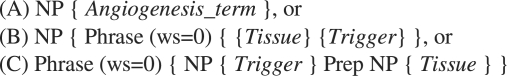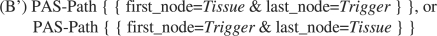Abstract
Motivation: Understanding key biological processes (bioprocesses) and their relationships with constituent biological entities and pharmaceutical agents is crucial for drug design and discovery. One way to harvest such information is searching the literature. However, bioprocesses are difficult to capture because they may occur in text in a variety of textual expressions. Moreover, a bioprocess is often composed of a series of bioevents, where a bioevent denotes changes to one or a group of cells involved in the bioprocess. Such bioevents are often used to refer to bioprocesses in text, which current techniques, relying solely on specialized lexicons, struggle to find.
Results: This article presents a range of methods for finding bioprocess terms and events. To facilitate the study, we built a gold standard corpus in which terms and events related to angiogenesis, a key biological process of the growth of new blood vessels, were annotated. Statistics of the annotated corpus revealed that over 36% of the text expressions that referred to angiogenesis appeared as events. The proposed methods respectively employed domain-specific vocabularies, a manually annotated corpus and unstructured domain-specific documents. Evaluation results showed that, while a supervised machine-learning model yielded the best precision, recall and F1 scores, the other methods achieved reasonable performance and less cost to develop.
Availability: The angiogenesis vocabularies, gold standard corpus, annotation guidelines and software described in this article are available at http://text0.mib.man.ac.uk/~mbassxw2/angiogenesis/
Contact: xinglong.wang@gmail.com
1 INTRODUCTION
1.1 Background and motivation
Biological processes (i.e. bioprocesses) occur in living organisms and the regulation of them is crucial to control and maintain the life cycles of the organisms. A bioprocess may consist of any number of chemical reactions or other types of biological events that may result in maintenance, changes or transformations of the organism. In drug discovery, it is important to understand bioprocesses and how they are regulated under normal conditions and dysregulated in disease. Regulation of bioprocesses may involve modulating their frequency, rate or extent, through the control of gene expression, protein modification or interaction with a protein, substrate molecule or larger structures. Scientists often need to gather facts that come from clear scientific evidence of how, when modulated, an existing or potential drug target affects critical pathophysiological processes leading to either the disease cure, prevention or amelioration of symptoms in the clinical setting. Typically, a bank of preclinical evidence is developed using cell lines, model organisms and clinical samples associating a target with key bioprocesses (and so disease phenotype). However, this process is very expensive and time consuming (Kola and Landis, 2004). To avoid unnecessary duplication of research, scientists must first review external activity in their area of interest in order to determine what questions remain unanswered, and to derive information to support or contest hypotheses. Laboratory resources may then be more efficiently directed to explore those questions. One important source of this information is published biomedical articles. However, given the vastness of the literature and an accelerating publication rate, manual techniques and conventional information retrieval techniques are unable to deliver timely, reliable, exhaustive and specific results. In addition, the scientific and publication process is not static in nature, but instead a continuous one.
Text mining technology has been increasingly popular to support knowledge discovery, hypothesis generation and to manage the mass of biological literature (Ananiadou et al., 2010; Hunter and Cohen, 2006), and text mining has shown promises for finding key biological entities (e.g. Smith et al., 2008), relationships among proteins (e.g. Krallinger et al., 2008a) and for establishing functional annotations (e.g. Alex et al., 2008).
As far as we know, there has been limited work in text mining to extract bioprocesses. One reason is due to its complexity. A bioprocess often involves a series of bioevents, where an event expresses a change of state of a cell or tissue, and such events are often used to refer to the bioprocess they participate in. In this article, we systematically investigate the extraction of bioprocess-related terms and events, including the definition of the task, the construction of a gold standard corpus for learning and evaluation, and the proposal of a number of approaches to identifying bioprocesses. We then compare the methods in terms of their performance results, as well as the amount of manual supervision required, as both of the factors are main considerations when deploying a new text mining system in practice.
Our work focuses on an exemplar biological process, angiogenesis. Angiogenesis is a key physiological process involving the growth of new blood vessels from pre-existing vessels, and it is vital in growth and development of tissues and organs. It is also a crucial step in the transition of tumours from a dormant state to a malignant one. Therefore, the identification of gene products involved in regulating angiogenesis, and pharmacological agents that have angiogenesis inhibitory effects, has been one of the main lines of research for treatment of solid tumours, and hence mining facts related to angiogenesis is a crucial step towards this goal.
1.2 Related Work
The recognition of specific biological processes in unstructured text has received relatively less attention in the biomedical text mining community. However, researchers have attempted to mine general bioprocess information from other knowledge sources such as ontology and biological data. For example, Hvidsten et al. (2003) proposed a systematic supervised learning model to predicting bioprocess by analysing microarray data. Their method benefited from the functional annotation of genes in the Gene Ontology (GO) (http://www.geneontology.org/GO.doc.shtml#biological_process). The method was evaluated on genes coding for proteins known to be involved in bioprocesses using cross-validation. Koike et al. (2004) reported work on finding relations between biological functions and genes and gene products. Their method first used a named entity recognition programme to annotate genes, gene products and biological function terms as defined in GO, and then extracted relations between the entities and biofunctions by analysing syntactic structures of the sentences. As noted by Koike et al., the terms for bioprocesses in GO were insufficient for automatic extraction in terms of recall. They experimented with a number of techniques to augment the functional terms, including mining-related terms using high co-occurrence counts, retrieving similar terms having similar collocations with GO terms, etc. However, neither of the approaches described above was able to find more complex bioprocess expressions such as events.
Recently, research has been conducted on the extraction of biomolecular events. In particular, the BioNLP 2009 shared task (Kim et al., 2009) attracted much attention and interesting solutions. The shared task provided annotated data for several types of gene-related bioevents, such as gene expression, transcription and regulation, and participants were asked to identify the event type and the word that triggers the event (i.e. trigger). The results of the shared task showed that event extraction was challenging: the best performing system achieved an overall F1-score of 51.95% (Björne, et al., 2009). After the shared tasks, researchers have proposed methods that further improved the state of the art. For example, Miwa et al. (2010) proposed a method that detects, in sequence, the event trigger and the edges linking the participants and the trigger, and then finds the best combination of the edges to form a complex event.
The BioNLP 2009 shared task did not include cellular or tissue bioevents, which play a central role in bioprocesses and are the focus of our work. The GENIA corpus (Kim et al., 2008), however, contains annotated examples of ‘cellular physiological process’, which is similar to our event definition. However, this type of event refers to a broad category and is not linked to any specific bioprocess, whereas we would like to extract events that closely relate to a given bioprocess such as angiogenesis. Section 2.1 provides more discussion on the similarity and difference between GENIA and our event definitions.
Also, GENIA style event annotation required both biological and linguistic expertise and a lengthy annotation process (Kim et al., 2008). Despite the fact that the GENIA annotation has been proven highly useful, people interested in mining bioevents in a new domain may hesitate to follow GENIA's strategy due to its high cost factor. This contributed to our effort in exploring methods taking into account not only performance scores, but also development cost.
2 METHODS
2.1 Task definition
Generally speaking, angiogenesis descriptions appear in text in one of the following two forms: angiogenesis terms and angiogenesis events, where a term refers to the name, synonym and other lexical variants of angiogenesis, and an event refers to a cellular or tissue bioevent or reaction, taking place as part of the bioprocess. For example, ‘angiogenesis’ and ‘angiogenic’ are angiogenesis terms, and phrase ‘capillary endothelial cell proliferation’, indicating a change of state of ‘capillary endothelial cell’, is an angiogenesis event. In order to examine how often angiogenesis bioprocess is expressed in the form of events, we created a corpus of 262 medline abstracts (http://www.nlm.nih.gov/databases/databases_medline.html), where angiogenesis terms and events, as well as several types of bioentities that were considered to closely relate to angiogenesis bioprocess, were manually annotated. Table 1 shows the annotation markables and their counts of occurrence in this corpus: 36.5% (479 out of 1313) of the angiogenesis mentions are events, which highlights the importance of event recognition for bioprocess extraction. The corpus was also used to develop supervised machine learning systems and for evaluation. Section 2.5 gives detail on the annotation and the learning systems.
Table 1.
Annotated markables and their counts in the angiogenesis corpus
| Markable | Counts |
|---|---|
| Angiogenesis term | 834 |
| Angiogenesis event | 479 |
| Gene or gene product | 2901 |
| Tissue | 2190 |
| Cell | 1065 |
The definition of angiogenesis event largely follows that of ‘cellular physiological process’, a type of event defined in GO and also in the GENIA ontology (Kim et al., 2006). However, our events are restricted to the domain of angiogenesis, and do not contain explicit links to their trigger words and participants. In GENIA definition (Kim et al., 2008), a participant is the bioentity that is involved in an event, and each event must attach to a trigger word denoting the action that caused the changes. For example, in GENIA annotation, phrase ‘capillary endothelial cell proliferation’ would have ‘proliferation’ marked as the trigger word and ‘capillary endothelial cell’ as the participant. In contrast, our annotation would regard the entire phrase as an angiogenesis event, where trigger words and participants would not be explicitly linked to the event. Section 2.5 elaborates the difference between our event annotation and the GENIA project, and Table 3 shows some illustrative examples. This decision significantly simplified the annotation process, and hence reduced the annotation cost. In summary, we define an angiogenesis bioevent as either:
the change of state of a cell, cell component or tissue that is specific to angiogenesis. (e.g. ‘vascular sprouting’) or
the change of state of some property of the entities mentioned above (e.g. ‘increase in vascular density’).
As mentioned in the above definition, the key entities taking part in angiogenesis are cell, cell components and tissues. For simplicity, they are referred to as tissues, tissue terms or tissue entities in the rest of the article. The term bioentity, on the other hand, is used to refer to genes, gene products and the tissue entities as defined above. Also, a trigger or trigger word is the word which denotes the action that caused the changes of state in an event.
Table 3.
Comparison of the annotations of GENIA's ‘cellular physiological process’ and the angiogenesis event
| Angiogenesis corpus | GENIA corpus | |
|---|---|---|
| 1 | ‘MEK5 signaling modulates endothelial cell migration and focal contact turnover.’ | ‘MEK5 signaling modulates endothelial cellmigration and focal contact turnover.’ |
| 2 | ‘… resulting in increased vascular proliferation but defective maturation.’ | ‘… resulting in increased vascular proliferation but defective maturation. |
Events are highlighted in bold font, entities are italicized and trigger words are underlined.
In our definition, the tissue terms are not restricted to nouns. In fact, our corpus contains many tissues composed of words of other parts of speech (POS), which would not be annotated as entities in other annotation projects, such as GENIA (Kim et al., 2008). This decision was made due to the observation that words of syntactic classes other than nouns (e.g. adjectives) were often as informative as nouns in indicating the presence of a bioentity. For example, ‘vascular’, ‘arterial’ and ‘microvascular’ are adjective tissue entities that frequently occur in our corpus denoting something related to, affecting, or consisting of blood vessels. We treated angiogenesis terms the same way. The following sections propose methods for identifying angiogenesis terms and events, and evaluation results.
2.2 Dictionary-based method
One solution to the extraction of angiogenesis terms and events is using vocabularies. There are, however, limited lexical resources available for biological process terms. While GO and MeSH (http://www.nlm.nih.gov/mesh/) contain branches for biological processes, the information provided for each specific process is very limited. For example, the angiogenesis term in GO (GO:0001525) is annotated with only one synonym: ‘blood vessel formation from pre-existing blood vessels’and its descendent terms also look like definitions, e.g. ‘angiogenesis involved in wound healing’ (GO:0060978). Such terms and synonyms are insufficient to help computer programmes mine angiogenesis terms and events, which highlighted the lack of ontological support for extracting bioprocesses from text. Therefore, we manually built three vocabularies: angiogenesis terms, tissues and triggers, where the first vocabulary contains 10 variants of the names and synonyms of angiogenesis, the second consists of 27 cell and tissue entities related to angiogenesis and the third contains 39 derivation forms of verbs that are good indicators of angiogenesis events. For example, ‘angiogenesis’ and ‘angiogenic’ are terms in the first vocabulary, ‘vascular’ and ‘endothelial cell’ appear in the second and ‘development’ and ‘proliferation’ are examples from the third. The vocabularies are flat lists and do not contain any hierarchical information, and a domain expert spent 40 h developing the vocabularies. We then designed patterns to extract angiogenesis terms and events using the terms in the vocabularies.
The patterns are shown in Figure 1, where NP denotes a noun phrase, Prep is a preposition, Phrase is a container enclosing any other query components and ws denotes the maximum distance (i.e. number of words) allowed between the components in a phrase. We applied pattern (A) to find angiogenesis terms, and based on the definition described in Section 2.1, patterns (B) and (C) were used for recognizing angiogenesis events. In more detail, pattern (B) finds noun phrases in which a tissue modifies a trigger next to it (i.e. ws = 0); and pattern (C) recognizes phrases where a tissue modifies a trigger as a preposition phrase and all components should occur adjacent to each other. We chose these two particular syntactic structures to model angiogenesis events based on domain experts' knowledge and observations. For example, according to pattern C, phrase ‘development of endothelial cell’ will be tagged as an angiogenesis event, because ‘development’ is a trigger word, ‘of’ a preposition and ‘endothelial cell’ is a tissue. The patterns were applied at sentence level, and before applying the patterns, the documents were pre-processed using the following natural language processing (NLP) steps: sentence splitting, tokenization, POS tagging and chunking, as described in Alex et al. (2008).
Fig. 1.
Patterns for finding angiogenesis terms and events.
2.3 Pattern matching with syntactic relation
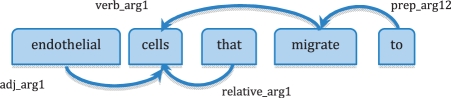
The patterns B and C defined in Section 2.2 are rather strict in that a trigger and a tissue must follow the designated word order to form an angiogenesis event. An angiogenesis event, however, can be expressed in many ways in text and the tissue and trigger may not follow a specific word order. For example, as shown in Figure 2, ‘endothelial cells that migrate to’ can be an angiogenesis event, where ‘endothelial cell’ is a tissue entity and ‘migrate’ a trigger. However, neither pattern B nor C would identify the event, because the trigger ‘migrate’ appears in a relative clause.
Fig. 2.
A parsed phrase in ENJU's predicate–argument representation.
One solution is to generalize patterns (B) and (C) in Figure 1 so that a syntactically related pair of tissue and trigger can be considered to be an angiogenesis event. We can employ a natural language parser to find syntactic relations between words. In our experiments, we used the ENJU HPSG parser (Miyao and Tsujii, 2008), which has been shown to yield good results on finding protein–protein interactions (Miyao et al., 2009) and species disambiguation (Wang et al., 2010), among other biomedical information extraction tasks. ENJU analyzes sentences and generates predicate–argument structures (PASs), each of which consists of a predicate, an argument and a relation between them. Figure 2 shows a phrase parsed by ENJU, where each arrowed line and the words it connects denote a PAS, and the direction of the line is from the predicate to the argument. For example, predicate ‘migrate’, argument ‘cells’ and relation ‘verb_arg1’ form a PASs.
A sentence parsed by ENJU can be represented as a graph, in which each node maps to a word and each edge to a PAS relation between the words. As shown in Figure 3, we define that, if the first node on a syntactic path is a tissue term, and the last node a trigger, then the sequence of words on this path is tagged as an event, and vice versa. Using pattern (B′) in Figure 3, the example shown in Figure 2 will be recognized as an event, because trigger word ‘migrate’ and tissue ‘endothelial cells’ are connected via the syntactic path ‘migrate’, ‘cells’ and ‘endothelial’. When constructing events with paths, the direction of PAS was not taken into account, and we also set the maximum number of edges allowed on a path to 5.
Fig. 3.
Patterns for finding angiogenesis events with PAS relations.
We also expanded the vocabularies by including the derivation forms of the tissues and triggers, because they appear in text not only as one POS, but also others with the same lexical root. For example, both ‘vascular development’ and ‘develops vasculature’ are angiogenesis events, where ‘vascular’ is an adjective derivation of ‘vasculature’ and ‘development’ a noun derivation of ‘develop’. We used the Lexical Variants Generation Tool from NIH (http://lexsrv3.nlm.nih.gov/LexSysGroup/Projects/lvg/current/web/) for generating the derivations.
2.4 Automatic vocabulary construction using domain-specific documents
In addition to the syntactic patterns, the tissue and trigger vocabularies play an important role in the dictionary-based approach. The vocabularies used in Sections 2.2 and 2.3 were manually developed, where the choice of terms can be subjective and highly dependent on the curator's domain knowledge. In an attempt to alleviate this problem, we adopted a method to automatically populate the vocabularies using domain-specific texts. This approach requires little human input, because domain-specific documents are relatively easy to obtain. For example, the review articles in the Nature special issue on angiogenesis (DeWitt, 2005) and the Wikipedia page on this subject (http://en.wikipedia.org/wiki/Angiogenesis) are readily available as angiogenesis-related texts.
Compared with other knowledge sources, domain-specific texts have received less attention for their applications to biomedical text mining. One application is keyphrase extraction, which can be cast as a classification task (Frank et al. 1999; Turney, 1999): each phrase in a document is either a keyphrase or not, and the problem is to correctly classify a phrase into one of the two categories, for which off-the-shelf supervised machine learning tools can be used. However, this method requires a set of training documents, where the keyphrases in each document must be manually identified. Alternatively, previous work tackled keyphrase extraction using statistical measures (e.g. Frantzi et al., 2000) using a single corpus. In contrast, our method extracts salient angiogenesis-related predicate–argument pairs by comparing the statistical language models built respectively on the PAS generated from two ENJU-parsed corpora: a domain-specific corpus (i.e. foreground corpus) and a general one (i.e. background corpus). Intuitively, angiogenesis-related tissues and trigger words occur more frequently in the foreground corpus than in the background one, and therefore it is possible to extract these terms from the key predicate–argument pairs. Using the patterns defined in Section 2.3, the automatically generated tissue and trigger vocabularies can then be used to construct angiogenesis events.
2.4.1 Capturing domain-specific keyphrases by comparing language models
A statistical language model assigns a probability to a sequence of n words P(w1,…, wn) by means of a probability distribution. In NLP, a simplifying assumption is often made such that the probability of a word given all the previous words can be approximated by the probability of the word given a number of previous words. For example, a bigram model approximates P(wi|w1i−1) by the conditional probability of the preceding word P(wi|wi−1), and similarly, a trigram model is the conditional probability of the preceding two words, i.e. P(wi|wi−1wi−2).
Tomokiyo and Hurst (2003) approached keyphrase extraction by comparing the KL divergence between the language model of a foreground corpus, and that of a background one, where KL divergence is a non-symmetric metric of the inefficiency of assuming that the distribution is q when the true distribution is p (Cover and Thomas, 1991). Let p(x) and q(x) be two probability mass functions, the KL divergence between p and q is defined in Equation (1).
| (1) |
They also defined the term inside the summation of Equation (1) as point-wise KL divergence, as shown in Equation (2). Intuitively, point-wise KL divergence quantifies the contribution of the phrase w to the expected loss of the entire distribution.
| (2) |
Tomokiyo and Hurst went on to score each n-gram in text according to its ‘phraseness’ and ‘informativeness’, where the former computes how much information would be lost if assuming the independence of each word by applying the unigram model, instead of the n-gram one, and the latter is how much we lose information by assuming w is drawn from the background model instead of the foreground one. More formally, ‘phraseness’ and ‘informativeness’ are respectively defined in Equations (3) and (4):
| (3) |
| (4) |
where,
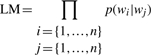 |
(5) |
and LMfgn is the n-gram model constructed from the foreground corpus, while LMbgn from the background one. The linear addition of ‘phraseness’ and ‘informativeness’ was used to score each n-gram, and the higher the score, the more salient the n-gram is in the foreground corpus. This strategy was shown to outperform some other keyphrase extraction methods, such as the likelihood ratios (Damerau, 1993).
This method could be used to find angiogenesis-related n-grams, given an angiogenesis-specific corpus and a general one. However, tissues and triggers are likely to contain one to several words, and calculating point-wise KL divergence for every n-gram, where, for example, n∈[1,4], is computationally expensive. More importantly, by definition, n-gram models only take into account strings of adjacent words, whereas as argued in Section 2.3, statistics of co-occurrences of words that are physically distant but syntactically close can be very useful to capture related concepts.
2.4.2 Comparing language models based on PAS relations
We also extract keyphrases by comparing language models computed on different corpora. However, instead of using n-gram language models, we adopted a language model based on pair-wise predicate–argument relations produced by the ENJU parser. There has been some research in incorporating syntactic and semantic relations produced by a natural language parser in language modelling. For example, Padó and Lapata (2007) built semantic vector space models using word syntactic relations, instead of word co-occurrence counts, and the models were shown to be comparable or superior to the state of the art, on single word priming, synonym detection and word sense disambiguation.
When using the PAS-based language models, ‘phraseness’ became irrelevant because the parser would have determined the relation between the predicate and argument. Consequently, we only needed to calculate the ‘informativeness’ of each argument and predicate pair (i.e. n=2), using Equation (4), where wi and wj, respectively, are an argument and a predicate, which have a direct PAS relation, as opposed to two neighbouring words as in a bi-gram language model.
In our experiments, the background corpus contained 1000 documents that were randomly selected from the collection of medline abstracts published between year 2000 and 2010. As to the foreground corpus, we experimented with three different document sets, in order to study how ‘domain-specific’ a foreground corpus needed to be. The first foreground corpus (i.e. RandomMedline) consisted of 250 randomly selected medline documents (~52k tokens) that contain the keyword ‘angiogenesis’. The second (i.e. AngioCorpus) was the 262 abstracts in our angiogenesis corpus (~58k tokens), retrieved using the patterns defined in Section 2.2. Note that in this experiment, we only used the ‘raw’ text but not the annotation (Section 2.5). Finally, the third corpus (i.e. ReviewArticles) contained the six full-text review articles from the angiogenesis special issue of Nature (DeWitt, 2005) and the Wikipedia page on angiogenesis (~53k tokens).
The background and foreground corpora were processed by ENJU to generate the PAS relations, and then the PAS-based language models were computed for each corpus, where Katz smoothing (Katz, 1987), reportedly to perform well on NLP tasks (Chen and Goodman, 1996), was applied to alleviate the data sparseness problem, and functional words such as prepositions and determiners, as well as words consisting of only digits and punctuation, were removed.
We then coupled each foreground corpus with the background one, and extracted a list of salient predicate–argument pairs using Equation (4), and the pairs were ranked according to their point-wise KL divergence scores. To assess the quality of the results, a domain expert manually reviewed the highest ranked 50 predicate–argument pairs, extracted using ReviewArticles as the foreground corpus. In this exercise, an argument–predicate pair was judged as relevant if it was either an angiogenesis-related entity (i.e. cell, tissue, gene or gene product) or an angiogenesis term or event. The result was promising: 32 phrases were judged as relevant and 12 possibly relevant. In other words, 88% of the automatically extracted argument–predicate pairs were relevant or possibly relevant. In addition, all phrases in the top 20 were considered relevant (14 out of 20) or possibly relevant (6 out of 20). Table 2 shows the top 10 angiogenesis-related predicate–argument pairs extracted in this experiment.
Table 2.
Top 10 angiogenesis-related phrases from ReviewArticles
| Rank | Predicate | Argument | Rank | Predicate | Argument |
|---|---|---|---|---|---|
| 1 | Vascular | Development | 6 | Vessel | Growth |
| 2 | Retinal | Vessels | 7 | Growth | Factor |
| 3 | Endothelial | Cells | 8 | Growing | Vessel |
| 4 | Dorsal | Aorta | 9 | Retinal | Angiogenesis |
| 5 | Retinal | Vascularization | 10 | Retinal | Development |
2.4.3 Event extraction
The automatically extracted list contains angiogenesis-related predicate–argument pairs, but in order to construct patterns for mining events as described in Section 2.3, we needed to acquire the vocabularies for tissue terms and trigger words. We consider a term as a tissue if it satisfies the following two conditions: (i) it belongs to a tissue dictionary; and (ii) it occurs in the predicate–argument list. Essentially, the conditions state that we want tissues (i.e. condition 1) that are relevant to the angiogenesis bioprocess (i.e. condition 2). We constructed the tissue dictionary using a subset of the UMLS ontology (http://http://www.nlm.nih.gov/research/umls/), which contains the following branches: body, body parts, organ, tissue, cell and cell components. UMLS was chosen because it is relatively comprehensive and integrated a number of biomedical ontology and vocabularies, such as MeSH (http://www.nlm.nih.gov/mesh/) and NCI thesaurus (http://ncit.nci.nih.gov/). We ‘flattened’ the dictionary so that it did not contain any hierarchy, and the final tissue dictionary contained 188 069 unique entries. Meanwhile, according to condition 2, a term must also match an argument or a predicate, which ranks among the top α (α=500 in our experiments) in the predicate-argument list. Note that a tissue entity can appear as either a predicate or an argument. For example, both ‘vascular’ and ‘vasculature’ are angiogenesis tissue entities, but the former is more likely to be a predicate as an adjective, whereas the latter, a noun, is more likely to be an argument.
Similarly, a trigger word is a verb that appears in the salient predicate–argument list. We needed a verb dictionary and for that purpose we examined lexical resources for the biological domain such as BioLexicon (Sasaki et al., 2008a). The coverage of the verb list in BioLexicon is small and does not include angiogenesis trigger words such as ‘proliferate’ and ‘migrate’. On the other hand, verbs in English dictionaries, such as WordNet, are too general. We then decided to build a verb dictionary by computing point-wise KL divergence scores for verb unigrams using Equation (4), on the foreground and background corpora, where n was set to 1 and w must be a verb as identified by a POS tagger (Alex et al., 2008). The verb list was then expanded to include the derivation forms using the NIH Lexical Variants Generation Tool. Finally, the top-ranked β terms (β = 150 in our experiments) in the intersection of the verb list and the predicate–argument list were selected as angiogenesis trigger words.
We then tagged angiogenesis events in text using the patterns defined in Figure 3. The only difference is that Tissue and Trigger were taken from the automatically constructed tissue and trigger lists, respectively, instead of the manually created ones. See the additional material to this article (http://text0.mib.man.ac.uk/~mbassxw2/angiogenesis/additional.html) for more discussion on how the parameters α and β affect the event extraction performance.
2.5 Learning from manual annotation
A supervised learning approach infers a model over training examples, where each example consists of a set of predefined features (e.g. word form and contextual information) and an output value. The trained model is then used to classify new instances. Supervised systems consistently excel as demonstrated in a range of evaluation challenges, such as BioCreative I (Hirschman et al., 2005) and II (Krallinger et al. 2008b) and the BioNLP shared tasks (e.g. Kim et al., 2009). Such methods do not rely on dictionaries. However, the availability of a training corpus is essential and therefore we hand-built a gold standard corpus for the identification of angiogenesis terms and events. In addition to training machine learning models, the corpus enabled systematic evaluation and comparison of the techniques proposed in this article.
2.5.1 Selecting documents for manual annotation
We first retrieved an initial pool of documents from the collection of medline abstracts that were published on and before October 2009. The patterns defined in Figure 1 were submitted as queries. More specifically, if an abstract contains a sentence that matches pattern (A), (B) or (C) (Fig. 1), then it will be retrieved and stored in the pool. We did not only use angiogenesis terms (e.g. ‘angiogenesis’) as queries, because documents that contain angiogenesis events were also of interest. We then randomly selected abstracts in several batches for annotation and in total the final annotated corpus contained 262 abstracts. Note that this retrieval procedure gave some advantage to the dictionary-based approach described in Section 2.2 in evaluation, because each document was guaranteed to contain at least an angiogenesis term or event that the dictionary-based method would be able to identify.
2.5.2 Manual annotation
Table 1 summarizes the annotation markables. The guidelines for annotating entities (i.e. gene or gene product, tissue and cell) and angiogenesis terms were relatively straightforward: every mention of the above entities should be annotated, and a mention of an entity can be either its full-name or abbreviation and acronym forms. As for angiogenesis events, we followed the definitions set in Section 2.1 and annotated the phrases that indicated the change of state of angiogenesis-related cells, cell components and tissues. Similar to GENIA's annotation (Kim et al., 2009), each event should contain a tissue term as a participant, and a trigger word indicating the action that changes the state of the participant or its biological property. Although this rule was not enforced due to limitation of the annotation tool, we found the majority of events contain a participating tissue entity (463 out of 479, or 97%), and those that do not have participants were mostly annotation errors. Different from the GENIA guidelines, a participating entity may consist of words of any POS, and triggers were not explicitly marked, in order to reduce annotation time. Table 3 shows two examples that illustrate the difference in our annotation and GENIA's. For the first example, we annotated phrase ‘endothelial cell migration’ as an angiogenesis event and ‘endothelial cell’ a cell entity, whereas according to the GENIA guidelines, ‘endothelial cell migration’ would be marked as a ‘cellular physiological process’ event, ‘endothelial cell’ as a cell entity and ‘migration’ a trigger word. As to the second example, GENIA annotators would not add any annotation, while we would annotate ‘vascular’ as a tissue entity, ‘vascular proliferation’ as one, and ‘vascular … maturation’ as the other event.
Our annotation guidelines also stated that the annotation was concerned with identifying what the author(s) intend to communicate in the text, and the annotators should not make any judgment as to the validity of the author's claims. During the annotation, the annotators may seek help from external resources and search engines, such as PubMed and Google. Also, when marking the entities and events, the annotators were permitted to nest them, but neither entities nor events were allowed to cross. Discontinuous co-ordinations such as ‘A and B cells’ were annotated as two nested entities ‘A and B cells’ and ‘B cells’. Three domain experts went through the abstracts and annotated angiogenesis terms, events and related bioentities. The annotation was carried out using the Callisto tool (http://callisto.mitre.org/) for its relative simplicity to use. In total, ~150 h was spent on annotation.
2.5.3 Quality control
To ensure the quality of annotation, we first went through a ‘pilot study’, in which 20 documents were doubly annotated. Inter-annotator agreement (IAA) for every type of markable was then calculated. The IAA results of angiogenesis terms were good (Table 4), indicating the annotators consistently agreed with each other. However, the IAA for angiogenesis events was not satisfactory (Table 5), which demonstrated the complexity and diversity in how angiogenesis events appear in text, and the fact that the task can be a challenge even for human annotators.
Table 4.
Angiogenesis term results (precision/recall/F1-score, in %)
| Method | Angiogenesis term |
|---|---|
| IAA | 82.35/67.47/74.14 |
| DictionaryBased | 71.94/59.52/65.15 |
| CRF | 94.68/91.00/92.80 |
Table 5.
Evaluation results (precision/recall/F1-score, in %)
| Method | Exact match | Boundary relaxed (± 2) |
|---|---|---|
| IAA | 35.00/58.33/43.75 | 45.00/75.00/56.25 |
| PatternBaseline | 33.43/21.91/26.47 | 49.43/32.40/39.14 |
| PatternExtended | 68.16/22.85/34.22 | 87.71/29.40/44.04 |
| CRF | 67.75/33.97/45.22 | 83.19/41.77/55.62 |
| CRF-entity | 71.06/40.93/51.94 | 88.64/51.05/64.79 |
| RandomMedline | 10.40/6.74/8.18 | 27.17/17.60/21.36 |
| AngioCorpus | 43.05/25.26/31.71 | 52.47/30.90/38.73 |
| ReviewArticles | 43.93/31.84/36.92 | 56.07/40.64/47.12 |
Nevertheless, we endeavored to improve the annotation consistency. During the pilot study, we reconciled the doubly annotated documents, found the causes of the discrepancies and re-emphasized the aforementioned annotation guidelines. Then the three annotators started two rounds of annotation, where 100 documents were annotated in the first round and 150 in the second. Double annotation was not performed in this phase due to resource constraints. However, to ensure annotation quality, for each document, a second annotator carried out validation after each round of annotation, and if in doubt, the two annotators were asked to agree on a gold standard through reconciliation. At the end of the annotation, two annotators revisited the 20 doubly annotated documents used for pilot study and updated the annotation. Then the reconciled 20 documents were added to the gold standard dataset. In total, the annotation project generated 270 unique abstracts. However, eight of which were discarded, because they were considered irrelevant to the domain, and consequently the final corpus contains 262 abstracts.
2.5.4 Supervised learning
We tackled both the entity and event recognition tasks with logistic regression models, which have been shown to be effective in handling large-scale classification problems (Andrew and Gao, 2007). In addition, an attractive feature of logistic regression models is that they produce probabilistic output that allows the information on the confidence of the decision to be used by subsequent components in the text processing pipeline. When the random variable to predict is a sequence, the logistic regression model is called linear chain Conditional Random Fields (CRFs) (Lafferty, 2001), which has demonstrated good results on a number of NLP tasks, ranging from POS tagging to chunking (e.g. Tsuruoka et al., 2009). In our experiments, we used CRFSuite (http://www.chokkan.org/software/crfsuite/), a fast implementation of CRF, for tagging the angiogenesis terms and events. In more detail, we converted the data to IOB2 representation (Ramhsaw and Marcus, 1995), where words that were not entities or events of interests received the tag O. For the words that formed an entity or event of semantic class x (e.g. angiogenesis event), the first word was tagged with B - x, and the remaining ones with I − x.
For tagging angiogenesis terms, we used the following features: unigram, bigram and trigram to the left and right of the current word, whether the current word was the head word of the current noun phrase, whether the term was seen in a noun phrase in the document title.
For extracting angiogenesis events, we tested two feature settings: the first was the same as described above, and the second exploited the gold standard annotation of genes, cells and tissues, in addition to the first feature set. These two settings respectively correspond to the CRF and CRF-entity systems in Table 5. We used the gold standard entities to estimate whether entity information was helpful for event identification. In more detail, both the semantic type and the text string of the gold standard entities were incorporated as features for classification. In practice, automatic systems (e.g. Hanisch et al., 2005; Sasaki et al., 2008b; Wilbur et al., 2007) can be used to generate the named entities. This way, the overall performance of event extraction may decrease. However, in order to focus on examining the performance of complex text mining tasks such as relation and event extraction, in experiments, it is a common practice to assume previous components produce gold standard annotations (e.g. Alex et al., 2008; Kim et al., 2009).
3 EVALUATION AND RESULTS
The performance of the systems was measured by precision, recall and F1-score (i.e. balanced precision and recall). To be considered correct, a system prediction must match not only the type of the entity or event, but also both boundaries. For angiogenesis events, sometimes boundaries are not crucial. For example, if the gold standard is ‘vascular endothelial cell proliferation’, then ‘endothelial cell proliferation’ is perhaps a good prediction, even if its left boundary does not match the gold standard. Therefore, we used an additional measure called approximate boundary matching, which allows the spans of the predicted events to slightly differ from the gold standard. A similar measure was also adopted in the BioNLP event evaluation tasks (Kim et al., 2009).
Table 4 compares three sets of results for tagging angiogenesis terms, where the CRF results were obtained by 5-fold cross-validation on the manually created gold standard data. The CRF model (Section 2.5.4) clearly outperformed DictionaryBased, which uses the manually compiled dictionary of angiogenesis terms (Section 2.2). Note that the IAA was calculated on a small set of 20 documents that were doubly annotated in the pilot annotation, and therefore it was possible that system performance exceeded IAA (Section 2.5.3).
Table 5 shows the results for angiogenesis event identification. Both PatternBaseline and PatternExtended exploited the manually compiled tissue and trigger vocabularies, but the former performed matches following simple patterns (Fig. 1), whereas the latter applied patterns incorporating ENJU's predicate–argument relations (Fig. 2). PatternExtended was a clear winner over PatternBaseline, which demonstrated that syntactic relations were useful. CRF and CRF-entity were supervised methods, and they were trained and tested by 5-fold cross-validation on the manually created corpus. As mentioned, the difference between the two systems is that CRF used only contextual word and n-gram as features, while CRF-entity also exploited the gold standard entity annotation. CRF-entity obtained the best results as measured by every metric, and the performance of CRF was also promising. Nevertheless, the two methods were the most expensive to develop, as they required high-quality training data, which were laborious and time consuming to produce, even though we significantly simplified the annotation guidelines as compared with other annotation projects such as GENIA.
The bottom three rows in Table 5 present the results of the method that automatically constructs tissue and trigger vocabularies by comparing PAS language models between a domain-specific corpus and a general one (Section 2.4). We experimented with three different domain-specific foreground corpora, and the distinct performance indicates that this method is sensitive to the choice of foreground corpus. The empirical results show that employing the collection of the angiogenesis review articles and Wikipedia page obtained the best results, which correlates with the fact that this foreground corpus contained more concentrated information regarding angiogenesis than the others. While using the manually constructed vocabularies achieved good precision (87.71%), it suffered from a poor recall (29.40%). On the other hand, the automatic method using ReviewArticles, yielded better recall and F1 scores, indicating its ability to discover a wider range of terms from the domain-specific documents.
4 CONCLUSIONS
This article presented solutions to a text mining task of automatically extracting terms and events describing specific bioprocesses. We examined angiogenesis, a bioprocess of blood vessel growth, and manually created two types of resources to assist the study: angiogenesis-related vocabularies and a gold standard corpus. In particular, the gold standard corpus consists of 262 medline abstracts, where angiogenesis terms and events, as well as genes, gene products, cells and tissues, were manually annotated. The statistics of the corpus shows that 36.5% of mentions of angiogenesis appear in text as an event, which previous bioprocess extracting techniques struggle to find.
We developed and compared a range of methods using the manually built vocabularies and gold standard corpus, and the experimental results showed that a CRF model outperformed the others: on detecting angiogenesis terms, it achieved an F1-score of 92.8%; on event recognition, the model yielded an F1 of 64.79% and a precision of 88.64%, when the restriction on event boundaries was relaxed. Nevertheless, the CRF model relied on the manually created gold standard, which domain experts spent 150 person/h to create. In contrast, the angiogenesis-specific vocabularies were less time consuming to develop (40 person/h), but the pattern matching approaches using the vocabularies obtained lower performance results. We also proposed a new method that automatically discovers angiogenesis-related tissue terms and trigger words, by comparing the language models built on the predicate–argument relations of two ENJU-parsed corpora: one contained angiogenesis-specific documents and the other general biomedical texts. This method required very little human supervision, and the pattern-based systems achieved better results when using the automatically built angiogenesis vocabularies than the manual ones. Overall, the relative low development cost of this new method indicates that it has better domain adaptability than the others, while achieving reasonable performance results.
ACKNOWLEDGEMENTS
We would like to thank Catriona Tate who helped on the annotation. The Oncology group at AstraZeneca provided the domain knowledge.
Funding: This work was funded by UK Biotechnology and Biological Sciences Research Council (BBSRC) under project Automated Biological Event Extraction from the Literature for Drug Discovery (reference number: BB/G013160/1). The UK National Centre for Text Mining is funded by UK Joint Information Systems Committee (JISC).
Conflict of Interest: none declared.
REFERENCES
- Alex B., et al. Assisted curation: does text mining really help? Pac. Symp. Biocomput. 2008;13:556–567. [PubMed] [Google Scholar]
- Andrew G., Gao J. Proceedings of the ICML. New York, NY, USA: ACM; 2007. Scalable training of L1-regularized log-linear models; pp. 33–40. [Google Scholar]
- Ananiadou S., et al. Event extraction for systems biology by text mining the literature. Trends Biotechnol. 2010;28:381–390. doi: 10.1016/j.tibtech.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Björne J., et al. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing: Shared Task. PA, USA: The Association for Computational Linguistics; 2009. Extracting complex biological events with rich graph-based feature sets. [Google Scholar]
- Chen S.F., Goodman J.T. Proceedings of the 34th Annual Meeting of the Association of Computational Linguistics. San Francisco, CA, USA: Morgan Kaufmann; 1996. An empirical study of smoothing techniques for language modeling; pp. 310–318. [Google Scholar]
- Cover T.M., Thomas J.A. Elements of Information Theory. New York, NY, USA: John Wiley; 1991. [Google Scholar]
- Damerau F. Generating and evaluating domain-oriented multi-word terms from texts. Informat. Process. Manag. 1993;29:433–447. [Google Scholar]
- DeWitt N. Nature Special Issue on Angiogenesis. Vol. 438. London, UK: Nature Publishing Group; 2005. (7070) [Google Scholar]
- Frank E., et al. Proceedings of International Joint Conference on Artificial Intelligence (IJCAI). San Francisco, CA, USA: Morgan Kaufmann; 1999. Domain-specific keyphrase extraction; pp. 668–673. [Google Scholar]
- Frantzi K., et al. Automatic recognition of multi-word terms. Int. J. Digit. Libr. 2000;3:117–132. [Google Scholar]
- Hanisch D., et al. ProMine: rule-based protein and gene entity recognition. BMC Bioinformatics. 2005;6(Suppl. 1):S14. doi: 10.1186/1471-2105-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman L., et al. Overview of the BioCreative: critical assessment of information extraction for biology. BMC. Bioinformatics. 2005;6(Suppl. 1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter L., Cohen K.B. Biomedical language processing: what's beyond PubMed. Mol. Cell. 2006;21:589–594. doi: 10.1016/j.molcel.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvidsten T.R., et al. Learning rule-based models of biological process from gene expression time profiles using gene ontology. Bioinformatics. 2003;19:1116–1123. doi: 10.1093/bioinformatics/btg047. [DOI] [PubMed] [Google Scholar]
- Katz S.M. Estimation of probabilities from sparse data for language model component of a speech recogniser. IEEE Trans. Acoust. Speech Signal Process. 1987;35:400–401. [Google Scholar]
- Kim J-D., et al. Technical Report TR-NLP-UT-2006-2. Tokyo, Japan: Tsujii Laboratory, University of Tokyo; 2006. GENIA ontology. [Google Scholar]
- Kim J.-D., et al. Corpus annotation for mining biomedical events from literature. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-10. Article no. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-D., et al. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing: Shared Task. PA, USA: The Association for Computational Linguistics; 2009. Overview of BioNLP'09 shared task on event extraction. [Google Scholar]
- Koike A., et al. Automatic extraction of gene/protein biological functions from biomedical text. Bioinformatics. 2004;21:1227–1236. doi: 10.1093/bioinformatics/bti084. [DOI] [PubMed] [Google Scholar]
- Kola I., Landis J. Can the pharmaceutical industry reduce attrition rate? Nat. Rev. Drug Discov. 2004;3:711–716. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Krallinger M., et al. Overview of the protein-protein interaction extraction task of BioCreative II. Genome Biol. 2008a;9(Suppl. 2):S4. doi: 10.1186/gb-2008-9-s2-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M., et al. Evaluation of text mining systems for biology: overview of the second BioCreative community challenge.Genome. Biol. 2008b;9(Suppl. 2):S1. doi: 10.1186/gb-2008-9-s2-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty J., et al. Proceedings of the 18th International Conference on Machine Learning. San Francisco, CA, USA: Morgan Kaufmann; 2001. Conditional random fields: Probabilistic models for segmenting and labeling sequence data; pp. 282–289. [Google Scholar]
- Miwa M., et al. Event extraction with complex event classification using rich features. J. Bioinformatics Comput. Biol. 2010;8:131–146. doi: 10.1142/s0219720010004586. [DOI] [PubMed] [Google Scholar]
- Miyao Y., Tsujii J. Feature forest models for probabilistic HPSG parsing. Comput. Linguist. 2008;34:35–80. [Google Scholar]
- Miyao Y., et al. Evaluating contributions of natural language parsers to protein-protein interaction extraction. Bioinformatics. 2009;25:394–400. doi: 10.1093/bioinformatics/btn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padó S., Lapata M. Dependency-based construction of semantic space models. Comput. Linguist. 2007;33:161–199. [Google Scholar]
- Ramhsaw L., Marcus M. Proceedings of the 3rd ACL Workshop on Very Large Corpora. San Francisco, CA, USA: Morgan Kaufmann; 1995. Text chunking using transformation based learning; pp. 82–94. [Google Scholar]
- Sasaki Y., et al. Proceedings of the Third International Symposium on Semantic Mining in Biomedicine (SMBM). Turku, Finland: Turku Centre for Computer Science; 2008a. BioLexicon: a lexical resource for the biology domain. [Google Scholar]
- Sasaki Y., et al. How to make the most of NE dictionaries in statistical NER. BMC Bioinformatics. 2008b;9(Suppl. 11):S5. doi: 10.1186/1471-2105-9-S11-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L., et al. Overview of BioCreative II gene mention recognition. Genome Biol. 2008;9(Suppl. 2) doi: 10.1186/gb-2008-9-s2-s2. Article no. S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomokiyo T., Hurst M. Proceedings of the ACL Workshop on Multiword Expressions. PA, USA: The Association for Computational Linguistics; 2003. A language model approach to keyphrase extraction. [Google Scholar]
- Tsuruoka Y., et al. Proceedings of ACL-IJCNLP. PA, USA: The Association for Computational Linguistics; 2009. Stochastic gradient descent training for L1-regularized log-linear models with cumulative penalty; pp. 477–485. [Google Scholar]
- Turney P.D. Technical Report ERB-1057. Ottawa, Ontario, Canada: National Research Council, Institute for Information Technology; 1999. Learning to extract keyphrases from text. [Google Scholar]
- Wang X., et al. Disambiguating the species of biomedical named entities using natural language parsers. Bioinformatics. 2010;26:661–667. doi: 10.1093/bioinformatics/btq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur J., et al. Proceeding of the BioCreative II Workshop. Madrid, Spain: Spanish National Cancer Centre (CNIO); 2007. BioCreative II Gene Mention Task; pp. 7–16. [Google Scholar]