Abstract
The role of trivalent arsenic (As3+) on the regulation of the recently identified noncoding small RNAs, mainly microRNAs, has not been explored so far. In the present study, we provide evidence showing that As3+ is a potent inducer for the expression of miR-190 in human bronchial epithelial cells. The induction of miR-190 by As3+ is concentration dependent and associated with the expression of the host gene of miR-190, talin 2, a gene encoding a high-molecular-weight cytoskeletal protein. The elevated level of miR-190 induced by As3+ is capable of downregulating the translation of the PH domain leucine-rich repeat protein phosphatase (PHLPP), a negative regulator of Akt signaling. Such a downregulation is occurred through direct interaction of the miR-190 with the 3′-UTR region of the PHLPP mRNA, leading to a diminished PHLPP protein expression and consequently, an enhanced Akt activation and expression of vascular endothelial growth factor, an Akt-regulated protein. Overexpression of miR-190 itself is able to enhance proliferation and malignant transformation of the cells as determined by anchorage-independent growth of the cells in soft agar. Accordingly, the data presented suggest that induction of miR-190 is one of the key mechanisms in As3+-induced carcinogenesis.
Keywords: arsenite, microRNA, Akt, carcinogenesis
Environmental or occupational exposure to arsenic, especially the trivalent inorganic arsenic (As3+), is continually a major public health concern in a number of countries worldwide, including the United States (Heck et al., 2009). In the southwestern region of the United States, As3+ levels in the groundwater of some areas ranged from 680 to 1880 μg/l (∼9 to 25μM) (Camacho et al., 2011). Several population-based case-control studies suggested a strong association of As3+ exposure with small and nonsmall cell lung cancers (Heck et al., 2009) and other cancers (Celik et al., 2008). It is particularly important to note that not only high levels of As3+ exposure but also low to moderate levels of As3+ exposure in certain regions of the world have been linked to an increased risk of lung cancer. The International Agency for Research on Cancer has classified As3+ as a group I carcinogen for lung cancer. In addition, several ecological and cohort studies also confirmed As3+ contribution to human lung cancers (Chen et al., 2004; Chiu et al., 2004; Hopenhayn-Rich et al., 1998). Thus, there is an urgent need to understand how lower dose and sustained As3+ exposure causes malignant transformation of the lung cells.
Previous studies by our group and others have shown that As3+ activated several kinases upstream of transcription factors, thereby, regulating the expression of genes important for cell cycle transition and cell proliferation (Cavigelli et al., 1996; Chen et al., 2003; Ye et al., 2005). Recently, a group of genes that encode small noncoding RNA, including microRNA (miRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and piwiRNA, have been identified. In this study, we explored the effect of As3+ on the expression of miRNAs.
Human miRNA genes are either located in intergenic areas or in introns of known genes. Under the regulation of RNA polymerase II, primary miRNA (pri-miRNA) is transcribed and then subjected to processing by the RNase enzyme Drosha in the nucleus to form precursor miRNA (pre-miRNA) with hairpin loops about 70 nt in length. The pre-miRNA is then exported to the cytoplasm by the nuclear membrane protein Exportin-5 and further processed by the RNase Dicer to generate mature miRNA. Following association with the miRISC protein complex, one strand of the mature miRNA binds to its complementary 3-UTR region of the target mRNA to repress translation or promote degradation of the target mRNA, depending on the degree of complementary binding (Beezhold et al., 2010).
Emerging evidence suggests that many miRNAs play a pivotal role in malignant transformation and carcinogenesis of cells. Examples of this include the overexpression of miR-155 in pancreatic cancer (Gironella et al., 2007) and lymphomas (Eis et al., 2005) as well as an increase of miR-221/222 in prostate cancer (Galardi et al., 2007). Human miRNA-190 is derived from an intron region of the talin 2 gene on chromosome 15. The proteins of the talin family include talin 1 and talin 2. They share several highly conserved domains, such as the rod domain and the FERM domain, that are important for interaction with F-actin, integrin, PIP5KI isoforms, and focal adhesion molecules (Critchley and Gingras, 2008; Monkley et al., 2001). Earlier data indicate that inhibition of miR-190 decreased growth of HeLa cells without an overall change in the apoptotic caspase activity (Cheng et al., 2005). An additional study shows that the level of miR-190 was increased in granulocytes from patients with primary myelofibrosis (PMF) (Guglielmelli et al., 2007). These results suggest a role for miR-190 in regulating cell cycle or proliferation rate.
In the present report, we demonstrate that As3+ is a potent inducer of human miR-190, correlating with an increased accumulation of its host gene product talin 2 protein. Through repressing expression of the PH domain leucine-rich repeat protein phosphatase (PHLPP), its target gene encoding a negative regulator of Akt signaling, increased level of miR-190 is able to enhance the activation of Akt, leading to an increased expression of vascular endothelial growth factor (VEGF). Overexpression of miR-190 itself is able to enhance proliferation and anchorage-independent growth of the cells in soft agar. Induction of miR-190, therefore, may provide a new mechanistic insight into the carcinogenicity of environmental or occupational exposure to As3+.
MATERIALS AND METHODS
Cell culture and reagents.
BEAS-2B cells were purchased from ATCC (Manassas, VA) and seeded in 6-well plates at a density of 5 × 105 cells/well. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) medium supplemented with 5% fetal bovine serum (FBS) for approximately 48 h before treatment with arsenic chloride (As3+) (Sigma, MO) for 6–12 h (unless otherwise specified). Cells were lysed with TRIzol reagent (Invitrogen, Carlsbad, CA) according to manufacturer's instructions for total RNA isolation and real-time PCR experiments. Antibodies against talin 2, β-catenin, PHLPP, TP53INP1, phospho-Akt, and nonphospho-Akt were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) or Abcam (Cambridge, MA). ELISA kit for the detection of VEGF was purchased from Thermo Scientific (Rockford, IL). Dual-Luciferase Assay Reagents were purchased from Promega (Madison, WI).
Western blotting.
Total cellular proteins were prepared with NuPage LDS sample buffer (Invitrogen) containing 50μM DTT. The cell lysates were run on 6% SDS-PAGE gels and then transferred to polyvinylidene difluoride membranes (Invitrogen). Membranes were first probed with the primary antibodies at a dilution of 1:2000. The secondary antibody with AP tag was then applied at a dilution of 1:5000. Densitometry analysis of the Western blotting data was performed using a Molecular Biosystems scanner (San Diego, CA).
Real-time PCR.
The miRNA array was performed using the Cancer microRNA qPCR Array with QuantiMir from System Biosciences (Mountain View, CA) according to the manufacturer's instruction. RNA was isolated from untreated cells as well as cells treated with 20μM As3+ for 6 h using either TRIzol reagent or a MirVana kit from Ambion (Austin, TX). The real-time PCR primers and probes for the detection of talin 2 mRNA were designed using Roche online ProbeFinder program (http://www.universalprobelibrary.com) by selecting Automatically Select an intron spanning assay and G6PD as the preferred reference gene. The designated amplicon will be specific for talin 2 mRNA only without contamination of the genomic DNA sequences. The primer sequences are: left primer, 5′-TGGTCAAATCGGCCTCAG-3′ and right primer, 5′-GACCACTGTCCGTCGTCTG-3′. Probe #49 was used for the talin 2 real-time PCR. Expression of some individual miRNAs was also analyzed using TaqMan microRNA Assays from Applied Biosystems (Foster City, CA). Reverse transcription and real-time steps were performed according to the manufacturer's specifications from total RNA isolates. Either miR-30c or snoRNA U47 was used as the endogenous control for all real-time PCR experiments of miRNAs.
Transfection of the cells with miRNA mimic.
To validate the role of miR-190 on its target mRNAs, BEAS-2B cells (1 × 105/ml) were seeded in 6-well tissue culture plates and transfected with 50nM pre-miR negative control (mock), miR-190 mimic, or 100nM anti-miR-190 (miR-190In) in triplicates using a reverse transfection procedures as suggested by the manufacturer. The Pre-miR(tm) miRNA Starter Kit that contains chemically modified pre-miR negative control, pre-miR-190 mimic, and pre-miR-190In were purchased from Ambion. The pre-miR negative control used in mock transfection is a pre-miRNA molecule with random sequence that does not target any known mRNA in the human cells. The transfected cells were first cultured in serum-free medium at 37°C for 24 h and then cultured with 5% FBS for an additional 24 h. Various concentrations of As3+ were added 12 h before the end of the culture. Protein levels of potential miR-190 targets were determined by Western blotting as described above.
Reporter vector construction.
To construct a luciferase reporter vector for the talin 2 promoter, the genomic DNA encompassing the 1095 bp human talin 2 promoter region were amplified with PCR primers: left primer, 5′-ATCGGCTAGCCACCATGCCAGGCTAATTTT-3′ and right primer, 5′-CAGTCTCGAGACTCGACACGCATCGTACAC-3′. The left and right primers designed to amplify the region from genomic DNA had the restriction enzyme sites for NheI and XhoI included, respectively, to aid insertion of the cloned region into the vector. The talin 2 promoter fragment was then cloned into the pGL3-basic vector from Promega.
For the construction of the PHLPP 3′-UTR miRNA-targeting reporter vector, the genomic DNA containing a 911 bp region of the human PHLPP 3′-UTR was amplified with left primer, 5′-GATCGAGCTCCAAGAGTCTCCCAGGCTCAC-3′ and right primer, 5′-GCTAAAGCTTTCCATTTGTGCATTCTGCTT-3′. For the purpose of deleting the miR-190-binding site in the 3′-UTR region of the PHLPP mRNA, an additional left primer with sequence 5′-GTCAGAGCTCATGTAAAGACAAAGAACAAAAGGTTTA-3′ was introduced in a separate PCR reaction, which generates a deleted 3′-UTR by removing the sequence containing the binding site of miR-190. The amplified fragments were cloned into the pMIR-REPORT vector (Ambion) utilizing the SacI and HindIII restriction enzyme sites. Chemically competent Escherichia coli (Invitrogen) were transformed with the vectors following the manufacturers recommendations. DNA sequencing was performed on the purified vectors to confirm proper insertion of the fragments.
Reporter gene activity assay.
The indicated reporter vectors were transfected along with a pRL-TK Renilla luciferase vector for normalization into BEAS-2B cells in 24-well tissue culture plates using Lipofectamine 2000 (Invitrogen), at concentrations of 200 and 10 ng/well, respectively, and 1.56 μl of the transfection reagent in a total volume of 500 μl. After 12 h, 500 μl of DMEM supplemented with 5% FBS was added, and the cells were incubated for an additional 12 h. Following appropriate treatments or additional cotransfections with miR-190 oligos, a Dual-Luciferase Reporter assay (Promega) was performed according to the manufacturer's specifications.
Generation of stable cell lines.
Stable cell lines overexpressing either miR-190 or a control oligo were generated by transfecting miRNASelect pEP-hsa-miR-190 and pEP-miR null expression vectors (Cell Biolabs, San Diego, CA) into BEAS-2B cells using the same general protocol as the luciferase reporter gene assay. The transfected cells were subjected to selection by puromycin at 1 μg/ml for 3 days.
Cell proliferation and carcinogenic transformation assays.
Control and miR-190 stably expressing cells were seeded into a 96-well plate at 5 × 103 cells/well, cultured for 16 h followed by serum starvation for 12 h. An assay for proliferation was then performed using Cell Titer 96 AQueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer's specifications. The plate was then read at 490 nm at 1, 2, 3, and 4 h time points. Carcinogenic transformation of the cells was determined by seeding the cells in soft agar. Briefly, the stably transfected cells were suspended in a 0.33% agar solution above a 0.5% agar layer containing 2-mercaptoethanol and penicillin/streptomycin in wells of 6-well plates. The plates were incubated at 37°C and 5% CO2 in a humidified incubator without feeding for 21 days. The colonies were counted and imaged on days 14 and 21.
Enzyme-linked immunosorbant assay.
The protein concentrations of VEGF in supernatants from BEAS-2B cells receiving various treatments were determined using a DuoSet ELISA development system (R&D systems, Minneapolis, MN). The cells were transfected with the pre-miR-190 or pre-miR-190In oligos as described earlier. The supernatants were collected and the ELISA was performed according to the manufacturer's specifications. The supernatants were either diluted 1:1 or not diluted before incubation with the ELISA plate. Visualization was achieved using appropriate volumes of solution A and solution B from the Substrate Reagent Pack (R&D systems). The reaction was quenched with sulfuric acid and then read on a plate reader with a wavelength of 450 nm.
Statistics.
Both SigmaPlot 9.0 and Microsoft Excel were used in statistical analyses for the quantitative data. The data were expressed as mean ± SD.
RESULTS
Microarray of miRNA Expression
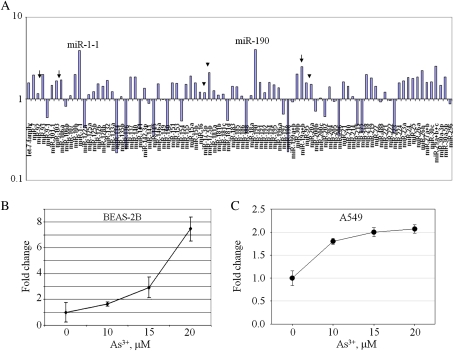
Environmental or occupational exposure to As3+ poses a great risk of developing multiple types of cancer including cancers of the lung, skin, liver, and bladder. Little is known about the effect of As3+ on the expression of miRNAs. One possible mechanism for cancer development induced by As3+ is the deregulation of miRNA, especially those oncogenic miRNAs. In order to determine which miRNAs are deregulated by As3+ exposure, a microarray experiment was performed using Cancer microRNA qPCR Array that contains 95 different miRNAs. Total RNA used in this analysis was isolated from the bronchial epithelial cell line, BEAS-2B, exposed to 20μM As3+ for 6 h. The array showed an upregulation of 67 miRNAs as well as a downregulation of 28 other miRNAs (Fig. 1A). Among the upregulated miRNAs by As3+, the highest miRNAs induced are the miR-1-1 and miR-190. A fourfold induction of miR-1-1 and fivefold induction of miR-190 by As3+ was noted (Supplementary Table 1). The capability of As3+ on the induction of miRNAs in this array was additionally supported by the parallel upregulation of miRNAs within miRNA clusters that share the same transcriptional unit, such as the miR-106-363 cluster (miR-92, miR-106a, and miR-19a, indicated by arrows in Fig. 1) and the miR-17-92 cluster (miR-17-3p, miR-17-5p, and miR-20a, indicated by arrowheads in Fig. 1). Of the upregulated miRNAs, miR-190 was an attractive candidate for follow-up study because of its involvement in cell growth and proliferation (Cheng et al., 2005).
FIG. 1.
As3+ regulates expression of miRNAs. (A) A real-time PCR-based miRNA array using Cancer miRNA Panel shows expression of miRNAs after As3+ exposure. Arrows indicate miRNAs in the miR-106-363 cluster; arrowheads indicate miRNAs in the miR-17-92 cluster. Data are representative of three experiments. (B) As3+ induces expression of miR-190. Real-time PCR was performed using total RNA extracted from the BEAS-2B cells treated with the indicated concentrations of As3+ for 12 h. Data are means ± SD of five experiments. (C) As3+ induces miR-190 in A549 cells. Cells were treated as in B. Data are means ± SD of three experiments.
To validate the array data, we next performed real-time PCR using total RNA isolated from BEAS-2B cells treated with increasing concentrations of As3+ for 12 h. A concentration-dependent increase of miR-190 was seen in the cells treated with As3+ (Fig. 1B). As3+ at concentrations of 10, 15 and 20μM induced a twofold, threefold, and sevenfold increase of miR-190 expression in the BEAS-2B cells, respectively. Moreover, in addition to BEAS-2B cells, we also observed induction of miR-190 by As3+ in other two different cell lines, A549 lung cancer cell line (Fig. 1C) and SAEC, the primary human small airway epithelial cell line (Supplementary Fig. S1). A threefold induction of miR-190 was noted in SAEC cells treated with lower concentrations of As3+, 2.5–5μM, for 48 h.
As3+ Induces miR-190 through Activating Talin 2 Gene
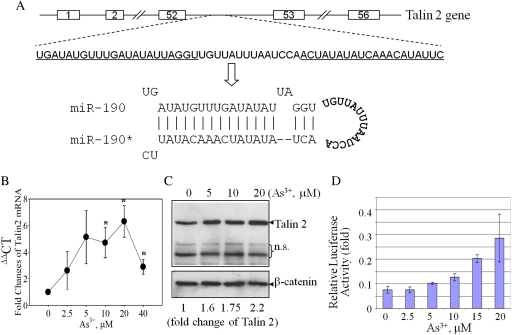
Noncoding RNAs, including miRNAs, are transcribed from either intergenic loci or intronic DNA of genes. A database search for the genomic context surrounding the miR-190 using the online Ensemble genome information system (Flicek et al., 2008) revealed that miR-190 is located on chromosome 15 within intron 52 of the talin 2 gene (Fig. 2A). The talin family proteins, including talin 1 and talin 2, are important molecules for the spreading and mobility of the cells by regulating the function of integrins and the assembly of the stress fibers (Zhang et al., 2008). To determine whether an increase in expression of miR-190 correlates with expression of the talin 2 mRNA and protein, real-time PCR, and Western blotting were performed (Figs. 2B and 2C). In agreement with the data for miR-190 expression, As3+ induced talin 2 mRNA accumulation in a concentration-dependent manner (Fig. 2B), which correlated with an increased levels of the talin 2 protein (Fig. 2C). The peak induction of the talin 2 mRNA occurred in the cells treated with 20μM As3+ for 12 h (Fig. 2B). Semiquantification of the Western blotting data by densitometry scanning showed that As3+ at concentrations of 10 and 20μM induced a 1.5- to 2-fold increase of the talin 2 protein (Fig. 2C, the numbers at the bottom).
FIG. 2.
Talin 2 expression in response to As3+. (A) Schematic diagram of the talin 2 gene and pre-miR-190 within the intron 52 region of the talin 2 gene. Underlined sequences indicate the sequences that form pre-miR-190 duplex through complementarily association of the inverted repeat region. The asterisk (*) indicates passage strand of the miR-190 duplex. (B) Real-time PCR analysis of talin 2 mRNA production in response to As3+. (C) Talin 2 protein was induced by a 12-h exposure of the cells with As3+ at the indicated concentrations. A loading control was made by Western blotting using antibody against β-catenin. n.s.: nonspecific bands. The bottom numbers show semiquantification of the talin 2 protein expression by densitometry. Data are representative of three experiments. (D) The talin 2 promoter reporter gene activity is activated by As3+ exposure. Data is normalized by calculating the ratio between Firefly luciferase activity versus Renilla luciferase activity.
To further assess the capability of As3+ on the induction of the talin 2 gene, a luciferase reporter gene activity assay was conducted by using a reporter vector containing the proximal 1 kb region of the talin 2 promoter. Transfection of BEAS-2B cells with the talin 2 reporter gene and followed by exposure to As3+ showed a concentration-dependent induction of talin 2 promoter activity (Fig. 2D). All of these data clearly indicate that the expression of miR-190 in response to As3+ is associated with the expression of its host gene. These results also suggest that regulation of miR-190 by As3+ is very likely at the level of transcription.
miR-190 Targets PHLPP
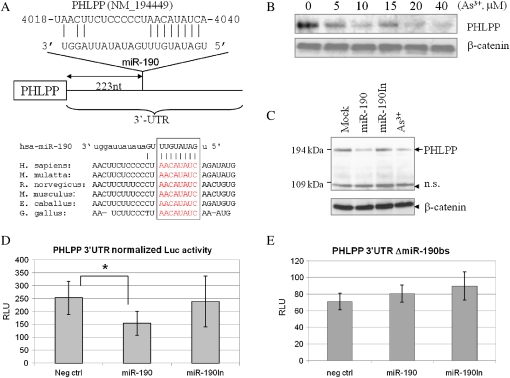
As3+ is an established human carcinogen and some miRNAs have been previously demonstrated as oncogenic miRNAs that can suppress expression of the tumor suppressors or activate cell growth signaling (Shi et al., 2008). The information available on the role of miR-190 in human carcinogenesis is currently very limited. We postulate that miR-190 may be partially responsible for the carcinogenicity of As3+ by affecting certain oncogenic signals. In order to identify potential targets of miR-190, we adopted a computational approach by using several of the available prediction algorithms based on evolutionary conservation of target sites across species, including miRDB (Wang, 2008; Yang et al., 2008), PicTar (Krek et al., 2005), and Targetscan (Lewis et al., 2003). One limitation of this approach is that each of these algorithms predicts hundreds of possible targets for miR-190 or other miRNAs. To circumvent this problem, we focused on the targets important for cell proliferative or presurvival signals that were predicted by all of these programs. Among the predicted candidate targets, 3′-UTRs of human PHLPP and tumor protein 53-induced nuclear protein 1 (TP53INP1) contained regions that matched the seed sequences of miR-190. The predicted miR-190-binding sites are found 223 and 217 nt downstream of the stop code for PHLPP (Fig. 3A) and TP53INP1 (Supplementary Fig. S2A) mRNAs, respectively. The binding site of miR-190 is well conserved among PHLPP mRNAs from other species, including monkey, rat, mouse, horse, and chicken, which is indicative for the importance of miR-190 on the regulation of PHLPP (Fig. 3A). PHLPP is known to target phosphorylated serine 473 specifically for dephosphorylation and inactivation of Akt kinase, leading to an increased apoptotic potential, and thereby causing tumor suppression (Gao et al., 2005). We had previously demonstrated that As3+ is able to activate Akt in BEAS-2B cells (Zhang et al., 2006). Accordingly, it is very likely that As3+-induced Akt activation is achieved through the downregulation of PHLPP by miR-190, leading to a sustained Akt activation and an increase in cell growth and proliferation, resulting in cell transformation. Western blotting examining the level of the PHLPP protein in the cells treated with As3+ clearly indicated a concentration-dependent repression of the PHLPP protein (Fig. 3B), suggesting a potential link between As3+-induced miR-190 and Akt activation.
FIG. 3.
miR-190 targets PHLPP. (A) Schematic diagram of the miR-190-binding site at the 3′-UTR of PHLPP mRNA. Lower panel shows alignment of the predicted miR-190-binding sites to 3-UTR of PHLPP from different species (Homo sapiens, Macaca mutatta, Rattus norvegicus, Mus musculus, Eguus caballus, and Gallus gallus). (B) Western blotting shows concentration-dependent reduction of the PHLPP protein in the cells treated with As3+. (C) Overexpression of miR-190 downregulates PHLPP. The BEAS-2B cells were mock transfected, or transfected with a miR-190, miR-190In, or treated with 20μM of As3+. The protein level of PHLPP was determined by Western blotting. (D) Overexpression of miR-190, but not miR-190In, downregulates the activity of the PHLPP 3′-UTR reporter gene. (E) Overexpression of miR-190 has no effect on the PHLPP 3′-UTR reporter without the miR-190-binding site.
Transfection of the Cells with miR-190 Precursor Suppresses PHLPP Expression
To verify the above observation indicating that As3+-induced expression of miR-190 can repress the translation of the PHLPP protein, the cells were directly transfected with a mock transfection, miR-190, and miR-190In, respectively. As depicted in Figure 3C, PHLPP protein levels were reduced in the cells transfected with miR-190 by 60–70% relative to the mock-transfected cells (Fig. 3C, lanes 1 and 2). No inhibitory effect of the miR-190In on PHLPP was observed (Fig. 3C, lane 3). Again, treatment of the cells with As3+ decreased expression of the PHLPP protein (Fig. 3C, lane 4).
To confirm that miR-190-mediated repression of PHLPP expression is achieved through interacting with the potential miR-190-binding site in the PHLPP 3′-UTR region, the entire 3′-UTR of PHLPP mRNA was cloned downstream of a luciferase gene in the pMIR-REPORT vector. The BEAS-2B cells were cotransfected with this reporter vector, pRL-TK Renilla luciferase vector, and miR-190 or miR-190In. We observed that the cotransfection of the miR-190 suppressed the luciferase activity of the vector with the PHLPP 3′-UTR by about 40% (Fig. 3D). In contrast, the miR-190In did not show inhibition on the PHLPP 3′-UTR reporter activity. Moreover, deletion of the miR-190-binding site within the PHLPP 3′-UTR abrogated the repressive ability of the miR-190 on the reporter gene activity (Fig. 3E), demonstrating specificity of the target sequence for PHLPP. Thus, these data provide strong evidence suggesting that As3+-induced miR-190 is crucial for the downregulation of the PHLPP protein.
Expression of miR-190 Enhances Akt Phosphorylation and VEGF Production
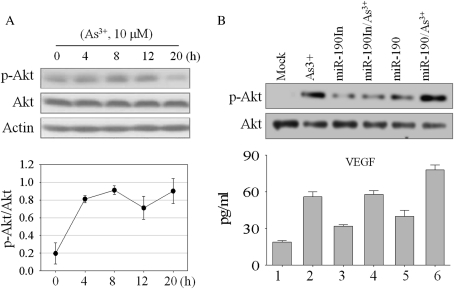
Having found that miR-190 represses expression of PHLPP, a negative regulator of Akt kinase by dephosphorylation of the serine 473 (Ser473)-phosphorylated, but not threonine 308 (Thr308)-phosphorylated, Akt (Gao et al., 2005), we investigated Ser473 phosphorylation of Akt in the cells treated with 20μM As3+ in the presence or absence of overexpressed miR-190 or miR-190In. Western blotting assays using antibody specific for Ser473 phosphorylation of Akt was performed followed by a densitometry quantification of Akt phosphorylation. As indicated in Figure 4A, 10μM As3+ activated Akt in a time-dependent manner with a peak occurring at 8 h, as judged by the Ser473 phosphorylation of Akt. Although decreased after As3+ treatment for 12 and 20 h, the activation of Akt at these time points is still evident (compare lanes 4 and 5 with lane 1, Fig. 4A). In an additional experiment using cells treated with As3+ for 12 h in the absence or presence of miR-190, both As3+ and miR-190 induced Akt phosphorylation notably (Fig. 4B, comparing lanes 2 and 5 with lane 1). Akt activation by As3+ was substantially enhanced in the cells overexpressing miR-190 (Fig. 4, lane 6). In contrast, this effect was not seen in the cells transfected with the miR-190In (Fig. 4B, lanes 3 and 4).
FIG. 4.
Overexpression of miR-190 enhances As3+-induced Akt phosphorylation and VEGF generation. (A) BEAS-2B cells treated with 10μM As3+ for the indicated hours, and Akt phosphorylation at Ser473 was determined by Western blotting. The bottom panel shows relative levels of Akt phosphorylation as determined by the ratio of phospho-Akt versus nonphospho-Akt that were determined by densitometry scanning of three independent Western blotting experiments. (B) BEAS-2B cells were transfected with the miR-190 and miR-190In, respectively. Phosphorylation of Akt at Ser473 was analyzed by Western blot. The bottom panel shows VEGF expression following miRNA transfection and As3+ exposure by ELISA. Data shown is representative of three experiments.
To determine if the effect of miR-190 on Akt phosphorylation has any downstream effects, an ELISA experiment for the expression of VEGF, a protein whose expression is known to be induced by the activation of Akt (Jiang et al., 2000), was performed. Results from the ELISA show that both As3+ and miR-190 are able to induce expression of VEGF. Combination of As3+ and miR-190 further enhanced VEGF induction (Fig. 4B, lane 6, the bottom panel). Enforced expression of miR-190In has very marginal effect on the basal VEGF expression (Fig. 4B, bottom panel, lane 3). Furthermore, miR-190In appears to be unable to potentiate As3+-induced VEGF expression (lane 4). These results indicate that miR-190 is indeed able to stimulate Akt activation and VEGF expression induced by As3+.
miR-190 Enhances Cell Proliferation and Carcinogenic Transformation
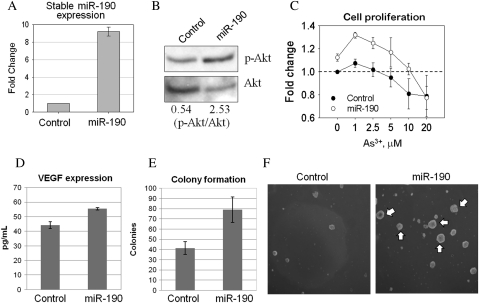
The observations that miR-190 induced by As3+ represses PHLPP and consequently activates Akt imply that this miRNA is an important regulator of critical cellular events associated with carcinogenesis, such as proliferation and transformation. To determine if overexpression of miR-190 leads to substantial changes in cell proliferation, colony formation, and/or other properties associated with malignant transformation and carcinogenesis, stably transfected BEAS-2B cell lines were generated by expressing miR-190 or a control vector. The stable miR-190 overexpressing cells exhibited an approximate ninefold increase of miR-190 expression compared with control transfected cells (Fig. 5A). A substantial enhancement of Akt activation in the cells stably expressing miR-190 was observed (Fig. 5B). To determine how miR-190 expression affects cells, we next measured cell growth by an MTS-based cell proliferation assay. As depicted in Fig. 5C, cells stably expressing miR-190 showed a significant increase in proliferation in either the absence or presence of different concentrations of As3+. In agreement with the results from transient transfection, VEGF expression was increased in the stable miR-190 expression cells (Fig. 5D). To determine directly whether overexpression of miR-190 enhances malignant transformation of the cells, we next performed anchorage-independent cell growth assay by seeding the cells in soft agar. Colony formation by the cells stably expressing miR-190 was substantially enhanced. The number of colonies was increased more than 50% when compared with cells stably expressing the control miRNA (Fig. 5E). Remarkably, cells stably expressing miR-190 formed large colonies (pointed by white arrows, Fig. 5F), whereas cells stably expressing the control miRNA formed only small aggregates of the cells (Fig. 5F). These results suggest that miR-190 is an oncogenic miRNA.
FIG. 5.
Stable expression of miR-190 enhances cell proliferation and carcinogenic transformation. (A) The levels of miR-190 in the cells stably transfected with a control miRNA and miR-190 were determined by real-time PCR. (B) Levels of Akt activation in the stably transfected cells were determined by Western blotting using antibody against Ser473-phosphorylated Akt (top panel) or the nonphospho-Akt (bottom panel). The numbers at the bottom of the lower panel indicate ratios of phospho-Akt versus nonphospho-Akt. (C) The stably transfected BEAS-2B cells were cultured for 16 h followed by As3+ treatment with the indicated concentrations for an additional 12 h. Cell proliferation was determined as described in the “Experimental Procedures.” (D) VEGF levels were determined by ELISA in the stably transfected cells. (E) Quantification of the colony numbers of the stably transfected cells in soft agar. (F) Anchorage-independent growth of the stably transfected cells was determined by the colony formation in soft agar. White arrows indicate large colonies formed by the cells stably transfected with miR-190.
DISCUSSION
As3+ is tumorigenic as demonstrated in both experimental animal and epidemiological studies of human populations in some specific environmental or occupational settings. The detailed mechanism of As3+-induced malignant transformation of the cells, however, remains to be fully established. A number of intracellular signaling pathways can be perturbed in response to As3+ exposure, including redox balance, kinase activation, transcription factor activity, DNA damage repair, and cell cycle. All of these pathways are either directly or indirectly linked to cell growth, apoptosis, and transformation. The regulatory role of As3+ on the expression and function of miRNA has not been explored yet. The discovery in this report that As3+ induces miR-190 expression, therefore, may provide an additional mechanistic insight into As3+ exposure and human cancer.
Unlike other well-studied miRNAs, such as let-7, miR-155, and miR-21, the function of miR-190 is largely unsubstantiated. In HeLa cells, inhibition of miR-190 by antisense RNA compromised cell growth without effect on cell apoptosis (Cheng et al., 2005). The level of miR-190 was significantly increased in the granulocytes isolated from patients with primary myelofibrosis relative to granulocytes from normal subjects (Guglielmelli et al., 2007). PMF is the rarest and most severe form of the Ph− myeloproliferative disorders, including PMF, polycythemia vera, and essential thrombocythemia. Because PMF is characterized by aberrant proliferation of the megakaryocytes in the bone marrow, it is believed that such a disorder is very likely due to deregulation of the stem-cell niche or deficiency in stem-cell differentiation. Indeed, a recent study using human embryonic stem cells and differentiated embryoid bodies indicated a significant upregulation of miR-190 expression in embryoid bodies and embryonic stem cells relative to the adult cells (Ren et al., 2009). Furthermore, an association of miR-190 overexpression with clonal malignant transformation has been observed in hematopoietic stem cells (Lataillade et al., 2008), suggesting potential linkage of miR-190, cancer stem cells, and cancer development.
Additional evidence suggesting the role of miR-190 played in cancer development has recently been provided by miRNA profiling in pancreatic cancer tissue samples and pancreatic cancer cell lines (Zhang et al., 2009). Real-time PCR data indicate that miR-190 was one of the eight most highly expressed miRNAs in pancreatic cancer, whereas its expression in normal pancreatic tissues was hardly detectable. An average of 21-fold increase of miR-190 was noted in 17 pancreatic cancer tissues relative to the case-matched normal pancreatic tissues. Similarly, miR-190 had been previously implicated as a signature miRNA in B-cell chronic lymphocytic leukemias (CLL) (Calin et al., 2004). As compared with the normal CD5+ B cells, the level of miR-190 in CLL was substantially increased. It was believed that this increase in miR-190 expression in CLL is possibly a result of genomic rearrangement because the host gene of miR-190, talin 2, is located in the common fragile sites that are prone to breakage and rearrangement in cancers. Increased expression of miR-190 has most recently been demonstrated in several other types of tumors, including bladder cancer (Ichimi et al., 2009), breast cancer (Lowery et al., 2009), lung cancer (Navon et al., 2009), a subset of HBV-positive hepatocellular carcinoma (Ura et al., 2009), and colorectal cancer (Ng et al., 2009). Intriguingly, a miR-190 homolog has also been identified in the Bombyx mori (silkworm) genome, and its expression has been implicated as an important process for the normal development of B. mori (Cao et al., 2008). All of these observations clearly indicate that miR-190 is a cell growth–regulating miRNA. Overexpression of miR-190, thus, is oncogenic.
miR-190 is an intronic miRNA whose gene is located in the 52nd intron of the talin 2 gene. The observed association of miR-190 expression with its host gene, talin 2, in the present study was complementarily supported by observations indicating coordination between talin 2 gene transcription and miR-190 biogenesis in neuron (Zheng et al., 2010). In mouse or rat hippocampal neurons, activation of the μ-opioid receptor by its agonist, fentanyl, reduced both talin 2 gene transcription and miR-190 expression in a manner of extracellular signal-regulated kinase–mediated phosphorylation of YY1. Phosphorylation of YY1, a transcription factor, resulted in an impairment of YY1 association with and activation on the talin 2 promoter, leading to a decreased transcription of talin 2 gene and consequently, reduced biogenesis of miR-190. It is still unclear how intronic miRNAs are transcribed and processed. Several lines of evidence implicated interdependency of pre-mRNA splicing and Drosha-dependent pri-miRNA processing (Shomron and Levy, 2009), whereas other studies revealed a possible mutual competition between pre-mRNA splicing and pri-miRNA processing (Kim and Kim, 2007).
Emerging evidence shows that oncogenic miRNAs contribute to the initiation and progression of cancer largely through regulating the expression of genes involved in the control of cell lineage development, cell proliferation, cell apoptosis, and kinase activation in the cells. For example, both miR-17 and miR-20a are able to negatively regulate expression of the E2F1 transcription factor to alter the dynamics of cell cycle progression (O'Donnell et al., 2005). The potential targets of miR-190 identified in the present report, PHLPP and TP53INP1, have been previously demonstrated as important regulators for protein kinase Akt signaling and cell apoptosis, respectively (Gao et al., 2005; Okamura et al., 2001; Ito et al., 2006; Jiang et al., 2006). PHLPP is a PH domain-containing serine phosphatase, which specifically dephosphorylates phosphorylated Ser473 of Akt and inactivates Akt, leading to cell apoptosis (Gao et al., 2005). It has been well established that full activation of Akt requires both Thr308 phosphorylation by PDK1 and Ser473 phosphorylation by mTORC 2 or DNA-PK. Accordingly, loss of PHLPP expression will prolong the Ser473 phosphorylation and activation of Akt. Indeed, in some breast and colon cancers, it was believed that loss of PHLPP expression is responsible for the sustained Akt activation, tumorigenesis, and metastasis (Liu et al., 2009; Qiao et al., 2007). Thus, induction of miR-190 by As3+ and the consequent PHLPP downregulation by miR-190 can provide the cells with a self-sustaining growth signal due to loss of a negative regulator of the Akt kinases. This may also partially explain observations indicating that As3+ is a potent inducer of Akt activation in bronchial epithelial cells and fibroblast cells as reported by ourselves and others (Ouyang et al., 2006; Zhang et al., 2006). Downregulation of TP53INP1 by As3+-induced miR-190, on the other hand, would cause the cells to be insensitive to proapoptotic signals. All of these effects of miR-190 are induced by As3+, and therefore, may be causatively important in malignant transformation and tumorigenesis of the cells.
In summary, we have provided the first evidence showing that As3+ is capable of inducing miR-190 expression associated with an appreciable protein accumulation of its host gene talin 2. Elevated miR-190 in response to As3+ is responsible for the downregulation of PHLPP and TP53INP1, two important tumor suppressors. Reduced PHLPP expression will consequently cause a prolonged or potentiated Akt activation and expression of its downstream targets, such as VEGF, leading to either a malignant transformation of the normal cells or tumorigenesis of the transformed cells. We believe that these discoveries may shed new light on the carcinogenic mechanism of environmental and occupational As3+ exposure. Further experiments are in progress to determine additional authentic targets of the miR-190 and how miR-190 or its host gene, talin 2, is transcriptionally regulated by As3+.
FUNDING
National Institutes of Health (5RO1ES017217-02 to F.C.)
Supplementary Material
Acknowledgments
We thank Ms Yongju Lu at the West Virginia University for technique assistance in soft agar cell transformation assay. We also thank the members in the Pathology and Physiology Branch of the National Institute for Occupational Safety and Health for critical reading.
References
- Beezhold KJ, Castranova V, Chen F. Microprocessor of microRNAs: regulation and potential for therapeutic intervention. Mol. Cancer. 2010;9:134. doi: 10.1186/1476-4598-9-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho LM, Gutierrez M, Alarcon-Herrera MT, Villalba Mde L, Deng S. Occurrence and treatment of arsenic in groundwater and soil in northern Mexico and southwestern USA. Chemosphere. 2011;83:211–225. doi: 10.1016/j.chemosphere.2010.12.067. [DOI] [PubMed] [Google Scholar]
- Cao J, Tong C, Wu X, Lv J, Yang Z, Jin Y. Identification of conserved microRNAs in Bombyx mori (silkworm) and regulation of fibroin L chain production by microRNAs in heterologous system. Insect Biochem. Mol. Biol. 2008;38:1066–1071. doi: 10.1016/j.ibmb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Cavigelli M, Li WW, Lin A, Su B, Yoshioka K, Karin M. The tumor promoter arsenite stimulates AP-1 activity by inhibiting a JNK phosphatase. EMBO J. 1996;15:6269–6279. [PMC free article] [PubMed] [Google Scholar]
- Celik I, Gallicchio L, Boyd K, Lam TK, Matanoski G, Tao X, Shiels M, Hammond E, Chen L, Robinson KA, et al. Arsenic in drinking water and lung cancer: a systematic review. Environ. Res. 2008;108:48–55. doi: 10.1016/j.envres.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Chen CL, Hsu LI, Chiou HY, Hsueh YM, Chen SY, Wu MM, Chen CJ. Ingested arsenic, cigarette smoking, and lung cancer risk: a follow-up study in arseniasis-endemic areas in Taiwan. JAMA. 2004;292:2984–2990. doi: 10.1001/jama.292.24.2984. [DOI] [PubMed] [Google Scholar]
- Chen F, Castranova V, Li Z, Karin M, Shi X. Inhibitor of nuclear factor kappaB kinase deficiency enhances oxidative stress and prolongs c-Jun NH2-terminal kinase activation induced by arsenic. Cancer Res. 2003;63:7689–7693. [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu HF, Ho SC, Yang CY. Lung cancer mortality reduction after installation of tap-water supply system in an arseniasis-endemic area in Southwestern Taiwan. Lung Cancer. 2004;46:265–270. doi: 10.1016/j.lungcan.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Critchley DR, Gingras AR. Talin at a glance. J. Cell Sci. 2008;121:1345–1347. doi: 10.1242/jcs.018085. [DOI] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol. Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmelli P, Tozzi L, Pancrazzi A, Bogani C, Antonioli E, Ponziani V, Poli G, Zini R, Ferrari S, Manfredini R, et al. MicroRNA expression profile in granulocytes from primary myelofibrosis patients. Exp. Hematol. 2007;35:1708–1718. doi: 10.1016/j.exphem.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Heck JE, Andrew AS, Onega T, Rigas JR, Jackson BP, Karagas MR, Duell EJ. Lung cancer in a U.S. population with low to moderate arsenic exposure. Environ. Health Perspect. 2009;117:1718–1723. doi: 10.1289/ehp.0900566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopenhayn-Rich C, Biggs ML, Smith AH. Lung and kidney cancer mortality associated with arsenic in drinking water in Cordoba, Argentina. Int. J. Epidemiol. 1998;27:561–569. doi: 10.1093/ije/27.4.561. [DOI] [PubMed] [Google Scholar]
- Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int. J. Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- Ito Y, Motoo Y, Yoshida H, Iovanna JL, Takamura Y, Miya A, Kuma K, Miyauchi A. Decreased expression of tumor protein p53-induced nuclear protein 1 (TP53INP1) in breast carcinoma. Anticancer Res. 2006;26:4391–4395. [PubMed] [Google Scholar]
- Jiang BH, Zheng JZ, Aoki M, Vogt PK. Phosphatidylinositol 3-kinase signaling mediates angiogenesis and expression of vascular endothelial growth factor in endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1749–1753. doi: 10.1073/pnas.040560897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang PH, Motoo Y, Garcia S, Iovanna JL, Pebusque MJ, Sawabu N. Down-expression of tumor protein p53-induced nuclear protein 1 in human gastric cancer. World J. Gastroenterol. 2006;12:691–696. doi: 10.3748/wjg.v12.i5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lataillade JJ, Pierre-Louis O, Hasselbalch HC, Uzan G, Jasmin C, Martyre MC, Le Bousse-Kerdiles MC. Does primary myelofibrosis involve a defective stem cell niche? From concept to evidence. Blood. 2008;112:3026–3035. doi: 10.1182/blood-2008-06-158386. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G, et al. MicroRNA signatures predict estrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monkley SJ, Pritchard CA, Critchley DR. Analysis of the mammalian talin2 gene TLN2. Biochem. Biophys. Res. Commun. 2001;286:880–885. doi: 10.1006/bbrc.2001.5497. [DOI] [PubMed] [Google Scholar]
- Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009;4:e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of colorectal cancer patients: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, Monden M, Nakamura Y. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell. 2001;8:85–94. doi: 10.1016/s1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Li J, Ma Q, Huang C. Essential roles of PI-3K/Akt/IKKbeta/NFkappaB pathway in cyclin D1 induction by arsenite in JB6 Cl41 cells. Carcinogenesis. 2006;27:864–873. doi: 10.1093/carcin/bgi321. [DOI] [PubMed] [Google Scholar]
- Qiao M, Iglehart JD, Pardee AB. Metastatic potential of 21T human breast cancer cells depends on Akt/protein kinase B activation. Cancer Res. 2007;67:5293–5299. doi: 10.1158/0008-5472.CAN-07-0877. [DOI] [PubMed] [Google Scholar]
- Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J. Transl. Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi XB, Tepper CG, deVere White RW. Cancerous miRNAs and their regulation. Cell Cycle. 2008;7:1529–1538. doi: 10.4161/cc.7.11.5977. [DOI] [PubMed] [Google Scholar]
- Shomron N, Levy C. MicroRNA-biogenesis and Pre-mRNA splicing crosstalk. J. Biomed. Biotechnol. 2009;2009:594678. doi: 10.1155/2009/594678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- Ye J, Li A, Liu Q, Wang X, Zhou J. Inhibition of mitogen-activated protein kinase kinase enhances apoptosis induced by arsenic trioxide in human breast cancer MCF-7 cells. Clin. Exp. Pharmacol. Physiol. 2005;32:1042–1048. doi: 10.1111/j.1440-1681.2005.04302.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Jiang G, Cai Y, Monkley SJ, Critchley DR, Sheetz MP. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat. Cell. Biol. 2008;10:1062–1068. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bhatia D, Xia H, Castranova V, Shi X, Chen F. Nucleolin links to arsenic-induced stabilization of GADD45alpha mRNA. Nucleic Acids Res. 2006;34:485–495. doi: 10.1093/nar/gkj459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J. Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chu J, Zeng Y, Loh HH, Law PY. Yin Yang 1 phosphorylation contributes to the differential effects of mu-opioid receptor agonists on microRNA-190 expression. J. Biol. Chem. 2010;285:21994–22002. doi: 10.1074/jbc.M110.112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.