancestral allele could be reliably designated. Although less divergent than with the COI data, the ANT network also showed a distinction in two groups of sequences separated by 4 mutational steps (Fig. 3a). None of these four mutations corresponded to non-synonymous changes. Finally, the 22 amino acids deletion found in 5 alleles (H_4, H_14, H_39, H_43, H_50) was also retrieved (represented by a dot line in Fig. 3a). McDonald-Kreitman neutrality tests could not be performed between these groups, as there was no fixed difference between them. BI analysis showed that one of the groups (hereafter called Group A) occupied a basal position within the resulting tree, while a second group (Group B) formed a monophyletic, derived clade supported by a posterior probability of 1 (Fig. 3b). Within group B, the five alleles with a 22 amino acid deletion also formed a monophyletic clade (posterior probability = 1; Fig. 3b). When the sequence fragment corresponding to the deletion was removed from the analyses, these 5 alleles still grouped together, indicating that their phylogenetic relationship was independent from the indel presence. The alleles containing the deletion were found in all studied basins, not showing any apparent geographic pattern (Table S2, Figure 3a).
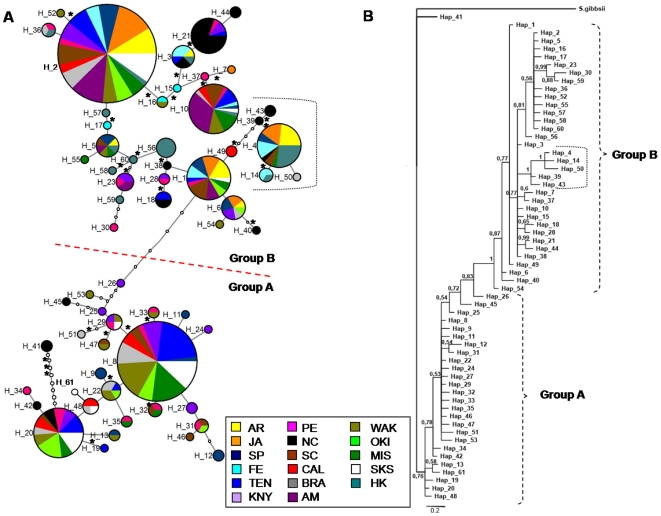
Figure 3. Network and phylogeny for ANT.
a) Median-joining allele network for Styela plicata using ANT results. Area of circles is proportional to the number of individuals found for each allele. Partitions inside the circles represent the proportion of each population within each allele. Small circles represent missing alleles. Lines between circles represent one mutational step and non-synonymous substitutions are indicated with an asterisk; b) Phylogeny of partial ANT gene sequences using Bayesian inference. The congeneric species Styela gibbsii was used as an outgroup. Posterior probabilities are indicated when >0.5. The dot line mark the clade corresponding to sequences with a 22 amino acid deletion.
The private allele H_41 from North Carolina appeared genetically distinct from all the others in both the network and the BI analyses (Fig. 3a). This sample was re-extracted and sequenced de novo, but the same resulting sequence was obtained. The Mediterranean populations only presented alleles from Group A of ANT, while the remaining populations presented alleles from both groups (especially, those populations from the Pacific Ocean). This pattern explains the lower genetic diversity found in the Mediterranean basin compared with that of the other oceans. Group B seems to be a highly successful derived clade that has spread in most populations. Interestingly, in all localities in which there was an excess of heterozygotes (negative Fis), there was also a higher than expected proportion of individuals having one allele of each group (A or B; 0.75 observed vs. 0.49 expected frequency). This is especially noteworthy in the Pacific populations, where we found twice the number of “mixed” genotypes than expected. The only exception was for North Carolina, which had a significant deficit of heterozygotes and less than expected genotypes with an allele from each group.
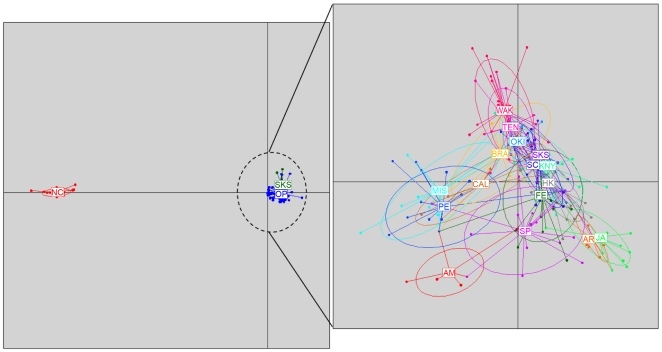
Finally, DAPC analyses were performed combining results obtained for COI and ANT. In order to avoid cluttering of populations, a first DAPC was performed with 3 groups: the North Carolina population (significantly different from the rest in previous analyses), the Sakushima population (the only natural substratum population) and the remaining populations. The PCA components retained explained 98.6% of the total variance observed. The scatterplot of the first two components of the DA (Fig. 4) showed that the first axis separates North Carolina from the rest, which form a tight cluster, while the second axis slightly sets apart the Sakushima population, although with a clear overlap of the inertia ellipses. We then repeated the analysis removing the North Carolina population and considering all populations as separate groups. 99.2% of the total variance was explained by the retained components of the PCA. The populations appeared mixed in the space of the first two axes of the discriminant analysis (Fig. 4), although the first axis separated slightly Misaki, Port Elizabeth and Manly on one extreme, and the two Mediterranean populations at the other end. The rest of the populations clustered tightly together, with the natural substratum population (Sakushima) appearing in a central position.
Figure 4. Discriminant analysis of principal components (DAPC).
Left: plot of the first two principal components obtained in the DAPC analysis considering three groups: the North Carolina population (NC), the Sakushima Island population (SKS) and other populations (OP). Right: plot of the DAPC results analyzing all populations as individual groups, except North Carolina, which was not analyzed (see text). Population codes as in Table 1. Labels are placed at the centre of dispersion for each group, further delineated by inertia ellipses. Dots represent individuals.
Discussion
Several remarkable features emerged from the recovered distribution of the genetic variability. First, there is a divergence in lineages for both markers, each featuring two groups of sequences. Second, the genetic pool is well mixed at the basin level, with little or no phylogeographic signal remaining. Third, many population pairs are genetically different, regardless of the geographic distance among them. Finally, there seems to be an effect of selection on the genetic makeup of this species, as illustrated by the highly divergent population of North Carolina and the intra-individual distribution of both groups of ANT sequences.
The most parsimonious explanation for the presence of two groups of sequences for COI (group 1 and 2) and ANT (group A and B) is that they have arisen concomitantly in a past fragmentation event within the native area of the species. We cannot, however, exclude an independent origin of these genetic splits. At present, the distribution of the groups obtained with the two markers is totally unrelated. Sequences of the Group A for ANT were found in ascidians having mitochondrial sequences of both lineages (Groups 1 and 2), and in direct proportion to their relative abundances. The same trend was observed for individuals having sequences of Group B for ANT (Table S3). If the differentiation of ANT and COI in different lineages occurred simultaneously in allopatric regions, the link between these markers was lost long ago. Mitochondrial genes are inherited maternally, while nuclear genes can be shuffled repeatedly through sexual reproduction. Thus, the lack of congruence found in the distribution of both markers could be due to frequent contact between individuals from different lineages coupled with genetic drift. A greater sensitivity of mitochondrial genes to genetic drift has been previously reported [88], and may explain the differences observed between mitochondrial and nuclear markers [e.g. 88]–[90]. In addition, no geographic pattern was observed in the distributions of the lineages observed for both markers. Even in the putative native area of S. plicata (NW Pacific), we found sequences of the two groups of COI and ANT in the same populations and, for ANT, even in the same individual.
Barros et al. [54] found nine COI haplotypes for Styela plicata, 8 belonging to our Group 1 and one to our Group 2. Based on this divergent haplotype, these authors suggested that there could be a cryptic species within what is known as Styela plicata. Our results did not lend support to this hypothesis, as the nuclear marker showed a distribution unrelated to these two groups of mitochondrial sequences. Furthermore, when comparing our mitochondrial sequences with other species of the genus, the resulting genetic divergence was much higher than that found between our two COI groups (3.27% between our groups, 21.12% between S. plicata and S. gibbsii; 22.7% with S. clava, and 20% with S. montereyensis). The divergent sequences of S. plicata reported from Australia (Lake Conjola) by Pérez-Portela et al. [107] (GenBank accession numbers FJ528633-34 for COI and FH897323 for 18S rRNA) were likely the result of sample mislabelling (Pérez-Portela, pers. comm.). We sequenced 4 further specimens from the same locality and verified that they all had typical S. plicata COI sequences (i.e., Haplotype 5).
Although the native range of Styela plicata is not known with certainty, the prevailing hypothesis is that it comes from the NW Pacific area [36], [54]. S. plicata would have then dispersed to other tropical and warm-water regions by ship fouling, likely since the early transoceanic navigation times [36]. Our results indicated that at present the genetic pool of S. plicata is well mixed among basins, with most genetic variability found within populations. Moreover, high genetic variability and the putatively most ancient alleles have not only been found in the NW Pacific populations (e.g. Sakushima, Hong Kong) but also in other oceanic basins (e.g. North East Pacific, Atlantic and Indian Ocean; see also David et al. [91]). Thus, we could not find any clear genetic signal in favour (or against) the hypothesis on the NW Pacific origin of this species. The only potential trend observed in our data was for the Mediterranean basin. The Mediterranean populations presented the lowest values for all diversity indexes, and only displayed group 1 for COI and group B for ANT. However, these findings should be interpreted with caution, as only two Mediterranean localities were included in this study. Lack of resolution for assessing native areas was also found in studies with other ascidian species that are believed to be ancient colonizers (e.g. Ciona intestinalis [38]). On the other hand, species that have spread more recently still have a genetic signature of their introduction history (e.g. Botryllus schlossei [41], [42], Microcosmus squamiger [92], Styela clava [45]).
Long-distance dispersal of introduced marine species across oceans probably occurs via major shipping routes while further spread at a local scale may take place through local traffic and recreational boating [13], [34], [42], [91], [93]. Our results indicate that many populations of S. plicata are well differentiated from others in terms of allele frequencies. This observation is in agreement with results obtained for other ascidians inhabiting harbours and marinas [37], [41], [44], [ but see 38 for an exception]. As expected when anthropogenic transport is the vector of dispersal, genetic differentiation among S. plicata populations was unrelated to geographic distance. Some distant populations (e.g. Hong Kong and Ferrol) were genetically similar, while closer populations such as Knysna and Port Elizabeth (South Africa) were significantly divergent. The stochasticity of main transport events through international ship traffic could determine the observed patterns among basins. However, our sampling design was inappropriate to assess the degree of connectivity among closely located populations (i.e. post-border dispersion, [34]). Thus, it still remains necessary to evaluate the role of small-scale processes in colonization dynamics, and to assess the importance of recreational boating in spreading introduced species.
Low genetic diversity caused by a founder effect or a bottleneck is not always the benchmark for introductory events [28], [94], [95]. In fact, recurrent introductions typically lead to highly diverse populations, especially if they receive migrants from native populations that are genetically structured [26], [30], [44], [96], [97]. Here, we found that genetic diversity indexes varied according to the studied population, with overall values ranging from moderate to high for both markers. Some exceptions were these populations where only one or two mitochondrial haplotypes were present (i.e. Arenys de Mar, Tenerife, Manly).
Besides recurrent introductions through ship transport, population differentiation could also be due to selection. Here, we found uneven abundances for each major group obtained for COI (Group 1 and 2) and ANT (Group A and B). For COI, haplotypes from Group 1 were considerably more frequent and diverse than haplotypes from Group 2. It is possible that these groups stand for differential adaptive capabilities of the individuals to stressful environments. This adaptive capability does not need to be directly linked to our studied gene (non-significant McDonald-Kreitman test), but to other mitochondrial genes. Differential adaptation to environmental factors (e.g., temperature, salinity) of mitochondrial sequences within one species is not a rare phenomenon, and has been described in many species [98]–[104].
For the ANT gene, selection may be favouring heterozygotes that have an allele of each group (A and B). In fact, the excess of heterozygotes found in most populations is due to the number of individuals with an allele each of A and B. Accordingly, the number of individuals with both alleles from the same group (A or B) was lower than expected. Homozygotes for the basal Group A occurred ca. 5 times less than expected based on allele frequencies. Thus, it is possible that populations that originally had only one group of ANT sequences were seeded with arriving individuals featuring the other group. The mingling of both groups may have favoured the heterozygotes with an allele from each group, and if this combination had an adaptive value, enhanced the fitness of those individuals. As for the COI lineages, this new adaptive capability to the environment is not necessarily linked to the ANT gene itself. Admixture between lineages can foster the emergence of novel genetic combinations with different physiological attributes and invasive characteristics [30]. In contrast to our results, solitary ascidians inhabiting artificial structures usually have a general deficit of heterozygotes [38], [44], [105].
Early invasions should not be considered “naturalized,” rather, their impacts, potential for further spread, and degree of integration in local processes and interactions should be assessed. A throughout knowledge of introduced species is required to understand and interpret the present-day structure, function, and conservation of marine communities [7], [35], [36]. Our genetic study of an ancient wanderer has uncovered signatures of deep divergences and recent mixing, with a phylogeographic signal mostly blurred. Current evolutionary processes may include adaptive changes and low and stochastic connectivity among established populations. More studies on S. plicata's biological cycle, interactions with other marine species, and local-scale genetic structure are necessary to understand the biology, ecology and post-border dispersal of this species and prevent ecosystem alterations.
Supporting Information
Haplotype frequencies observed for the COI gene. Numbers in bold are private haplotypes.
(DOC)
Allele frequencies observed for the ANT gene. Sequences with a 22 amino acid deletion are indicated with an asterisk.
(DOC)
ANT allelic phase and COI haplotypes for each individual analyzed.
(DOC)
Acknowledgments
We are grateful to R. Pérez-Portela, A. Villamor, J. Bishop, M. Rius, M. Lilly, P. Erwin, R. Da Rocha, T. Iseto, E. Hirose, M. Yoshida, Y. Saito, T. Nishikawa, S. M. Arellano, P.Y. Qian and P. Miranda for kindly providing samples for this study. A.R. Davis collected samples from Lake Conjola (Australia) to verify results from previous studies. We thank O. Wangensteen, N. Massana, A. Garcia, C. Palacín, C. Dalmau, E. Calahorro, E. Arias and C. Torres for assistance in the field. R. Pérez-Portela helped with the analyses. We would also like to thank L. Jost for useful comments on the D estimator. P. Erwin kindly reviewed the manuscript for English grammar.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was funded by the Marie Curie International Reintegration Grant FP7-PEOPLE-2010-RG 277038 (within the 7th European Community Framework Program), by the Spanish Government projects CTM2010-22218 and CTM2010-17755, and by an APIF fellowship of the University of Barcelona to MCP. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vermeij GJ. An agenda for invasion biology. Biol Conserv. 1996;78:3–9. [Google Scholar]
- 2.Cohen AN, Carlton JT. Accelerating invasion rate in a highly invaded estuary. Science. 1998;279:555–558. doi: 10.1126/science.279.5350.555. [DOI] [PubMed] [Google Scholar]
- 3.Mack MC, D'Antonio CM. Impacts of biological invasions on disturbance regimes. Trends Ecol Evol. 1998;13:195–198. doi: 10.1016/S0169-5347(97)01286-X. [DOI] [PubMed] [Google Scholar]
- 4.Mack RN, Simberloff D, Lonsdale WM, Evans H, Clout M, et al. Biotic invasions: causes, epidemiology, global consequences, and control. Ecol Appl. 2000;10:689–710. [Google Scholar]
- 5.Mooney HA, Cleland EE. The evolutionary impact of invasive species. Proc Natl Acad Sci USA. 2001;98:5446–5451. doi: 10.1073/pnas.091093398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crooks JA. Characterizing ecosystem-level consequences of biological invasions: the role of ecosystem engineers. Oikos. 2002;97:153–166. [Google Scholar]
- 7.Grosholz E. Ecological and evolutionary consequences of coastal invasions. Trends Ecol Evol. 2002;17:22–27. [Google Scholar]
- 8.Lafferty KD, Kuris AM. Biological control of marine pests. Ecology. 1996;77:1989–2000. [Google Scholar]
- 9.Bax N, Carlton JT, Mathews-Amos A, Haedrich RL, Howarth FG, et al. The control of biological invasions in the world's oceans. Conserv Biol. 2001;15:1234–1246. [Google Scholar]
- 10.Lodge DM, Williams S, MacIsaac HJ, Hayes KR, Leung B, et al. Biological invasions: Recommendations for US policy and management. Ecol Appl. 2006;16:2035–2054. doi: 10.1890/1051-0761(2006)016[2035:birfup]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Papaconstantinou C. The spreading of lessepsian fish migrants into the Aegean Sea Greece. Sci Mar. 1990;54:313–316. [Google Scholar]
- 12.Carlton JT, Geller JB. Ecological roulette - the global transport of nonindigenous marine organisms. Science. 1993;261:78–82. doi: 10.1126/science.261.5117.78. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz GM, Carlton JT, Grosholz ED, Hines AH. Global invasions of marine and estuarine habitats by non-indigenous species: Mechanisms, extent, and consequences. Am Zool. 1997;37:621–632. [Google Scholar]
- 14.Galil BS. A sea under siege - alien species in the Mediterranean. Biol Invasions. 2000;2:177–186. [Google Scholar]
- 15.Orensanz JM, Schwindt E, Pastorino G, Bortolus A, Casas G, et al. No longer the pristine confines of the world ocean: A survey of exotic marine species in the southwestern Atlantic. Biol Invasions. 2002;4:115–143. [Google Scholar]
- 16.Castilla JC, Guinez R, Caro AU, Ortiz V. Invasion of a rocky intertidal shore by the tunicate Pyura praeputialis in the Bay of Antofagasta, Chile. Proc Natl Acad Sci USA. 2004;101:8517–8524. doi: 10.1073/pnas.0401921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zenetos A. Trend in aliens species in the Mediterranean. An answer to Galil, 2009 〈〈Taking stock: inventory of alien species in the Mediterranean Sea〉〉. Biol Invasions. 2010;12:3379–3381. [Google Scholar]
- 18.Carlton JT. Mans Role in Changing the Face of the Ocean - Biological Invasions and Implications for Conservation of Near-Shore Environments. Conserv Biol. 1989;3:265–273. [Google Scholar]
- 19.Blakeslee AMH, McKenzie CH, Darling JA, Byers JE, Pringle JM, et al. A hitchhiker's guide to the Maritimes: anthropogenic transport facilitates long-distance dispersal of an invasive marine crab to Newfoundland. Divers Distrib. 2010;16:879–891. [Google Scholar]
- 20.Zibrowius H. Ongoing modification of the Mediterranean marine fauna and flora by the establishment of exotic species. Mésogée. 1991;51:83–107. [Google Scholar]
- 21.Glasby TM, Connell SD, Holloway MG, Hewitt CL. Nonindigenous biota on artificial structures: could habitat creation facilitate biological invasions? Mar Biol. 2007;151:887–895. [Google Scholar]
- 22.Tyrrell MC, Byers JE. Do artificial substrates favor nonindigenous fouling species over native species? J Exp Mar Biol Ecol. 2007;342:54–60. [Google Scholar]
- 23.Dafforn KA, Johnston EL, Glasby TM. Shallow moving structures promote marine invader dominance. Biofouling. 2009;25:277–287. doi: 10.1080/08927010802710618. [DOI] [PubMed] [Google Scholar]
- 24.Carman M, Hoagland K, Green-Beach E, Grunden D. Tunicate faunas of two North Atlantic-New England islands: Martha's Vineyard, Massachusetts and Block Island, Rhode Island. Aquatic Inv. 2009;4:65–70. [Google Scholar]
- 25.Bulleri F, Chapman MG. The introduction of coastal infrastructure as a driver of change in marine environments. J Appl Ecol. 2010;47:26–35. [Google Scholar]
- 26.Holland BS. Genetics of marine bioinvasions. Hydrobiologia. 2000;420:63–71. [Google Scholar]
- 27.Grosberg R, Cunningham CW. Genetic structure in the sea. From populations to communities. In: Bertness MD, Gaines SD, Hay ME, editors. Marine Community Ecology. Sinauer Associates, Inc., Sunderland, Massachusetts; 2001. pp. 61–84. [Google Scholar]
- 28.Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, et al. The population biology of invasive species. Annu Rev Ecol Syst. 2001;32:305–332. [Google Scholar]
- 29.Féral JP. How useful are the genetic markers in attempts to understand and manage marine biodiversity? J Exp Mar Biol Ecol. 2002;268:121–145. [Google Scholar]
- 30.Geller JB, Darling JA, Carlton JT. Genetic perspectives on marine biological invasions. Annu Rev Mar Sci. 2010;2:367–393. doi: 10.1146/annurev.marine.010908.163745. [DOI] [PubMed] [Google Scholar]
- 31.Govindarajan AF, Halanych KK, Cunningham CW. Mitochondrial evolution and phylogeography in the hydrozoan Obelia geniculata (Cnidaria). Mar Biol. 2005;146:213–222. [Google Scholar]
- 32.Darling JA, Blum MJ. DNA-based methods for monitoring invasive species: a review and prospectus. Biol Invasions. 2007;9:751–765. [Google Scholar]
- 33.Estoup A, Guillemaud T. Reconstructing routes of invasion using genetic data: why, how and so what? Mol Ecol. 2010;19:4113–4130. doi: 10.1111/j.1365-294X.2010.04773.x. [DOI] [PubMed] [Google Scholar]
- 34.Goldstien SJ, Schiel DR, Gemmell NJ. Regional connectivity and coastal expansion: differentiating pre-border and post-border vectors for the invasive tunicate Styela clava. Mol Ecol. 2010;19:874–885. doi: 10.1111/j.1365-294X.2010.04527.x. [DOI] [PubMed] [Google Scholar]
- 35.Carlton JT. Community assembly and historical biogeography in the North Atlantic Ocean: the potential role of human-mediated dispersal vectors. Hydrobiologia. 2003;503:1–8. [Google Scholar]
- 36.Carlton JT. Rilov G, Crooks JA, editors. Deep Invasion Ecology and the Assembly of Communities in Historical Time. Biological Invasions in Marine Ecosystems: Ecological, Management, and Geographic Perspectives: Ecological Studies. 2009. pp. 13–56.
- 37.Turon X, Tarjuelo I, Duran S, Pascual M. Characterising invasion processes with genetic data: an Atlantic clade of Clavelina lepadiformis (Ascidiacea) introduced into Mediterranean harbours. Hydrobiologia. 2003;503:29–35. [Google Scholar]
- 38.Zhan AB, MacIsaac HJ, Cristescu ME. Invasion genetics of the Ciona intestinalis species complex: from regional endemism to global homogeneity. Mol Ecol. 2010;19:4678–4694. doi: 10.1111/j.1365-294X.2010.04837.x. [DOI] [PubMed] [Google Scholar]
- 39.Lambert CC, Lambert G. Persistence and differential distribution of nonindigenous ascidians in harbors of the Southern California Bight. Mar Ecol Prog Ser. 2003;259:145–161. [Google Scholar]
- 40.Lambert G. Invasive sea squirts: A growing global problem. J Exp Mar Biol Ecol. 2007;342:3–4. [Google Scholar]
- 41.Lejeusne C, Bock DG, Therriault TW, MacIsaac HJ, Cristescu ME. Comparative phylogeography of two colonial ascidians reveals contrasting invasion histories in North America. Biol Invasions. 2011;13:635–650. [Google Scholar]
- 42.Lopez-Legentil S, Turon X, Planes S. Genetic structure of the star sea squirt, Botryllus schlosseri, introduced in southern European harbours. Mol Ecol. 2006;15:3957–3967. doi: 10.1111/j.1365-294X.2006.03087.x. [DOI] [PubMed] [Google Scholar]
- 43.Turon X, Nishikawa T, Rius M. Spread of Microcosmus squamiger (Ascidiacea : Pyuridae) in the Mediterranean Sea and adjacent waters. J Exp Mar Biol Ecol. 2007;342:185–188. [Google Scholar]
- 44.Dupont L, Viard F, Davis MH, Nishikawa T, Bishop JDD. Pathways of spread of the introduced ascidian Styela clava (Tunicata) in Northern Europe, as revealed by microsatellite markers. Biol Invasions. 2010;12:2707–2721. [Google Scholar]
- 45.Goldstien SJ, Dupont L, Viard F, Hallas PJ, Nishikawa T, et al. Global Phylogeography of the Widely Introduced North West Pacific Ascidian Styela clava. PLoS ONE. 2011;6(2):e16755. doi: 10.1371/journal.pone.0016755. doi: 10.1371/journal.pone.0016755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert G. A global overview of ascidian introductions and their possible impact on the endemic fauna. In: Sawada H, Yokosawa H, Lambert CC, editors. The Biology of Ascidians. Tokio: Springer-Verlag; 2001. pp. 249–257. [Google Scholar]
- 47.McDonald J. The invasive pest species Ciona intestinalis (Linnaeus, 1767) reported in a harbour in southern Western Australia. Mar Pollut Bull. 2004;49:868–870. doi: 10.1016/j.marpolbul.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 48.Ramsay A, Davidson J, Bourque D, Stryhn H. Recruitment patterns and population development of the invasive ascidian Ciona intestinalis in Prince Edward Island, Canada. Aquatic Inv. 2009;4:169–176. [Google Scholar]
- 49.Locke A, Hanson JM, MacNair NG, Smith AH. Rapid response to non-indigenous species. 2. Case studies of invasive tunicates in Prince Edward Island. Aquatic Inv. 2009;4:249–258. [Google Scholar]
- 50.Lejeusne C, Bock DG, Therriault TW, MacIsaac HJ, Cristescu ME. Comparative phylogeography of two colonial ascidians reveals contrasting invasion histories in North America. Biol Invasions. 2011;13:635–650. [Google Scholar]
- 51.Hewitt CL, Campbell ML, Thresher RE, Martin RB, Boyd S, et al. Introduced and cryptogenic species in Port Phillip Bay, Victoria, Australia. Mar Biol. 2004;144:183–202. [Google Scholar]
- 52.Carlton JT. Species invasions: Insights into ecology, evolution, and biogeography. Bio Science. 2006;56:694–695. [Google Scholar]
- 53.Abbott DP, Lambert CC, Lambert G, Newberry A, Carlton JT. Carlton JT, editor. Chordata: Ascidiacea. The Light & Smith manual: intertidal invertebrates from central California to Oregon. 2007. pp. 949–964. Fourth edition, completely revised and expanded.
- 54.Barros R, Rocha R, Pie M. Human-mediated global dispersion of Styela plicata (Tunicata, Ascidiacea). Aquatic Inv. 2009;4:45–57. [Google Scholar]
- 55.Van Name WG. The north and south american ascidians. B Am Mus Nat Hist. 1945;84:1–463. [Google Scholar]
- 56.Sims LL. Osmoregulatory Capabilities of 3 Macrosympatric Stolidobranch Ascidians, Styela clava Herdman, Styela plicata (Lesueur), and Styela montereyensis (Dall). J Exp Mar Biol Ecol. 1984;82:117–129. [Google Scholar]
- 57.Thiyagarajan V, Qian PY. Effect of temperature, salinity and delayed attachment on development of the solitary ascidian Styela plicata (Lesueur). J Exp Mar Biol Ecol. 2003;290:133–146. [Google Scholar]
- 58.Naranjo SA, Carballo JL, García-gómez JC. Effects of enviromental stress on ascidian populations in Algeciras Bay (southern Spain). Possible marine bioindicators? Mar Ecol Prog Ser. 1996;144:119–131. [Google Scholar]
- 59.Sabbadin A. Il ciclo biologico di Ciona intestinalis (L.), Molgula manhattensis (de Kay) e Styela plicata (Lesueur) nella laguna veneta. Arch Oceanogr Limnol. 1957;XI:1–28. [Google Scholar]
- 60.Yamaguchi M. Growth and reproductive-cycles of marine fouling ascidians Ciona intestinalis, Styela plicata, Botrylloides violaceus, and Leptoclinum mitsukurii at Aburatsubo-Moroiso Inlet (Central Japan). Mar Biol. 1975;29:253–259. [Google Scholar]
- 61.Sciscioli M, Lepaore E, Tursi A. Relationship between Styela plicata (Les.)(Tunicata) settlement and spawning. Mem Biol Mar Oceanogr. 1978;8:65–75. [Google Scholar]
- 62.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech. 1994;3:294–299. [PubMed] [Google Scholar]
- 63.Jarman SN, Ward RD, Elliott NG. Oligonucleotide primers for PCR amplification of coelomate introns. Mar Biotechnol. 2002;4:347–355. doi: 10.1007/s10126-002-0029-6. [DOI] [PubMed] [Google Scholar]
- 64.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 65.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 66.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 68.Martin DP, Lemey P, Lott M, Moulton V, Posada D, et al. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. 2004. Laboratire Génome, Populations, Interactions, CNRS UMR 5171. Université de Montpellier II, Montpellier (France)
- 70.Jost L. GST and its relatives do not measure differentiation. Mol Ecol. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- 71.Chao A, Shen T-J. SPADE (Species Prediction and Diversity Estimation). 2009. http://chao.stat.nthu.edu.tw/softwareCE.html.
- 72.Narum SR. Beyond Bonferroni: Less conservative analyses for conservation genetics. Conserv Genet. 2006;7:783–787. [Google Scholar]
- 73.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 74.Rousset F. Genetic Differentiation and estimation of gene flow from F-Statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics. 2010;11:94. doi: 10.1186/1471-2156-11-94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 77.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Mol Biol Evol. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- 80.Harpending HC. Signature of ancient population-growth in a low-resolution mitochondrial-DNA mismatch distribution. Hum Biol. 1994;66:591–600. [PubMed] [Google Scholar]
- 81.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 82.Templeton AR, Boerwinkle E, Sing CF. A cladistic-analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping.1. Basic theory and an analysis of alcohol-dehydrogenase activity in Drosophila. Genetics. 1987;117:343–351. doi: 10.1093/genetics/117.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Templeton AR, Sing CF. A cladistic-analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping.4. Nested analyses with cladogram uncertainty and recombination. Genetics. 1993;134:659–669. doi: 10.1093/genetics/134.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 85.Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 86.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 87.Mcdonald JH, Kreitman M. Neutral mutation hypothesis test - Reply. Nature. 1991;354:116. doi: 10.1038/354114e0. [DOI] [PubMed] [Google Scholar]
- 88.Shaw PW, Arkhipkin AI, Al Khairulla H. Genetic structuring of Patagonian toothfish populations in the Southwest Atlantic Ocean: the effect of the Antarctic Polar Front and deep-water troughs as barriers to genetic exchange. Mol Ecol. 2004;13:3293–3303. doi: 10.1111/j.1365-294X.2004.02327.x. [DOI] [PubMed] [Google Scholar]
- 89.Darling JA, Bagley MJ, Roman J, Tepolt CK, Geller JB. Genetic patterns across multiple introductions of the globally invasive crab genus Carcinus. Mol Ecol. 2008;17:4992–5007. doi: 10.1111/j.1365-294X.2008.03978.x. [DOI] [PubMed] [Google Scholar]
- 90.Drew JA, Allen GR, Erdmann MV. Congruence between mitochondrial genes and color morphs in a coral reef fish: population variability in the Indo-Pacific damselfish Chrysiptera rex (Snyder, 1909). Coral Reefs. 2010;29:439–444. [Google Scholar]
- 91.David GK, Marshall DJ, Riginos C. Latitudinal variability in spatial genetic structure in the invasive ascidian, Styela plicata. Mar Biol. 2010;157:1955–1965. [Google Scholar]
- 92.Rius M, Pascual M, Turon X. Phylogeography of the widespread marine invader Microcosmus squamiger (Ascidiacea) reveals high genetic diversity of introduced populations and non-independent colonizations. Divers Distrib. 2008;14:818–828. [Google Scholar]
- 93.Wasson K, Zabin CJ, Bedinger L, Diaz MC, Pearse JS. Biological invasions of estuaries without international shipping: the importance of intraregional transport. Biol Conserv. 2001;102:143–153. [Google Scholar]
- 94.Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dupont L, Viard F, David P, Bishop JDD. Combined effects of bottlenecks and selfing in populations of Corella eumyota, a recently introduced sea squirt in the English Channel. Divers Distrib. 2007;13:808–817. [Google Scholar]
- 96.Simon-Bouhet B, Garcia-Meunier P, Viard F. Multiple introductions promote range expansion of the mollusc Cyclope neritea (Nassariidae) in France: evidence from mitochondrial sequence data. Mol Ecol. 2006;15:1699–1711. doi: 10.1111/j.1365-294X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- 97.Roman J, Darling JA. Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol Evol. 2007;22:454–464. doi: 10.1016/j.tree.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 98.Bastrop R, Jurss K, Sturmbauer C. Cryptic species in a marine polychaete and their independent introduction from North America to Europe. Mol Biol Evol. 1998;15:97–103. doi: 10.1093/oxfordjournals.molbev.a025919. [DOI] [PubMed] [Google Scholar]
- 99.Gerber AS, Loggins R, Kumar S, Dowling TE. Does nonneutral evolution shape observed patterns of DNA variation in animal mitochondrial genomes? Annu Rev Genet. 2001;35:539–566. doi: 10.1146/annurev.genet.35.102401.091106. [DOI] [PubMed] [Google Scholar]
- 100.Schizas NV, Coull BC, Chandler GT, Quattro JM. Sympatry of distinct mitochondrial DNA lineages in a copepod inhabiting estuarine creeks in the southeastern USA. Mar Biol. 2002;140:585–594. [Google Scholar]
- 101.Rawson PD, Burton RS. Functional coadaptation between cytochrome c and cytochrome c oxidase within allopatric populations of a marine copepod. Proc Natl Acad Sci USA. 2002;99:12955–12958. doi: 10.1073/pnas.202335899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kelly DW, MacIsaac HJ, Heath DD. Vicariance and dispersal effects on phylogeographic structure and speciation in a widespread estuarine invertebrate. Evolution. 2006;60:257–267. [PubMed] [Google Scholar]
- 103.Roman J. Diluting the founder effect: cryptic invasions expand a marine invader's range. Proc R Soc B. 2006;273:2453–2459. doi: 10.1098/rspb.2006.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Folino-Rorem NC, Darling JA, D'Ausilio CA. Genetic analysis reveals multiple cryptic invasive species of the hydrozoan genus Cordylophora. Biol Invasions. 2009;11:1869–1882. [Google Scholar]
- 105.Dupont L, Viard F, Dowell MJ, Wood C, Bishop JDD. Fine- and regional-scale genetic structure of the exotic ascidian Styela clava (Tunicata) in southwest England, 50 years after its introduction. Mol Ecol. 2009;18:442–453. doi: 10.1111/j.1365-294X.2008.04045.x. [DOI] [PubMed] [Google Scholar]
- 106.Petit RJ, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conserv Biol. 1998;12:844–855. [Google Scholar]
- 107.Pérez-Portela R, Bishop JDD, Davis AR, Turon X. Phylogeny of the families Pyuridae and Styelidae (Stolidobrachiata, Ascidiacea) inferred from mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol. 2009;50:560–570. doi: 10.1016/j.ympev.2008.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Haplotype frequencies observed for the COI gene. Numbers in bold are private haplotypes.
(DOC)
Allele frequencies observed for the ANT gene. Sequences with a 22 amino acid deletion are indicated with an asterisk.
(DOC)
ANT allelic phase and COI haplotypes for each individual analyzed.
(DOC)