Abstract
Our current natural product program utilizes new actinomycetes originating from unexplored and underexplored ecological niches, employing cytotoxicity against a selected panel of cancer cell lines as the preliminary screen to identify hit strains for natural product dereplication, followed by mechanism-based assays of the purified natural products to discover potential anticancer drug leads. Three new linear polyketides, actinopolysporins A (1), B (2), and C (3), along with the known antineoplastic antibiotic tubercidin (4), were isolated from the halophilic actinomycete Actinopolyspora erythraea YIM 90600, and the structures of the new compounds were elucidated on the basis of spectroscopic data interpretation. All four compounds were assayed for their ability to stabilize the tumor suppressor Programmed Cell Death Protein 4 (Pdcd4), which is known to antagonize critical events in oncogenic pathways. Only 4 significantly inhibited proteasomal degradation of a model Pdcd4-luciferase fusion protein, with an IC50 of 0.88 ± 0.09 μM, unveiling a novel biological activity for this well-studied natural product.
Many prokaryotic organisms inhabit “extreme environments” where the chemical or physical conditions differ significantly from those found in habitats that support more abundant and varied life forms. An extreme environment is one that humans would consider severe or uninhabitable, through extremes of temperature, pH, salinity, pressure, and even radiation. Such environments are inhabited by diverse populations of microorganisms, most of which are able to live only under the relevant extreme condition.1,2 The selective pressures of severe pH, temperature, salinity, and pressure have led to organisms apparently adapted to their environment. These extremophiles possess, and presumably benefit from, new enzymes, which thereby enable the biosynthesis of novel molecules.3 The diversity of proteins and secondary metabolites characteristic of the extremophiles have not been well studied. Indeed, this avenue for natural products discovery remains largely unexplored. Accordingly, we initiated recently a program designed to discover new natural products from new extremophilic actinomycetes originating from unexplored and underexplored ecological niches for anticancer drug discovery.4 Both extracts and purified compounds were tested in a high-throughput screening (HTS) assay designed to identify stabilizers of the tumor suppressor Programmed Cell Death Protein 4 (Pdcd4).5
Strain YIM 90600, isolated from Baicheng salt field in Xingjiang Province, in the northwest of China, has been identified as a new member of the genus Actinopolyspora on the basis of polyphasic taxonomy.6 YIM 90600 is a moderately halophilic actinomycete displaying optimal growth characteristics at 37 °C in pH 7–8 medium containing 10–15% (w/v) NaCl. We previously reported that fermentation of YIM 90600 under these conditions gave rise to new erythromycin analogues.4 Further investigation of extracts from large-scale fermentation cultures of strain YIM 90600 revealed the presence of additional new metabolites. We report here the isolation, structure elucidation, and preliminary biological activity assays of three new linear polyketides, actinopolysporins A (1), B (2), C (3) and the previously known antineoplastic antibiotic tubercidin (4).7 The carbon backbones of 1–3 are rare among known natural products, and their biosynthesis could be envisioned to proceed through a common polyketide pathway from condensation of an acetate or a propionate starter unit with four or five methyl malonate extender units, followed by varying modifications. Biological activity assays have focused on the ability of these natural products to stabilize Pdcd4, a tumor suppressor that has been shown to exert its antitumor function by inhibiting translation.5
RESULTS AND DISCUSSION
Recent efforts have indicated that YIM 90600 is a novel species of Actinopolyspora, for which the name Actinopolyspora erythraea sp. nov. has been proposed.6 This strain was cultivated in a NaCl-based medium for 28 days with vigorous shaking.4 The fermentation broth was extracted with EtOAc, the mycelia were extracted with acetone, and the combined extracts were subjected to multiple rounds of silica gel, LH-20, and C18 chromatography to afford pure compounds 1–4.
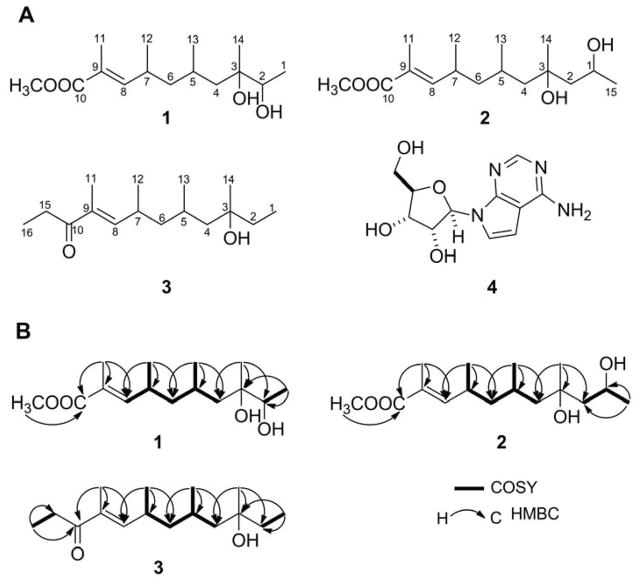
Compound 1 was isolated as a colorless oil, for which HRMALDIMS established its molecular formula as C15H28O4, thereby indicating two degrees of unsaturation. The IR spectrum supported the presence of hydroxy (3410 cm−1), carbonyl (1709 cm−1), and olefinic (1620 cm−1) moieties. Analysis of the NMR spectra (Table 1) revealed chemical shifts indicative of a carboxyl moiety (δC 169.2) conjugated with a double bond, a tri-substituted double bond [(δH 6.56, δC 148.7) and (δC 126.4, s)], a methoxy (δH 3.73, δC 52.0), one tertiary methyl group (δH 1.16, δC 24.2), four secondary methyl groups [(δH 1.14, δC 17.9), (δH 1.86, δC 12.8), (δH 0.99, δC 20.9), (δH 0.93, δC 22.4)], and a secondary alcohol methine group (δH 3.56, δC 75.3). gHSQC NMR experiments supported the presence of six methyls, two methylenes, and four methine carbons, while the remaining quaternary carbons were identified from the gHMBC experiment. Both COSY correlations as well as the MS data were supportive of the linearity of its carbon scaffold. On the basis of the gHMBC correlations from the six methyl groups to the corresponding carbons, the complete carbon connectivity was established (Figure 1). The absence of any correlation between CH3-11 and H-8 in the NOESY spectrum supported the (E)-double bond geometry. This assignment was substantiated by the observed upfield 13C NMR chemical shift of CH3-11 (δC 12.8), resulting from the γ-gauche interaction shielding of the CH3-11 by C-7. Compound 1 has been named actinopolysporin A.
Table 1.
Summary of 1H and 13C NMR Data for Actinopolysporins A (1), B (2), and C (3) in CDCl3a
| position | 1 (J in Hz) | 2 (J in Hz) | 3 (J in Hz) | |||
|---|---|---|---|---|---|---|
| δH (mult.) | δC | δH (mult.) | δC | δH (mult.) | δC | |
| 1 | 1.14, d (6.5) | 17.9, q | 4.22, m | 65.8, d | 0.94, t (7.0) | 8.54, q |
| 2a | 3.56, q (6.5) | 75.3, d | 1.64, dd (14.5, 11.0) | 48.3, t | 1.58, m | 35.6, t |
| 2b | 1.41 (overlapped) | 1.41, m | ||||
| 3 | 75.4, s | 74.6, s | 73.8, s | |||
| 4a | 1.53, dd (14.5, 6.0) | 42.7, t | 1.31, dd (14.0, 6.7) | 52.5, t | 1.36, dd (14.6, 5.8) | 49.0, t |
| 4b | 1.15 (overlapped) | 1.43 (overlapped) | 1.46 (overlapped) | |||
| 5 | 1.62 (overlapped) | 26.8, d | 1.56 (overlapped) | 27.0, d | 1.57 (overlapped) | 27.3, d |
| 6a | 1.63 (overlapped) | 46.2, t | 1.42 (overlapped) | 46.7, t | 1.55 (overlapped) | 46.7, t |
| 6b | 1.19, m | 1.22, m | 1.30, m | |||
| 7 | 2.63, m | 31.4, d | 2.60, m | 31.3, d | 2.72, m | 31.6, d |
| 8 | 6.56, brd (10.5) | 148.7, d | 6.52, brd (10.0) | 148.5, d | 6.43, brd (9.8) | 148.4, d |
| 9 | 126.4, s | 126.5, s | 135.7, s | |||
| 10 | 169.2, s | 169.2, s | 203.3, s | |||
| 11 | 1.86, d (1.5) | 12.8, q | 1.86, d (1.5) | 12.8, q | 1.86, brs | 11.8, q |
| 12 | 0.99, d (6.5) | 20.9, q | 0.98, d (6.5) | 20.8, q | 1.06, d (6.5) | 20.9, q |
| 13 | 0.93, d (6.5) | 22.4, q | 0.94, d (6.5) | 22.1, q | 0.99, d (6.5) | 22.1, q |
| 14 | 1.16, s | 24.2, q | 1.28, s | 26.9, q | 1.19, s | 27.0, q |
| 15 | 1.20, d (6.0) | 24.7, q | 2.72 (overlapped) | 30.7, t | ||
| 16 | 1.15, t (7.0) | 9.25, q | ||||
| OMe | 3.73, s | 52.0, q | 3.74, s | 52.0, q | ||
Assignments were made on the basis of 1H–1H COSY, gHSQC, and gHMBC experiments.
Figure 1.
Actinopolysporins A (1), B (2), C (3) and tubercidin (4) isolated from Actinopolyspora erythraea YIM 90600: A, Structures of 1, 2, 3, and 4 and B, key gCOSY and gHMBC correlations for 1, 2, 3 supporting their structural assignments.
Compound 2 (actinopolysporin B) was also isolated as a colorless oil, with a molecular formula of C16H30O4 as determined by HRMALDIMS analysis. Comparison of the spectroscopic data for 2 with those of 1 revealed these compounds to be quite similar except for an additional saturated methylene moiety (δC 48.3, t). gHMBC correlations of CH3-14 with this methylene at C-2 and 1H–1H COSY correlations of H2-2 with H-1 supported the assignment of C-2 as a methylene moiety (Figure 1).
Compound 3 (actinopolysporin C), a colorless oil, was similarly subjected to HRMALDIMS analysis, establishing its molecular formula as C16H30O2. Comparison of the 1H NMR data of 3 with those of 1 showed general similarities. An obvious difference found between 3 and 1 was the additional two triplet methyl signals (δH 0.94, t, J = 7.0 Hz and 1.15, t, J = 7.0 Hz) and the absence of the proton signals of an oxygenated methine and a methoxyl group characteristic of 1. In addition, an α,β-unsaturated ketone group (δC 203.3) was evident in the 13C NMR spectrum of 3. The two triplet methyl signals exhibited obvious gHMBC cross-peaks with those of the C-10 ketone and hydroxylated C-3, respectively, hence supporting the final structural assignment of compound 3 (Figure 1).
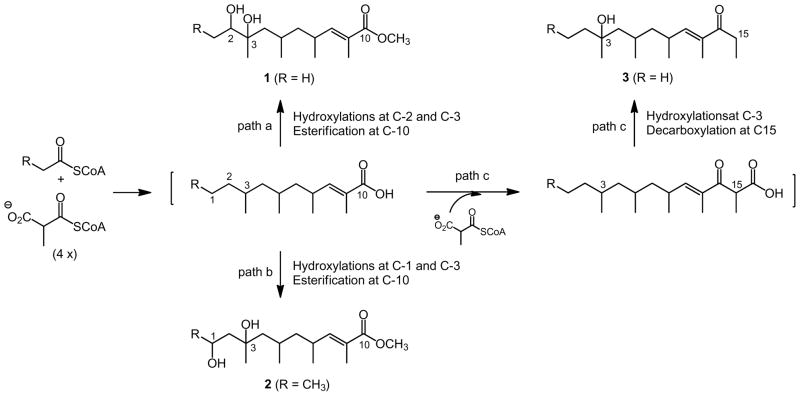
The linear carbon scaffolds represented by 1–3 are rare among known natural products. We previously reported the isolation and structural elucidation of several analogues of the 14-membered macrolide erythromycin, as well as a linear shunt metabolite for erythromycin biosynthesis from YIM 90600, supporting the processive mechanism for polyketide biosynthesis.4 While bearing some structural similarity, 1–3 are unlikely to be derived from the erythromycin biosynthesis pathway. Instead, another polyketide biosynthetic process, capable of incorporating multiple starter units and performing a varying number of chain elongation steps as well as carrying a plethora of tailoring modifications of the nascent polyketide intermediate to imbue additional structural diversity, could be envisaged for 1–3 biosynthesis in YIM 90600; Streptomyces species are known to encode multiple polyketide biosynthetic machineries.8 Thus, a common pathway could be proposed for 1–3 from condensation of an acetate or propionate starter unit with four or five methyl malonate extender units, followed by varying modifications: (i) an acetate starter unit upon four cycles of condensations with the methylmalonyl CoA extender unit followed by hydroxylations at C-2 and C-3 and esterification at C-19 would afford 1 (Figure 2, path a); (ii) a propionate starter unit upon four cycles of condensations with the methylmalonyl CoA extender unit followed by hydroxylations at C-1 and C-3 and esterification at C-10 would yield 2 (Figure 2, path b); and (iii) an acetate starter unit upon five cycles of condensation with the methylmalonyl CoA extender unit followed by hydroxylation at C-3 and decarboxylation at C-15 would give rise to 3 (Figure 2, path c). These features emphasize once again the plasticity and promiscuity of the polyketide biosynthetic machinery, supporting the approach of applying combinatorial biosynthesis strategies and methods to polyketide biosynthetic machinery for producing natural product compound structural diversity.9
Figure 2.
A common pathway proposed for actinopolysporin A (1), B (2), and C (3) biosynthesis in Actinopolyspora erythraea YIM 90600.
In addition to the three actinopolysporins, compound 4 was isolated as a white powder, for which its identity as the deazanucleoside tubercidin (Figure 1) was readily established by comparison of the MS and NMR data obtained with those reported in the literature.10–12 Tubercidin (4) was first isolated from Streptomyces tubercidicus;10 this compound has since been isolated from many other organisms, including the blue-green alga Tolypothrix byssoidea11 and the marine sponge Caulospongia biflabellata.12 Actinopolyspora erythraea YIM 90600 now joins a growing list of producers of 4. The significant antineoplastic activity of 4 has been attributed to interaction with a myriad of molecular targets. By virtue of its structural similarity to adenine, 4 is incorporated into DNA and inhibits multiple polymerases, thereby inhibiting DNA replication as well as RNA and protein synthesis.
Our current natural product program, aimed at discovering potential anticancer drugs, utilizes new actinomycetes originating from exotic ecological niches, employing cytotoxicity against a selected panel of cancer cell lines as the preliminary screen to identify hit strains for natural product dereplication, followed by mechanism-based assays of the purified natural products to discover potential leads.13 The combined extracts from YIM 90600 showed potent activity in the preliminary cytotoxicity screen, thereby motivating our subsequent effort on large-scale fermentation and natural product isolation and structural elucidation.4
We previously described a luciferase-based reporter construct to monitor Pdcd4 stability and developed a new high-throughput screen (HTS) compatible assay for identifying stabilizers of Pdcd4, a tumor suppressor that was shown to exert its antitumor function via inhibition of translation.14 Pdcd4 interacts with and inhibits the activity of the RNA helicase eIF4A, a component of the eukaryotic initiation factor 4F (eIF4F) translation-initiation complex.5 Pdcd4-dependent translation inhibition directly correlates with inhibited transformation, as well as compromised cell migration and invasion.15,16 Furthermore, Pdcd4 overexpression was shown to limit tumor formation,17 while depletion of Pdcd4 increased tumorigenesis in vivo.18 Loss of Pdcd4 protein in response to tumor promoters and mitogens has been attributed to phosphorylation-dependent proteasomal degradation. In detail, Pdcd4 bears a phosphorylation-dependent degradation (PDD) domain that houses both a p70S6K1 phosphorylation and a β-transducing repeat containing protein 1 (β-TrCP1)-recognition motif. Phosphorylation within this PDD target domain by p70S6K1 allows for binding of the E3-ubiquitin ligase β-TrCP and subsequent proteasomal degradation in response to tumor-promoting conditions.18,19
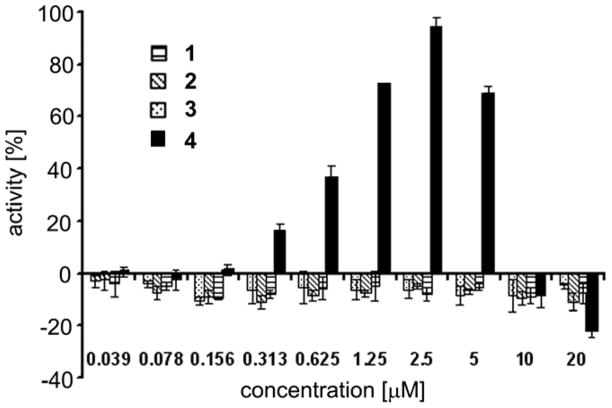
We subjected our current natural product collection to this HTS in search for Pdcd4 stabilizers. Of the four compounds isolated in this study, 1–3 were found to completely lack Pdcd4 stabilizing ability as shown in Figure 3. Hence, luciferase activity was not recovered by compounds 1–3 from 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced down-regulation. The loss of luciferase activity was similar for all three natural products over the range of concentrations spanning from 39 nM to 20 μM and all were comparable to cells treated with TPA-only controls (Figure 3). In contrast, co-treatment with 4 resulted in a significantly increased luciferase signal in comparison to TPA-only treated cells. Dose response analysis revealed Pdcd4 stabilizing activity of 4 with an IC50 of 0.88 ± 0.09 μM (Figure 3). Maximal activity reached 94.2 ± 3.6% at 2.5 μM. At higher doses (> 2.5 μM), increasing cell toxicity led to a decreased luciferase signal.14 These findings highlight 4 as the first example of an adenosine derivative able to stabilize Pdcd4 by interfering with its phosphorylation-dependent proteasomal degradation, revealing a potentially significant activity previously unrecognized for 4, a well-established but not yet fully understood anticancer clinical candidate. Importantly, stabilization of Pdcd4 under tumorigenic conditions is predicted to result in tumor suppressive effects. Based on its eIF4A inhibitory effect, stabilization of Pdcd4 by small molecules should selectively inhibit the translation of mRNAs with structured 5′-untranslated regions,5 which are characteristic of many proto-oncogenes. Consequently, stabilizers of Pdcd4 may specifically inhibit tumor-associated changes in translation. Based upon the efficacy of 4 in rescuing Pdcd4 from tumor promotor-induced degradation, in combination with the novelty of translation inhibition as a mode of action, we suggest advancing 4 for further development in the treatment and possible prevention of various cancers.
Figure 3.
Summary of Pdcd4 stabilization data for 1–4. The % activity of the fusion protein values are relative to a DMSO control in which slow proteasomal degradation of the fusion protein parallels that established for intact Pdcd4. These data correlate to an IC50 of 0.88 ±0.09 μM for Pdcd4 stabilization (and protection) by compound 4.
In conclusion, the extremophilic halophile YIM 90600 has been recently classified as a new Actinopolyspora erythraea sp. nov. species.6 In our continued effort to discover novel natural products from new extremophilic actinomycetes, we have previously discovered the new erythromycin analogues, erythronolides H and I,4 and now we report three additional new linear polyketides, actinopolysporins A, B, and C, as well as the known antitneoplastic antibiotic tubercidin (Figure 1). As new targets are discovered and become available in HTS format, libraries of purified natural products present an opportunity for the discovery of specific small molecule probes or potential drug leads. We discovered that 4 is a potent inhibitor of Pdcd4 protein degradation and thus a potential lead to develop novel stabilizers of the tumor suppressor Pdcd4. These results support the idea that, owing to their unique environments, extremophilic microbes can contribute in a significant way to the array of structural diversity available through secondary metabolism to drug discovery efforts. Extremophilic actinomycetes, in particular, warrant significant attention as producers of biologically active leads with unique modes of action.
EXPERIMENTAL SECTION
General Experimental Procedures
Optical rotations were measured in CHCl3 with a Perkin-Elmer 241 polarimeter at the sodium D line (589 nm). 1H and 13C NMR spectra were recorded at 25 °C with a Varian Unity INOVA 500 instrument operating at 500 MHz for 1H and 125 MHz for 13C nuclei. The chemical shifts were referenced to residual solvent signals: δH 7.26 and δC 77.29 for CDCl3. 1H-1H gCOSY (mixing time = 80 ms), gHSQC (1JCH = 140 Hz), and gHMBC (2–3JXH = 8.0 Hz) spectra were performed using standard Varian pulse sequences. High-resolution mass spectral data were acquired with an IonSpec HiResMALDI FT–mass spectrometer with a 7 tesla superconducting magnet. Semi-preparative HPLC was performed with a Varian liquid chromatograph system equipped with an Apollo C18 column (5 μm, 10 × 25 mm, Grace Davison Discovery Sciences, Deerfield, IL). Column chromatography was performed either on silica gel (230–400 mesh, Natland International, Research Triangle Park, NC), Lichroprep RP-18 gel (40–63 μm, Merck, Darmstadt, DE), or Sephadex LH-20 (Pharmacia, Kalamazoo, MI).
Isolation and Phylogenetic Identification of Actinopolyspora erythraea YIM 90600
Strain YIM 90600 was isolated from Baicheng salt field in Xinjiang Province, China, after 3 weeks of incubation at 37 °C on cellulose-casein multi-salt medium9 (microcrystalline cellulose 1%, casein 0.03%, KNO3 0.02%, K2HPO4 0.05%, CaCO3 0.002%, FeSO4 0.001%, NaCl 10%, MgCl2·6H2O 3%, KCl 2%, agar 1.5%). Multiple salts were sterilized separately before addition to the medium. The pH of the medium was adjusted to pH 7.5 with 1 N NaOH. The isolate was maintained on ISP 4 agar slants containing 10% (w/v) NaCl at 4 °C and as suspensions of mycelia fragments in 20% (v/v) glycerol. Morphological and physiological studies to identify the genus of the strain were carried out following a standard protocol.20 Assignment of YIM 90600 as Actinopolyspora erythraea sp. nov. on the basis of polyphasic taxonomy has been reported recently.6
Fermentation, Extraction, and Isolation
The inoculum was prepared by introducing the periphery of 7-day-old petri dish cultures of Actinopolyspora erythraea YIM 90600 into 250-mL baffled flasks containing 50 mL of the broth (soluble starch 0.5%, glucose 2%, peptone 0.2%, yeast extract 0.2%, soybean flour 1%, NaCl 10%, K2HPO4 0.05%, MgSO4·7H2O 0.05%, CaCO3 0.2% in tap water, pH 7.8, adjusted with 1 N NaOH), followed by shaking at 250 rpm and 28 °C for 5 days. The production fermentation was accomplished by adding the inoculum (50 mL) into 2-L baffled flasks containing 500 mL of the same culture broth, and then shaking under the same conditions for 28 days. The fermentation broth (14 L) was then centrifuged at 4000 rpm for 20 min. Mycelia were extracted with acetone (3 × 1 L), with the fermentation broth then extracted with EtOAc (3 × 4 L). Solvent removal in vacuo was followed by combination of the acetone and EtOAc extracts. The oily residue obtained (35 g) was chromatographed on silica gel using CHCl3-MeOH (100:0, 50:1, 20:1, 10:1, 5:1, 1:1 and 0:100, 2.5 L each) as the mobile phases, in a stepwise manner, to afford seven fractions, A–G. Fraction B (50:1) (500 mg) was chromatographed further over a Sephadex LH-20 column and eluted with methanol to yield three subfractions, A1–A3. Subfraction A2 was finally purified by semi-preparative HPLC to afford 1 (10.4 mg) and 2 (3.2 mg). Fraction C (20:1) (380 mg) was chromatographed over a C18 column, eluted in a step gradient manner with CH3OH-H2O, to render 3 (0.9 mg). Finally, 4 (15.0 mg) was obtained from fraction F (1:1) by repeated chromatography on silica gel and Sephadex LH-20 columns.
Actinopolysporin A (1)
colorless oil; (c 0.15, CHCl3); UV (MeOH) λmax log (ε) 240 (3.76) nm; IR νmax 3410, 1709, 1620, 1052, 960 cm−1; NMR data, see Table 1; HRMALDIMS (positive ion) m/z 295.1887 [M+Na]+ (calcd for C15H28O4, 295.1880).
Actinopolysporin B (2)
colorless oil; (c 0.10, CHCl3); UV (MeOH) λmax log (ε) 240 (3.80); NMR data, see Table 1; HRMALDIMS (positive ion) m/z 309.2040 [M+Na]+ (calcd for C16H30O4, 309.2036).
Actinopolysporin C (3)
colorless oil; (c 0.05, CHCl3); UV (MeOH) λmax log (ε) 251 (4.12); NMR data, see Table 1; HRMALDIMS (positive ion) m/z 277.2138 [M+Na]+ (calcd for C16H30O2, 277.2149).
Pdcd4 Stabilizing Activity Assay
Stabilization of Pdcd4 was assessed as previously described.14 In brief, HEK293 cells expressing a fusion protein comprised of a fragment of Pdcd4 containing the regulatory region (amino acids 39–91) and luciferase were plated (1000 cells/well, 40 μL/well) in 384-well opaque white plates and allowed to attach overnight (18 h). TPA (final concentration 10 nM) was added, followed (within 15 min) by test samples or controls. Following an eight-hour incubation, luciferase activity was measured 10–15 min after the addition of Steadylite Plus (Perkin-Elmer) reagent. Controls were DMSO only (no TPA), TPA only, and TPA + rapamycin (100 nM final). The activities of compounds were calculated using the following formula:
Supplementary Material
Acknowledgments
We thank the Analytical Instrumentation Center of the School of Pharmacy, UW–Madison, for support in obtaining the MS and NMR data. This project was supported in part by the Intramural Research Program of NIH, NCI, Center for Cancer Research and federal funds from NCI under contract HHSN261200800001E, NSF of China grants 2162028 and U0932601 and a grant from SRF for ROCS, Chinese Ministry of Education (all to L.X.Z.), the Chinese Ministry of Education 111 Project B08034 (to Y.D.), and NIH grants CA113297 and GM086184 (to B.S.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
Footnotes
Supporting Information. 1H and 13C NMR spectra of compounds 1–3. This material is available free-of-charge via the Internet at http://pubs.acs.org.
References
- 1.Aguilar A, Ingemansson T, Magnien E. Extremophiles. 1998;2:367–373. doi: 10.1007/s007920050080. [DOI] [PubMed] [Google Scholar]
- 2.Madigan MT, Oren A. Curr Opin Microbiol. 1999;2:265–269. doi: 10.1016/s1369-5274(99)80046-0. [DOI] [PubMed] [Google Scholar]
- 3.Wilson ZE, Brimble MA. Nat Prod Rep. 2009;26:44–71. doi: 10.1039/b800164m. [DOI] [PubMed] [Google Scholar]
- 4.Huang SX, Zhao LX, Tang SK, Jiang CL, Duan Y, Shen B. Org Lett. 2009;11:1353–1356. doi: 10.1021/ol900143j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaRonde-LeBlanc N, Santhanam AN, Baker AR, Wlodawer A, Colburn NH. Mol Cell Biol. 2007;27:147–156. doi: 10.1128/MCB.00867-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang SK, Wang Y, Klenk HP, Shi R, Lou K, Zhang YJ, Chen C, Ruan JS, Li WJ. Int J Syst Evol Microbiol. 2011;61:1693–1698. doi: 10.1099/ijs.0.022319-0. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki S, Marumo S. J Antibiot. 1960;13:360. [Google Scholar]
- 8.Nett M, Ikeda H, Moore BS. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Van Lanen SG, Shen B. Curr Opin Microbiol. 2006;9:252–260. doi: 10.1016/j.mib.2006.04.002. [DOI] [PubMed] [Google Scholar]; (b) Van Lanen SG, Shen B. Curr Opin Drug Discov Devel. 2008;11:186–195. [PubMed] [Google Scholar]
- 10.Smulson ME, Suhadolnik RJ. J Biol Chem. 1967;242:2872–2876. [PubMed] [Google Scholar]
- 11.Barchi JJ, Norton TR, Furusawa E, Patterson GML, Moore RE. Phytochemistry. 1983;22:2851–2852. [Google Scholar]
- 12.Biabani MF, Gunasekera SP, Longley RE, Wright AE, Pomponi SA. Pharm Biol. 2002;40:302–303. [Google Scholar]
- 13.(a) Huang SX, Powell E, Rajski SR, Zhao L, Jiang C, Duan Y, Xu W, Shen B. Org Lett. 2010;12:3525–3527. doi: 10.1021/ol1013526. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Powell E, Huang SX, Xu Y, Rajski SR, Wang Y, Peters N, Guo S, Xu HE, Hoffmann FM, Shen B, Xu W. Biochem Pharmacol. 2010;80:1221–1229. doi: 10.1016/j.bcp.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Yu Z, Zhao LX, Jiang CL, Duan Y, Wong L, Carver KC, Schuler LA, Shen B. J Antibiot. 2011;64:159–162. doi: 10.1038/ja.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Huang SX, Yu Z, Robert F, Zhao LX, Jiang Y, Duan Y, Pelletier J, Shen B. J Antibiot. 2011;64:164–166. doi: 10.1038/ja.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blees JS, Schmid T, Thomas CL, Baker AR, Benson L, Evans JR, Goncharova EI, Colburn NH, McMahon JB, Henrich CJ. J Biomol Screen. 2010;15:21–29. doi: 10.1177/1087057109351028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang HS, Matthews CP, Clair T, Wang Q, Baker AR, Li CC, Tan TH, Colburn NH. Mol Cell Biol. 2006;26:1297–306. doi: 10.1128/MCB.26.4.1297-1306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leupold JH, Yang HS, Colburn NH, Asangani I, Post S, Allgayer H. Oncogene. 2007;26:4550–4562. doi: 10.1038/sj.onc.1210234. [DOI] [PubMed] [Google Scholar]
- 17.Jansen AP, Camalier CE, Colburn NH. Cancer Res. 2005;65:6034–6041. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 18.Schmid T, Jansen AP, Baker AR, Hegamyer G, Hagan JP, Colburn NH. Cancer Res. 2008;68:1254–1260. doi: 10.1158/0008-5472.CAN-07-1719. [DOI] [PubMed] [Google Scholar]
- 19.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 20.Xu L-H, Li W-J, Liu Z-H, Jiang C-L. Actinomycete Systematics – Principles, Methods and Practice. Science Press; Beijing: 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.