Abstract
Nuclear factor erythroid 2–related factor 2 (Nrf2) is a transcription factor that promotes the transcription of cytoprotective genes in response to oxidative and electrophilic stresses. Most functions of Nrf2 were identified by studying biological models with Nrf2 deficiency, however, little is known about the effects of graded Nrf2 activation. In the present study, genomic gene expression profiles by microarray analysis were characterized with a “gene dose-response” model in livers of Nrf2-null mice, wild-type mice, Kelch-like ECH associating protein 1 (Keap1)-knockdown (Keap1-KD) mice with enhanced Nrf2 activation, and Keap1-hepatocyte knockout (Keap1-HKO) mice with maximum hepatic Nrf2 activation. Hepatic nuclear Nrf2 protein, glutathione concentrations, and known Nrf2 target genes were increased in a dose-dependent manner. In total, 115 genes were identified to be constitutively induced and 80 genes suppressed with graded Nrf2 activation. Messenger RNA of genes encoding enzymes in the pentose phosphate pathway and enzyme were low with Nrf2 deficiency and high with Nrf2 activation, indicating that Nrf2 is important for NADPH production. NADPH is the major reducing resource to scavenge oxidative stress, including regenerating glutathione and thioredoxin and is also used for anabolic pathways including lipid synthesis. High performance liquid chromatography-ultraviolet absorbance analysis confirmed that hepatic NADPH concentration was lowest in Nrf2-null mice and highest in Keap1-HKO mice. In addition, genes involved in fatty acid synthesis and desaturation were downregulated with graded Nrf2 activation. In conclusion, the present study suggests that Nrf2 protects against environmental insults by promoting the generation of NADPH, which is preferentially consumed by aiding scavenging of oxidative stress rather than fatty acid synthesis and desaturation.
Keywords: Nrf2, microarray, liver
Nuclear factor erythroid 2–related factor 2 (Nrf2) is a transcription factor that induces a battery of cytoprotective genes in response to oxidative/electrophilic insults. Under physiological conditions, Nrf2 is bound to its repressor, Kelch-like ECH associating protein 1 (Keap1), in the cytosol. Keap1 functions as an adapter protein that retains Nrf2 in the cytoplasm by interacting with the cytoskeleton, and it facilitates degradation of Nrf2 by binding with Cullin 3-based E3 ligase, a protein-protein complex that ubiquitinates Nrf2 protein (Cullinan et al., 2004). With the negative regulatory system of Keap1, Nrf2 protein has a very rapid turnover, with a half-life less than 20 min (Kobayashi et al., 2004), and thus Nrf2 protein is minimally detectable in unstressed conditions. There are numerous cysteine residues in Keap1 protein: murine and rat Keap1 have 25 cysteine residues and human Keap1 has 27 cysteine residues. The high cysteine content of Keap1 makes it an excellent sensor for oxidative/electrophilic stress. Yamamoto and coworkers therefore proposed that Nrf2-activating compounds directly modify the sulfhydryl groups of cysteines in Keap1 by oxidation, reduction, or alkylation, which would alter the conformation of Keap1 and cease the ubiquitination of Nrf2 (Kobayashi and Yamamoto, 2006). Once released from Keap1, Nrf2 translocates into the nucleus, heterodimerizes with a small musculoaponeurotic fibrosarcoma (Maf) protein, and promotes transcription of its target genes (Itoh et al., 1997). Consistent with this hypothesis, a number of Nrf2 activators have been reported to react with different cysteine residues of Keap1 (Hong et al., 2005; Luo et al., 2007).
After the Keap1-Nrf2 signal pathway was discovered, a number of natural and synthetic compounds were shown to activate Nrf2 and protect against oxidative stress-induced toxicity through Nrf2 activation. Potential therapeutic applications of Keap1-Nrf2 pathway have therefore been investigated. For example, curcumin was shown to protect against focal ischemia of cerebrum through upregulation of Nrf2, indicating Nrf2 might be suitable target for therapy of cerebral ischemia (Yang et al., 2009). Resveratrol was shown to increase transcription of GSH synthesis enzymes through activating Nrf2 (Zhang et al., 2009). Oltipraz was shown to protect against ANIT-induced cholestasis through Nrf2 activation, which indicated that pharmacological activation of Nrf2 might represent a therapeutic option for intrahepatic cholestasis (Tanaka et al., 2009). 2-Cyano-3,12-dioxooleana-1,9-dien-28-oic imidazolide (CDDO-Im), a synthetic triterpenoid, was shown to activate Nrf2 and protect against acetaminophen-induced liver injury (Reisman et al., 2009b). However, pharmacological activation of Nrf2 by chemicals is problematic, for they often have many off-target effects. For example, curcumin also activates AP-1, a transcription factor that promotes cell proliferation (Anand et al., 2008); resveratrol also activates SIRT-1 (Baur, 2010); oltipraz activates constitutive androstane receptor (CAR) and induces Cyp2b10 through CAR activation (Merrell et al., 2008); and CDDO-Im activates Smad and ERK and induces cell differentiation (Ji et al., 2006).
Nrf2-null mice have served as a valuable model to examine the Keap1-Nrf2 pathway. Compared with wild-type mice, Nrf2-null mice are more susceptible to liver injury by oxidative and electrophilic stress (Aleksunes and Manautou, 2007), acetaminophen (Enomoto et al., 2001), ethanol (Lamle et al., 2008), pentachlorophenol (Umemura et al., 2006), and high-fat diet (Tanaka et al., 2008). To investigate the effect of increased Nrf2 activation, Keap1-null mice were engineered. However, Keap1-null mice died at about 20 days of age (Wakabayashi et al., 2003), and thus a Keap1-hepatocyte knockout mouse (Keap1-HKO mouse) was engineered, utilizing the albumin-Cre loxP system. The Keap1-HKO mice have maximal activation of Nrf2 and Nrf2 target genes in liver and are more resistant to acetaminophen hepatotoxicity (Okawa et al., 2006). Later it was discovered that mice homozygous for Keap1 loxP sites (no Albumin-Cre transgene) have decreased or a “knockdown” of Keap1 (Keap1-KD), resulting in an intermediate activation of Nrf2 in multiple organs (Okada et al., 2008). Taking advantage of the fact that floxed Keap1 allele is hypomorphic, an animal of graded reduction of Keap1 expression in adult mice was generated (Taguchi et al., 2010).
Gene transcription profiles have been performed on the effect of Nrf2 in mouse macrophages (Woods et al., 2009), lung tissue (Blake et al., 2010), and embryonic fibroblasts (Malhotra et al., 2010), but there is a significant gap in our understanding of Nrf2 on genomic gene expression in liver. Liver is the primary organ for the detoxification of environmental chemicals, drugs, and endogenous toxicants. Thus, studying the function of Nrf2 in liver is of great importance. In the present study, we have utilized a “gene dose-response” approach including Nrf2-null mice lacking any functional Nrf2, wild-type mice having normal Nrf2 activation, Keap1-KD mice having enhanced Nrf2 activation, and Keap1-HKO mice having the highest Nrf2 activation in liver. Hepatic phenotypes as well as genomic gene expression profile were analyzed in this gene dose-response approach.
MATERIALS AND METHODS
Reagents.
Nrf2 antibody was purchased from Santa Cruz Biotechnology (sc-30915; Santa Cruz, CA). TATA-box binding protein (TBP) antibody was purchased from Abcam (ab51841; Cambridge, MA). All other chemicals, unless otherwise specified, were purchased from Sigma-Aldrich (St Louis, MO).
Animals and husbandry.
Eight-week-old male mice were used for this study. C57BL/6 mice were purchased from Charles River Laboratories, Inc. (Wilmington, MA). Nrf2-null mice were obtained from Dr Jefferson Chan (University of California, Irvine, CA) (Chan et al., 1996). Keap1-KD mice were supplied by Dr Masayuki Yamamoto (Tohoku University, Sendai, Japan). In an attempt to make a hepatocyte-specific Keap1-null mouse, utilizing a loxP, Alb-Cre system, a Keap1-KD mouse, in which Keap1 was decreased throughout the body, was engineered (Okada et al., 2008). Nrf2-null and Keap1-KD mice were backcrossed into the C57BL/6 background, and > 99% congenicity was confirmed by Jackson Laboratories (Bar Harbor, ME). Keap1-HKO mice were generated by crossing Keap1-KD mice and AlbCre+ mice, which express Cre only in hepatocytes. All the mice used in the study were bred at University of Kansas Medical Center and housed in a temperature-, light-, and humidity-controlled environment and had access to Teklad Rodent Diet #8604 (Harlan Laboratories, Madison, WI) and water ad libitum. The housing facility at the University of Kansas Medical Center is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care–accredited facility at the University of Kansas Medical Center. All the mice were euthanized in the morning (8:00–10:00 a.m.) under the fed state. All procedures were preapproved in accordance with Institutional Animal Care and Use Committee guidelines.
Nrf2 protein expression in hepatic nuclear extracts.
Nuclear extracts were prepared with the NE-PER nuclear extraction kit according to the manufacturer's protocol (Pierce Biotechnology, Rockford, IL). Protein concentrations were determined with a BCA Assay Kit from Pierce Biotechnology. Nuclear proteins (40 μg protein per lane) were electrophoretically resolved using polyacrylamide gels (4% stacking and 10% resolving). Gels were transblotted overnight at 4°C onto polyvinylidene fluoride membranes. Membranes were then washed with PBS-buffered saline containing 0.05% Tween-20 (PBS-T). Membranes were blocked for 1 h at room temperature with 5% nonfat milk in PBS-T. Blots were then incubated with primary antibody (1:1000 dilution in 2% nonfat milk in PBS-T) for 3 h at room temperature. Blots were washed in PBS-T and incubated with species-appropriate secondary antibody (anti-rabbit for Nrf2 and anti-mouse for TBP) conjugated with horseradish peroxidase (1:2000 dilution) in 2% nonfat milk in PBS-T buffer for 1 h at room temperature. Blots were then washed with PBS-T. Protein-antibody complexes were detected using an ECL chemiluminescent kit (Pierce Biotechnology) and exposed to x-ray film (Denville Scientific, Metuchen, NJ). Equal protein loading was confirmed by TBP. Nrf2 and TBP proteins migrated the same distance as proteins of approximately 110 and 37 kDa, respectively. Intensity of protein bands was quantified by Discovery Series Quantity One 1-D Analysis Software (Bio-Rad Laboratories, Hercules, CA). Individual blot densities were normalized to that of wild-type mice.
GSH quantification.
GSH concentrations were quantified by ultra high performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) according to a previous method with small modifications (Guan et al., 2003; New and Chan, 2008). Briefly, frozen liver tissue (50 mg) was diced, added to 200 μl of ice-cold KCl (1.15%), and homogenized over ice for 2 min. Tissue homogenates (50 μl) were transferred to a 1.5 ml snapcap conical-bottom centrifuge vials with glutathione ethyl ester (0.01 mg/ml, 20 μl) added as the internal standard (IS). Ellman's reagent (10mM, 100 μl) was added to the homogenate and vortexed, followed by addition of 30 μl of 5-sulfosalicylic acid (20%). After centrifuging at 11,600 g for 15 min, the supernatant was collected for LC/MS analysis. A Waters Acquity ultra performance LC system (Waters, Milford, MA) was used, and all chromatographic separations were performed using an Acquity UPLC BEH C18 column (1.7 μm, 100 × 2.1 mm I.D.). The mass spectrometer was a Waters Quattro Premier XE triple quadrupole instrument with an electrospray ionization (ESI) source (Waters). The multiple reaction monitoring transitions under ESI positive mode are as follows: m/z 504.8 > 272.9 for GSH-Ellman, 532.88 > 204.9 for IS-Ellman.
Total RNA isolation.
Total RNA was isolated using RNAzol B reagent (Tel Test, Inc., Friendswood, TX) according to the manufacturer's protocol. The concentration of total RNA in each sample was quantified spectrophotometrically at 260 nm. The integrity of each RNA sample was evaluated by formaldehyde-agarose gel electrophoresis before analysis.
Quantification of mRNA expression by RT-PCR assay.
Total RNA in mouse livers were reverse transcribed into complementary DNA (cDNA) by High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA), and the resulting cDNA were used for real-time PCR analysis using SYBR Green PCR Master Mix in 7900HT Fast Real-Time PCR System (Applied Biosystems). Oligonucleotide primers specific to mouse β-actin, Gclc, Nqo1, Me1, G6pdx, and Pgd were described in Supplementary Table 1.
Microarray and data analysis.
Gene expression in livers of Nrf2-null, WT, Keap1-KD, and Keap1-HKO mice was determined by the Affymetrix Mouse 430.20 arrays at the KUMC Microarray Core Facility. Biological replicates (n = 3) of each genotype were hybridized to individual arrays. Raw data CEL files were preprocessed using R in the Bioconducter by “affy” and Robust Multichip Averaging (gcRMA) packages (Irizarry et al., 2003). The mean probe intensities higher than log2100 in at least one group were selected for further analysis. Gene annotations were obtained using the mouse 4302 and “annaffy” packages. Differential gene expression was performed across all genotypes (p value < 0.05) by the Linear Models for Microarray Data (LIMMA) package. Probes that were shown to be differentially expressed were constructed in a Venn diagram which identifies the common and exclusively expressed genes of each genotype (Nrf2-null, WT, Keap1-KD, and Keap1-HKO). Two-way hierarchical clustering of pathway-specific gene expression was conducted using JMP 8.0 software (SAS Institute, Cary, NC) using agglomerative clustering for hierarchical clustering. The method starts with each point (gene) as its own cluster. At each step, the two clusters that are closest together are combined into a single cluster. This process continues until there is only one cluster containing all the points (genes). This kind of clustering is good for smaller data sets (a few hundred observations). The distance between two genes was calculated by the Ward method.
Pathway analysis.
Functional and pathway analysis of differentially expressed probe sets was performed using the Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com) and Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov) database. Pathway enrichment was determined by p value ≤ 0.05 and a minimum of four probes in the pathway. False-discovery rates are also reported.
NADPH quantification.
NADPH was determined by HPLC according to a previous method with minor modifications (Jones, 1981). Briefly, 50 mg of frozen tissues were homogenized in an extraction medium containing 0.5 M KOH, 50% ethanol (vol/vol), and 35% cesium chloride (wt/vol). The extracts were then centrifuged at 10,000 g for 15 min, and the supernatants were filtered through 0.2 μm Costar Spin-X HPLC microcentrifuge filters (Corning Inc., Corning, NY) and centrifuged at 16,000 g for 10 min. NADPH was eluted from a reverse-phase HPLC column (4.6 × 100 mm) using an LC20 HPLC system (Shimadzu, Japan) with a buffer system consisting of 100 mmol/l potassium phosphate (pH 6.0) (buffer A) and 100 mmol/l potassium phosphate containing 30% methanol (pH 6.0) (buffer B). The column was eluted with 100% buffer A from 0 to 5 min, 80% buffer A plus 20% buffer B from 5 to 10 min, and 100% buffer B from 10 to 15 min. Ultraviolet absorbance was monitored at 340 nm.
RESULTS
Characterization of Gene Dose-Response Model for Nrf2 Activation
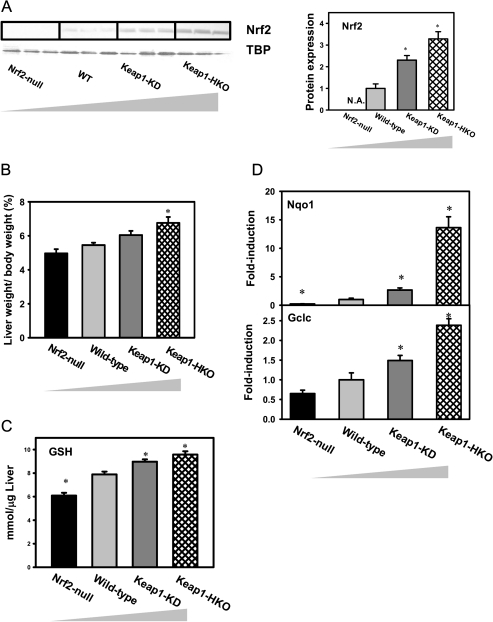
To confirm differential Nrf2 activation of the gene dose-response model, nuclear Nrf2 concentration, relative liver weight, hepatic GSH concentration, as well as messenger RNA (mRNA) expression of known Nrf2 target genes were characterized in livers of Nrf2-null, wild-type, Keap1-KD, and Keap1-HKO mice. Nuclear Nrf2 protein was undetectable in Nrf2-null mice, 1.4-fold higher in Keap1-KD mice, and 2.3-fold higher in Keap1-HKO mice than wild-type mice (Fig. 1A). The relative liver weight in Nrf2-null mice tended to be lower than wild-type mice but not statistically significant. The relative liver weight in Keap1-KD and Keap1-HKO mice was 11 and 24% higher than in wild-type mice, respectively (Fig. 1B). GSH concentrations in livers of Nrf2-null mice were 18% lower than in wild-type mice. In livers of Keap1-KD and Keap1-HKO mice, GSH concentrations were 7 and 14% higher than wild-type mice, respectively (Fig. 1C). Compared with wild-type mice, mRNA expression of Nqo1, a prototypical Nrf2 target gene, was 88% lower in Nrf2-null mice and 167 and 1262% higher in Keap1-KD and Keap1-HKO mice, respectively (Fig. 1D, upper panel). Compared with wild-type mice, mRNA of Gclc, another known Nrf2 target gene, was 36% lower in Nrf2-null mice and 49 and 138% higher in Keap1-KD and Keap1-HKO mice, respectively (Fig. 1D, lower panel). In summary, Nrf2 activation was low in Nrf2-null mice, normal in wild-type mice, enhanced in Keap1-KD mice, and greatest in Keap1-HKO mice.
FIG. 1.
Characterization of “gene dose-response” model. (A) Nuclear Nrf2 protein levels by Western blot analysis, (B) relative liver weight, (C) hepatic GSH concentrations by UPLC-MS/MS analysis, and (D) mRNA concentrations of known Nrf2 target genes by RT-PCR analysis in livers of Nrf2-null, wild-type, Keap1-KD, and Keap1-HKO mice. Data are presented as mean ± SEM of three animals per group for A and five animals per group for B–D. Asterisks (*) indicate statistically significant differences from wild-type mice (p < 0.05).
Transcriptional Profiling of Gene Dose-Response Model
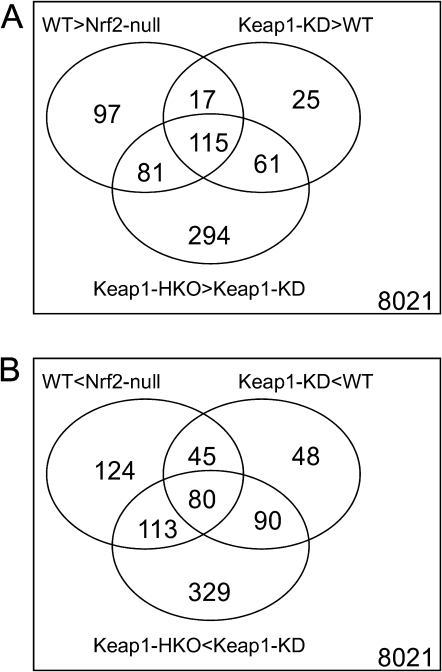
To investigate the effect of Nrf2 on gene transcription profiles in livers of mice, genomic mRNA expression was characterized by microarray. With approximately 45,101 probes, 8021 were detected with signal intensity higher than the threshold and were selected for further analysis. In the pool of 8021 probe sets, 1519 probes were identified to be differentially expressed among the gene dose-response groups based on LIMMA. Specifically, 310 probe sets were expressed higher in wild-type mice than in Nrf2-null mice, 294 were higher in Keap1-KD mice than in wild-type mice, and 550 were higher in Keap1-HKO mice than Keap1-KD mice. Focusing on the intersection of the three sets, 115 probes were identified to have dose-response behavior and were constitutively induced by Nrf2. In contrast, the signal intensity of 362 probe sets were lower in wild-type mice than in Nrf2-null mice, 263 were lower in Keap1-KD mice than in wild-type mice, and 612 were lower in Keap1-HKO mice than Keap1-KD mice. Focusing on the intersection of the three sets, 80 probe sets were identified to have dose-response behavior and were constitutively suppressed by Nrf2 (Fig. 2).
FIG. 2.
Transcriptional profiling of “gene dose-response” model to reveal Nrf2-dependent transcriptional targets. (A) Venn diagram of probe sets that had higher expression in wild-type mice than in Nrf2-null mice (WT > Nrf2-null), in Keap1-KD mice than in wild-type mice (Keap1-KD > WT), and in Keap1-HKO mice than in Keap1-KD mice (Keap1-HKO > Keap1-KD). (B) Venn diagram of probe sets that had lower expression in wild-type mice than in Nrf2-null mice (WT < Nrf2-null), in Keap1-KD mice than in wild-type mice (Keap1-KD < WT), and in Keap1-HKO mice than in Keap1-KD mice (Keap1-HKO < Keap1-KD).
Pathway Analysis of Genes Constitutively Induced or Suppressed with Nrf2 Activation
To categorize the functions of genes induced or suppressed by Nrf2, constitutively upregulated and downregulated probes sets were submitted separately to IPA and the DAVID. From the list of probe sets that were upregulated by Nrf2, core analysis using IPA revealed enrichment of Nrf2-mediated oxidative stress response, glutathione metabolism, and xenobiotic metabolism pathways (Table 1, upper panel). Downregulated pathways included PPARα/RXRα activation (Table 1, lower panel). Annotation clustering using DAVID analysis revealed enrichment of glutathione transferases, NADPH-mediated oxidation reduction, carboxylesterase, chemical and iron homeostasis, and cofactor biosynthesis in genes induced by Nrf2. The only annotation cluster enriched in genes suppressed by Nrf2 was involved in response to extracellular stimulus and nutrient (Table 2).
TABLE 1.
Functional Clustering of Genes Constitutively Induced or Suppressed with Nrf2 Activation Using IPA
| Category | Counta | %b | p Valuec |
| Clusters of genes induced through Nrf2 activation | |||
| Nrf2-mediated oxidative stress response | 14 | 7.7 | 5.84 × 10−12 |
| Glutathione metabolism | 10 | 10 | 2.78 × 10−11 |
| Xenobiotic metabolism signaling | 15 | 5.2 | 6.58 × 10−10 |
| Clusters of genes suppressed through Nrf2 activation | |||
| PPARα/RXRα activation | 4 | 2.2 | 5.05 × 10−3 |
Note. A canonical pathway–cased analysis was conducted using the IPA (Ingenuity Systems, www.ingenuity.com) to identify the pathways enriched in genes constitutively induced or suppressed with Nrf2 activation.
Probe number in a specific pathway.
The ratio of the number of genes that were differentially regulated by Nrf2 and map to the pathway, divided by the total number of genes that map to the canonical pathway, was calculated.
Fisher's exact test was used to calculate the p value determining that the probability of the association between the genes differentially regulated by Nrf2 and the canonical pathway was random.
TABLE 2.
Annotation Clustering of Genes Constitutively Induced or Suppressed with Nrf2 Activation Using DAVID Analysis
| Term | Enrichment score | Gene symbols |
| Top annotation cluster for the set of genes induced through Nrf2 activation | ||
| Glutathione transferase and glutathione metabolism | 8.2 | Gstm1, Gss, Gsta2, Gstm2, Gsta3, Gstm3, Gclc, Gstm4, Gsta4, Gpx4, Gstm6 |
| Oxidation reduction and NADPH bind enzymes | 6.0 | Cyp2g1, Xdh, Ptgr1, Htatip2, Ugdh, Coq7, Fth1, Akr1c13, Akr1a4, Cryl1, Fmo1, Gpx4, Aox1, Cyp2a5, Txnrd1, Nqo1, Srxn1 |
| Carboxylesterase | 3.9 | Ces2, Ces1, BC015286, Ces5 |
| Chemical and iron homeostasis | 3.2 | Xdh, Ftl1, Gclc, Fech, Ftl2, Hexa, Hexb, Bnip3, Afg3l2, Fth1, Abcg8, Anxa7 |
| Cofactor biosynthetic process | 2.4 | Gss, Fech, Gclc, Coq7, Gch1 |
| Top annotation cluster for the set of genes suppressed through Nrf2 activation | ||
| Response to nutrient and extracellular stimulus | 2.3 | Avpr1a, Adipor2, Cp, Apom, Cbs |
Note. DAVID (http://david.abcc.ncifcrf.gov) database was used to identify the pathways enriched in genes constitutively induced or suppressed with Nrf2 activation. Pathway enrichment was determined by the enrichment score > 2.
Regulation of Antioxidant System Genes by Nrf2
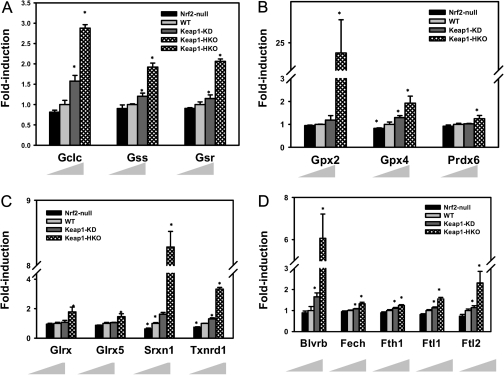
Figure 3 summarizes the antioxidant-associated enzymes that were significantly increased in the gene dose-response model. As shown in Figure 3A, Nrf2 activation increased transcription of genes involved in GSH biosynthesis (glutamate-cysteine ligase, Gclc and glutathione synthase, Gss) as well as GSH regeneration (glutathione reductase, Gsr). Compared with wild-type mice, mRNA of Gclc was induced 58% in Keap1-KD mice and 210% in Keap1-HKO mice and mRNA of Gss was induced 18% in Keap1-KD mice and 91% in Keap1-HKO mice. mRNA of Gsr increased 110% in Keap1-HKO mice over wild-type mice. Figure 3B indicates that Nrf2 activation increased the mRNA of genes involved in reduction of superoxide using GSH (glutathione peroxidase: Gpx2 and Gpx4) as well as using NADPH (peroxiredoxin, Prdx6). Especially, mRNA of Gpx2 and Gpx4 were increased in Keap1-HKO mice by 2260 and 105%, respectively. Figure 3C shows that Nrf2 activation increased transcription of genes involved in reduction of disulfide bonds between protein and GSH (glutaredoxin: Glrx1 and Glrx5), disulfide bonds with oxidized protein (sulfiredoxin: Srxn1), and disulfide bonds in oxidized thioredoxin (thioredoxin reductase: Txnrd1). The mRNA of Srxn1 increased 743% and Txnrd1 221% in Keap1-HKO mice. Figure 3D shows that Nrf2 activation increased transcription of genes involved in heme metabolism (biliverdin reductase B: Blvrb and ferrochelatase: Fech) and free ion storage (ferritin heavy chain: Fth1 and ferritin light chain: Ftl1 and Ftl2). In particular, the mRNA of Blvrb was increased in Keap1-HKO mice by 270% and Ftl2 171%.
FIG. 3.
Antioxidant genes that were induced in “gene dose-response” model. mRNA expression of genes involve in (A) GSH synthesis and regeneration, (B) reduction of protein disulfide bond, (C) reduction of hydrogen peroxide, and (D) bilirubin synthesis and ion storage in gene dose-response model. Data are presented as mean ± SEM of three animals per group. Asterisks (*) indicate statistically significant differences from wild-type mice (p < 0.05).
Regulation of NADPH Production Genes by Nrf2
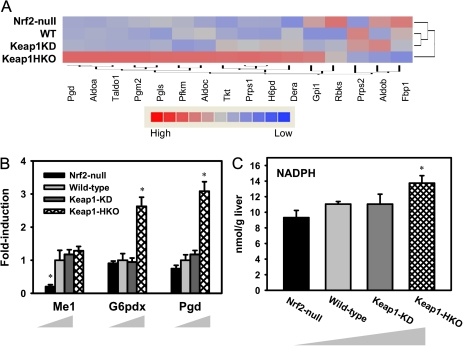
Figure 4 summarizes the effect of Nrf2 in producing NADPH in the Nrf2 gene dose-response model. The pentose phosphate pathway produces NADPH and generates 90% of the NADPH in the body. Figure 4A shows hierarchical clustering of genes involved in the pentose phosphate pathway. Of the 16 genes examined, 12 were highly expressed in Keap1-HKO mice, and only one gene was highly expressed in Nrf2-null mice. In addition, the expression of the gene that encodes malic enzyme (Me1) was low in Nrf2-null mice and high in wild-type, Keap1-KD, and Keap1-HKO mice (Fig. 4B). To validate the role of Nrf2 in increasing NADPH production, hepatic NADPH concentrations were determined by high performance liquid chromatography-ultraviolet absorbance (HPLC-UV) analysis. As shown in Figure 4C, NADPH concentrations tended to be lower in livers of Nrf2-null mice and were 24% higher in Keap1-HKO mice than wild-type mice.
FIG. 4.
NADPH production was induced in “gene dose-response” model. (A) Hierarchical clustering of genes involved in pentose phosphate pathway. Shading is based on the fold induction of mRNA of each gene compared with the wild-type mice. Genes involved in pentose phosphate pathway were selected according to KEGG category (KEGG: 00030). (B) mRNA expression of Me1, G6pdx, and Pgd by RT-PCR analysis and (C) hepatic NADPH concentration by HPLC-UV analysis in gene dose-response model. Data are presented as mean ± SEM three animals per group for A and five animals per group for B and C. Asterisks (*) indicate statistically significant differences from wild-type mice (p < 0.05).
Regulation of Lipid Biosynthesis Genes by Nrf2
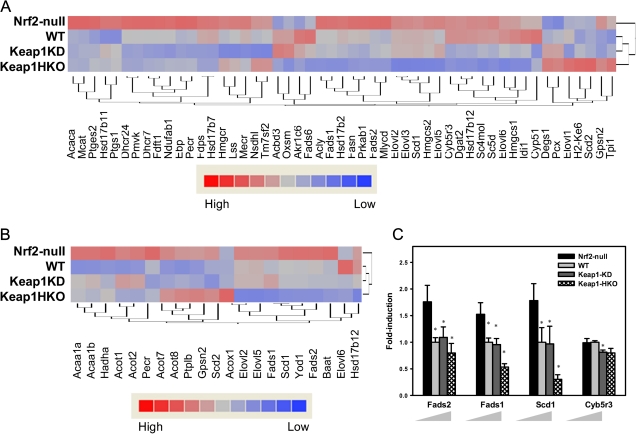
Figure 5A indicates hierarchical clustering of genes involved in lipid biosynthesis. Of 52 genes examined, 36 genes were expressed at higher levels in Nrf2-null mice and at lower levels in Keap1-HKO mice. In contrast, seven genes were expressed at higher levels in Keap1-HKO mice and lower levels in Nrf2-null mice. Specifically, Figure 5B shows hierarchical clustering of genes involved in fatty acid synthesis. Of 21 genes examined, 14 genes were expressed at higher levels in Nrf2-null mice and lower levels in Keap1-HKO mice. In contrast, only one gene was expressed at lower level in Nrf2-null mice and at higher level in Keap1-HKO mice. Figure 5C shows that genes involved in fatty acid desaturation were suppressed with Nrf2 activation. Fatty acid desaturase (Fads1 and Fads2) and stearoyl-CoA desaturase (Scd1) are desaturases that catalyze the addition of a double bond utilizing NADPH as the cofactor. The abundance of Fads1 mRNA was 54% higher in Nrf2-null mice and 46% lower in Keap1-HKO mice than wild-type mice. Similarly, the abundance of Fads2 mRNA was 52% higher in Nrf2-null mice and 30% lower in Keap1-HKO mice than wild-type mice and the abundance of Scd1 mRNA was 81% higher in Nrf2-null mice and 73% lower in Keap1-HKO mice than wild-type mice.
FIG. 5.
Lipid biosynthesis and fatty acid desaturation were suppressed in “gene dose-response” model. (A) Hierarchical clustering of genes involved in lipid biosynthesis. Genes were selected according to Gene Ontology category (GO:0008610). (B) Hierarchical clustering of genes involved in fatty acid biosynthesis. Shading is based on the fold induction of mRNA of each gene compared with the wild-type mice. Genes were selected according to Gene Ontology category (GO:0006633). (C) mRNA expression of genes involved in fatty acid desaturation. Data are presented as mean ± SEM of three animals per group. Asterisks (*) indicate statistically significant differences from wild-type mice (p < 0.05).
DISCUSSION
Liver is the primary organ in regard to multiple vital functions, including protein synthesis (e.g., albumin and clotting factors), glucose homeostasis, and lipid metabolism. Liver also plays the major role in detoxication of environmental chemicals, drugs, and endogenous toxicants. Nrf2 is the central regulator for increasing expression of genes that protect against oxidative and electrophilic stress. Thus, studying the function of Nrf2 in liver is of great importance. The present study used a gene dose-response model characterized by graded Nrf2 activation in livers of mice to systematically study the functions of Nrf2 in liver.
Continuous efforts have been made to identify Nrf2 target genes and thus the function of Nrf2. However, varied conclusions have been drawn using different models. For example, by comparing transcription profiles of the prostate of control mice and mice treated with soy isoflavones, which are natural Nrf2 inducers, Nrf2 was shown to be primarily involved in induction of phase I and phase II drug metabolism genes (Barve et al., 2008). By comparing transcription profiles of lung tissues in wild-type mice and Clara-cell-specific Keap1-KO mice, Nrf2 was shown to be important in induction of antioxidant genes (Blake et al., 2010). Chromatin-immunoprecipitation (ChIP-seq) of Nrf2 binding sites utilizing parallel sequencing of mouse embryonic fibroblasts of Nrf2-null mice and wild-type mice showed that Nrf2 is essential for basal expression of genes involved in cell proliferation (Malhotra et al., 2010). In the same study, by comparing mouse embryonic fibroblasts of wild-type mice and Keap1-null mice, genes induced by Nrf2 were enriched in antioxidative stress pathways. A proteomic expression profile of livers of Nrf2-null mice and wild-type mice found that the most profound changes were observed in proteins involved in lipid catabolism (Kitteringham et al., 2010). In the present study, by comparing genomic gene transcription profile in livers utilizing a gene dose-response model, it was determined that Nrf2-inducible genes are most enriched in protection against oxidative stress, glutathione metabolism, and xenobiotic metabolism. The various observations in these various studies may result from different biological samples used for comparison (cell lines vs. animal tissues) or different models to modulate Nrf2 activation (pharmacological vs. genetic).
With few exceptions, previously identified Nrf2 target genes involved in protection against oxidative stress were identified to be inducible in the gene dose-response model. GSH is a tripeptide that serves as the predominant cellular thiol resource and protects against oxidative and electrophilic stress by either directly scavenging reactive oxygen species or conjugating with electrophiles. It is well documented that Nrf2 activation can increase GSH content in both cell line models (Gao et al., 2010) and animal models (Reisman et al., 2009a). Consistent with the previous study, hepatic GSH concentrations increased in the gene dose-response model in a dose-dependent manner (Fig. 1C). GSH is mainly synthesized in liver, by a two-step reaction. The first and rate-limiting step is catalyzed by glutamate-cysteine ligase (Gcl), which is made up of a catalytic and a modifier subunit (Gclc and Gclm), that combine glutamate and cysteine into a dipeptide. The second step is catalyzed by glutathione synthase (Gss), which adds a glycine to the glutamate-cysteine dipeptide. In addition to de novo biosynthesis, GSH can be regenerated from GSSG by GSH reductase (Gsr). It is known that Gclc, Gss, and Gsr are Nrf2 target genes (Chan et al., 2001; Li et al., 2009), and the present study also shows that these genes were induced by Nrf2 in the gene dose-response model (Fig. 3A). Once hydrogen peroxide is formed, it can be detoxified through peroxiredoxin (Prdx) or reduced by GSH through GSH peroxidase (Gpx). In the present study, Prdx6, Gpx2, and Gpx4 were increased with Nrf2 activation (Fig. 3B). Similarly, induction of Gpx2 and Prdx6 are reported to be induced by Nrf2 in lung (Blake et al., 2010), and inorganic arsenic activated Nrf2 and increased Gpx4 expression in mouse embryonic stem cells (Chan et al., 2001). Failure to scavenge reactive oxygen species leads to oxidation of proteins and abnormal enzyme activity. Oxidized thiol groups in proteins can be reduced by glutaredoxin (Glrx) and thioredoxin. Oxidized thioredoxin can be regenerated by thioredoxin reductase (Txnrd), and oxidized peroxireduction can be regenerated by sulfiredoxin (Srxn). In the present study, Glrx1, Glrx5, Srxn1, and Txnrd1 were induced with Nrf2 activation, whereas only mRNA of Srxn1 and Txnrd1 were decreased in the absence of Nrf2 (Fig. 3C).
Bilirubin is another endogenous antioxidant and has been shown to efficiently scavenge peroxyl radicals (Stocker et al., 1987). Heme degradation generates biliverdin, and bilverdin reductase (Blvrb) converts the pro-oxidant biliverdin to the antioxidant bilirubin. Ferrochelatase (Fech) catalyzes the final step of heme biosynthesis. Both Blvrb and Fech are induced in the gene dose-response model (Fig. 3D), suggesting a role for Nrf2 in bilirubin metabolism. Ferritin is an iron-sequestering protein that prevents uncontrolled surges in the free intracellular concentration of ferric iron (Goven et al., 2010). The reduction of ferric ion by superoxide can generate reactive hydroxyl radicals via the Fenton reaction. Ferritin is composed of ferritin heavy chains (Fth) and ferritin light chains (Ftl). Fth1, Ftl1, and Ftl2 are induced by Nrf2 (Fig. 4), indicating an important role of Nrf2 in ferric ion scavenging.
The central role of Nrf2 in inducing genes encoding enzymes that directly synthesize GSH and scavenge oxidative stress is well recognized, however, its role in promoting NADPH production has received less attention. NADPH serves as the major reducing resource in the body, and many oxidoreduction reactions, including reducing oxidized GSH and thioredoxin, are performed by oxidizing NADPH into NADP+. Most of the cellular NADPH is generated by the pentose phosphate pathway, and small amounts of NADPH are generated by malic enzyme. In an earlier study, microarray analysis comparing intestinal gene expression profile between control mice and mice treated with sulforaphane identified glucose 6-phosphate dehydrogenase (G6pd), 6-phosphogluconate dehydrogenase (Pgd), and malic enzyme (Me1) as Nrf2 target genes and linked NADPH production to the Nrf2 signal pathway (Kirby et al., 2005). The effect of Nrf2 on these NADPH generating enzymes was later confirmed in mouse lung (Goven et al., 2010) and liver (Reisman et al., 2009c). Consistent with these observations, Pgd and Me1 were induced in mouse liver with Nrf2 activation in the present study. More importantly, the present study shows that the majority of genes encoding enzymes involved in the pentose phosphate pathway are induced by Nrf2. The induction of the mRNA of multiple genes, including Aldoa (aldolase A, fructose-bisphosphate), Taldo1 (transaldolase 1), Tkt (transketolase), Prps1 (phosphoribosyl pyrophosphate synthetase 1), and Dera (putative deoxyribose phosphate aldolase) strongly correlated with Nrf2 activation in the gene dose-response model (Fig. 4A), indicating these genes are novel Nrf2 target genes. To further confirm the role of Nrf2 in promoting NADPH production, hepatic NADPH concentrations were quantified in the gene dose-response model. Nrf2-null mice have less, whereas Keap1-HKO mice have more hepatic NADPH than wild-type mice (Fig. 4C). Higher NADPH concentrations in the Keap1-HKO mouse increase the availability of the cellular reducing resource. In contrast, lower NADPH concentrations render Nrf2-null mice less capable of reducing oxidative stress.
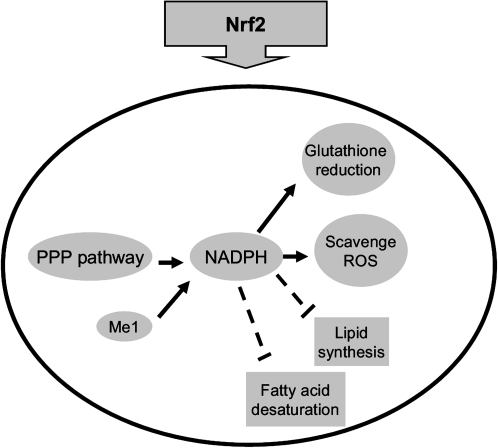
In contrast to the role of Nrf2 in reducing oxidative stress, conflicting data have been reported on the role of Nrf2 in lipid metabolism. One study reported that pharmacological activation of Nrf2 by CDDO-Im effectively prevented high-fat diet-induced increases in body weight, adipose mass, and hepatic lipid accumulation in wild-type mice but not in Nrf2-disrupted mice, indicating that the Nrf2 pathway is a novel target for the prevention of obesity (Shin et al., 2009). In contrast, another study showed that Nrf2-null mice have lower adipose tissue mass, greater formation of small adipocytes, and protects against weight gain and obesity by a high-fat diet (Pi et al., 2010), indicating that intact Nrf2 signaling facilitates obesity. Proteomic analysis comparing livers of wild-type mice and Nrf2-null mice showed that the majority of proteins upregulated in Nrf2-null mice are primarily involved in lipid metabolism with particular respect to lipogenesis (Kitteringham et al., 2010). Consistent with this, the present study shows that most genes involved in lipid biosynthesis were induced in Nrf2-null mice, and suppressed in Nrf2 activated mice, suggesting that Nrf2 negatively regulates lipogenesis (Fig. 5A). In addition, the mRNA expression of most genes involved in fatty acid synthesis and desaturation are much higher in Nrf2-null mice than the other three genotypes of mice, indicating that Nrf2 strongly suppresses the basal level of fatty acid synthesis and desaturation. It should be noted that NADPH, the major reducing power in the body, is used to reduce oxidative stress and provides the reducing equivalents for biosynthetic reactions, including lipid synthesis and fatty acid desaturation (Kuhajda et al., 1994; Shimomura et al., 1998). Specifically, expression of Fasn (fatty acid synthase) and Scd1 (stearoyl-CoA desaturase), which encode enzymes that use NADPH as the cofactor (Kuhajda et al., 1994; Ntambi et al., 1988), are suppressed by Nrf2 in a dose-dependent manner. Thus, Nrf2 regulation favors the use of NADPH to reduce oxidative stress rather than the biosynthesis of lipids (Fig. 6).
FIG. 6.
Proposed regulatory model for the role of Nrf2 in NADPH production and consumption. In Nrf2 activated cells, NADPH generation was promoted through pentose phosphate pathway and malic enzyme. Genes involved in antioxidant defense by using NADPH were induced and genes involved in lipid biosynthesis by using NADPH were suppressed.
In conclusion, the present study shows that Nrf2 induces expression of genes involved in antioxidant defense as well as NADPH generation. In addition, the absence of Nrf2 results in decreased expression of genes involved in lipid synthesis and fatty acid desaturation. Thus, Nrf2 activation increases the availability of NADPH for reducing oxidative stress decrease NADPH consumption for lipid biosynthesis.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci. oxfordjournals.org/.
FUNDING
National Institutes of Health (DK-081461, ES-09716, and RR-021940).
Supplementary Material
Acknowledgments
The authors would like to thank all the graduate students and postdoctoral fellows for technical support of the experiments as well as the critical revision of the manuscript. Nrf2-null mice were graciously provided by Dr Jefferson Chan (University of California-Irvine, Irvine, CA) and Keap1-kd mice by Dr Masayuki Yamamoto (Tohoku University, Aoba-ku, Sendai, Japan). Portions of this work were presented at the national meeting of the Society of Toxicology for which K.C.W. received the Carl C. Smith Second Place Award, 7–10 March 2011.
References
- Aleksunes LM, Manautou JE. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol. Pathol. 2007;35:459–473. doi: 10.1080/01926230701311344. [DOI] [PubMed] [Google Scholar]
- Anand P, Sundaram C, Jhurani S, Kunnumakkara AB, Aggarwal BB. Curcumin and cancer: an “old-age” disease with an “age-old” solution. Cancer Lett. 2008;267:133–164. doi: 10.1016/j.canlet.2008.03.025. [DOI] [PubMed] [Google Scholar]
- Barve A, Khor TO, Nair S, Lin W, Yu S, Jain MR, Chan JY, Kong AN. Pharmacogenomic profile of soy isoflavone concentrate in the prostate of Nrf2 deficient and wild-type mice. J. Pharm. Sci. 2008;97:4528–4545. doi: 10.1002/jps.21311. [DOI] [PubMed] [Google Scholar]
- Baur JA. Biochemical effects of SIRT1 activators. Biochim. Biophys. Acta. 2010;1804:1626–1634. doi: 10.1016/j.bbapap.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake DJ, Singh A, Kombairaju P, Malhotra D, Mariani TJ, Tuder RM, Gabrielson E, Biswal S. Deletion of Keap1 in the lung attenuates acute cigarette smoke-induced oxidative stress and inflammation. Am. J. Respir. Cell Mol. Biol. 2010;42:524–536. doi: 10.1165/rcmb.2009-0054OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto A, Itoh K, Nagayoshi E, Haruta J, Kimura T, O'Connor T, Harada T, Yamamoto M. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 2001;59:169–177. doi: 10.1093/toxsci/59.1.169. [DOI] [PubMed] [Google Scholar]
- Gao SS, Choi BM, Chen XY, Zhu RZ, Kim Y, So H, Park R, Sung M, Kim BR. Kaempferol suppresses cisplatin-induced apoptosis via inductions of heme oxygenase-1 and glutamate-cysteine ligase catalytic subunit in HEI-OC1 cell. Pharm. Res. 2010;27:235–245. doi: 10.1007/s11095-009-0003-3. [DOI] [PubMed] [Google Scholar]
- Goven D, Boutten A, Lecon-Malas V, Marchal-Somme J, Soler P, Boczkowski J, Bonay M. Induction of heme oxygenase-1, biliverdin reductase and H-ferritin in lung macrophage in smokers with primary spontaneous pneumothorax: role of HIF-1alpha. PLoS One. 2010;5:e10886. doi: 10.1371/journal.pone.0010886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Hoffman B, Dwivedi C, Matthees DP. A simultaneous liquid chromatography/mass spectrometric assay of glutathione, cysteine, homocysteine and their disulfides in biological samples. J. Pharm. Biomed. Anal. 2003;31:251–261. doi: 10.1016/s0731-7085(02)00594-0. [DOI] [PubMed] [Google Scholar]
- Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J. Biol. Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Ji Y, Lee HJ, Goodman C, Uskokovic M, Liby K, Sporn M, Suh N. The synthetic triterpenoid CDDO-imidazolide induces monocytic differentiation by activating the Smad and ERK signaling pathways in HL60 leukemia cells. Mol. Cancer Ther. 2006;5:1452–1458. doi: 10.1158/1535-7163.MCT-06-0136. [DOI] [PubMed] [Google Scholar]
- Jones DP. Determination of pyridine dinucleotides in cell extracts by high-performance liquid chromatography. J. Chromatogr. 1981;225:446–449. doi: 10.1016/s0378-4347(00)80293-5. [DOI] [PubMed] [Google Scholar]
- Kirby J, Halligan E, Baptista MJ, Allen S, Heath PR, Holden H, Barber SC, Loynes CA, Wood-Allum CA, Lunec J, et al. Mutant SOD1 alters the motor neuronal transcriptome: implications for familial ALS. Brain. 2005;128:1686–1706. doi: 10.1093/brain/awh503. [DOI] [PubMed] [Google Scholar]
- Kitteringham NR, Abdullah A, Walsh J, Randle L, Jenkins RE, Sison R, Goldring CE, Powell H, Sanderson C, Williams S, et al. Proteomic analysis of Nrf2 deficient transgenic mice reveals cellular defence and lipid metabolism as primary Nrf2-dependent pathways in the liver. J. Proteomics. 2010;73:1612–1631. doi: 10.1016/j.jprot.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Kuhajda FP, Jenner K, Wood FD, Hennigar RA, Jacobs LB, Dick JD, Pasternack GR. Fatty acid synthesis: a potential selective target for antineoplastic therapy. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamle J, Marhenke S, Borlak J, von Wasielewski R, Eriksson CJ, Geffers R, Manns MP, Yamamoto M, Vogel A. Nuclear factor-erythroid 2-related factor 2 prevents alcohol-induced fulminant liver injury. Gastroenterology. 2008;134:1159–1168. doi: 10.1053/j.gastro.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Li M, Chiu JF, Kelsen A, Lu SC, Fukagawa NK. Identification and characterization of an Nrf2-mediated ARE upstream of the rat glutamate cysteine ligase catalytic subunit gene (GCLC) J. Cell. Biochem. 2009;107:944–954. doi: 10.1002/jcb.22197. [DOI] [PubMed] [Google Scholar]
- Luo Y, Eggler AL, Liu D, Liu G, Mesecar AD, van Breemen RB. Sites of alkylation of human Keap1 by natural chemoprevention agents. J. Am. Soc. Mass Spectrom. 2007;18:2226–2232. doi: 10.1016/j.jasms.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell MD, Jackson JP, Augustine LM, Fisher CD, Slitt AL, Maher JM, Huang W, Moore DD, Zhang Y, Klaassen CD, et al. The Nrf2 activator oltipraz also activates the constitutive androstane receptor. Drug Metab. Dispos. 2008;36:1716–1721. doi: 10.1124/dmd.108.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New LS, Chan EC. Evaluation of BEH C18, BEH HILIC, and HSS T3 (C18) column chemistries for the UPLC-MS-MS analysis of glutathione, glutathione disulfide, and ophthalmic acid in mouse liver and human plasma. J. Chromatogr. Sci. 2008;46:209–214. doi: 10.1093/chromsci/46.3.209. [DOI] [PubMed] [Google Scholar]
- Ntambi JM, Buhrow SA, Kaestner KH, Christy RJ, Sibley E, Kelly TJ, Jr, Lane MD. Differentiation-induced gene expression in 3T3-L1 preadipocytes. Characterization of a differentially expressed gene encoding stearoyl-CoA desaturase. J. Biol. Chem. 1988;263:17291–17300. [PubMed] [Google Scholar]
- Okada K, Shoda J, Taguchi K, Maher JM, Ishizaki K, Inoue Y, Ohtsuki M, Goto N, Takeda K, Utsunomiya H, et al. Ursodeoxycholic acid stimulates Nrf2-mediated hepatocellular transport, detoxification, and antioxidative stress systems in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;295:G735–G747. doi: 10.1152/ajpgi.90321.2008. [DOI] [PubMed] [Google Scholar]
- Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem. Biophys. Res. Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- Pi J, Leung L, Xue P, Wang W, Hou Y, Liu D, Yehuda-Shnaidman E, Lee C, Lau J, Kurtz TW, et al. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J. Biol. Chem. 2010;285:9292–9300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman SA, Aleksunes LM, Klaassen CD. Oleanolic acid activates Nrf2 and protects from acetaminophen hepatotoxicity via Nrf2-dependent and Nrf2-independent processes. Biochem. Pharmacol. 2009a;77:1273–1282. doi: 10.1016/j.bcp.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman SA, Buckley DB, Tanaka Y, Klaassen CD. CDDO-Im protects from acetaminophen hepatotoxicity through induction of Nrf2-dependent genes. Toxicol. Appl. Pharmacol. 2009b;236:109–114. doi: 10.1016/j.taap.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman SA, Yeager RL, Yamamoto M, Klaassen CD. Increased Nrf2 activation in livers from Keap1-knockdown mice increases expression of cytoprotective genes that detoxify electrophiles more than those that detoxify reactive oxygen species. Toxicol. Sci. 2009c;108:35–47. doi: 10.1093/toxsci/kfn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Shimano H, Korn BS, Bashmakov Y, Horton JD. Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J. Biol. Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]
- Shin S, Wakabayashi J, Yates MS, Wakabayashi N, Dolan PM, Aja S, Liby KT, Sporn MB, Yamamoto M, Kensler TW. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur. J. Pharmacol. 2009;620:138–144. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R, Yamamoto Y, McDonagh AF, Glazer AN, Ames BN. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- Taguchi K, Maher JM, Suzuki T, Kawatani Y, Motohashi H, Yamamoto M. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol. Cell. Biol. 2010;30:3016–3026. doi: 10.1128/MCB.01591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Aleksunes LM, Cui YJ, Klaassen CD. ANIT-induced intrahepatic cholestasis alters hepatobiliary transporter expression via Nrf2-dependent and independent signaling. Toxicol. Sci. 2009;108:247–257. doi: 10.1093/toxsci/kfp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Aleksunes LM, Yeager RL, Gyamfi MA, Esterly N, Guo GL, Klaassen CD. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J. Pharmacol. Exp. Ther. 2008;325:655–664. doi: 10.1124/jpet.107.135822. [DOI] [PubMed] [Google Scholar]
- Umemura T, Kuroiwa Y, Kitamura Y, Ishii Y, Kanki K, Kodama Y, Itoh K, Yamamoto M, Nishikawa A, Hirose M. A crucial role of Nrf2 in in vivo defense against oxidative damage by an environmental pollutant, pentachlorophenol. Toxicol. Sci. 2006;90:111–119. doi: 10.1093/toxsci/kfj076. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- Woods CG, Fu J, Xue P, Hou Y, Pluta LJ, Yang L, Zhang Q, Thomas RS, Andersen ME, Pi J. Dose-dependent transitions in Nrf2-mediated adaptive response and related stress responses to hypochlorous acid in mouse macrophages. Toxicol. Appl. Pharmacol. 2009;238:27–36. doi: 10.1016/j.taap.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Zhang X, Fan H, Liu Y. Curcumin upregulates transcription factor Nrf2, HO-1 expression and protects rat brains against focal ischemia. Brain Res. 2009;1282:133–141. doi: 10.1016/j.brainres.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Zhang H, Shih A, Rinna A, Forman HJ. Resveratrol and 4-hydroxynonenal act in concert to increase glutamate cysteine ligase expression and glutathione in human bronchial epithelial cells. Arch. Biochem. Biophys. 2009;481:110–115. doi: 10.1016/j.abb.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.