Abstract
Insect symbioses lack the complexity and diversity of those associated with higher eukaryotic hosts. Symbiotic microbiomes are beneficial to their insect hosts in many ways, including dietary supplementation, tolerance to environmental perturbations and maintenance and/or enhancement of host immune system homeostasis. Recent studies have also highlighted the importance of the microbiome in the context of host pathogen transmission processes. Here we provide an overview of the relationship between insect disease vectors, such as tsetse flies and mosquitoes, and their associated microbiome. Several mechanisms are discussed through which symbiotic microbes may influence their host’s ability to transmit pathogens, as well as potential disease control strategies that harness symbiotic microbes to reduce pathogen transmission through an insect vector.
Symbiotic associations are ubiquitous in nature
Soon after birth all mammals begin a lifelong association with a complex and diverse microbial community that resides on the skin, vaginal mucosa, gastrointestinal (GI) tract and in the mouth. Advances in PCR-based technologies have greatly expanded our knowledge of these microbial systems, which largely represent mutually beneficial associations. The human gastrointestinal tract alone harbors over 500 distinct microbial taxa comprising an estimated 1014 microbes, significantly exceeding the number of cells in the human body [1]. Because the majority of these microbes cannot be cultivated in vitro, the functional bases of these interactions are difficult to dissect and have remained largely elusive. Nevertheless, findings from experimental systems using animals reared under aseptic conditions indicate that proper maintenance of the microbial consortium is essential for mammalian nutrient breakdown. Furthermore, the absence of microbial fauna, or modifications in the composition of the consortium, can reduce a host’s fitness and lead to autoimmune disease states such as inflammatory bowel disease, rheumatoid arthritis or Type I diabetes (reviewed in [2]). A symbiont product, polysaccharide A, produced by the prominent human symbiont Bacteroides fragilis protects animals from experimental colitis induced by Helicobacter hepaticus, a commensal bacterium with pathogenic potential [3, 4].
In contrast to higher eukaryotes, which are colonized by a multitude of commensal organisms representing members of five of the six kingdoms of life, insects harbor a significantly less diverse community of microbial symbionts. The reduced complexity of the insect microbiome enables investigations that aim to understand the contribution(s) individual symbionts make towards host physiological processes. Furthermore, the insect systems are easier to maintain than are the vertebrate models because generation times are relatively short and husbandry practices less costly. In particular in the case of Drosophila, extensive mutant lines exist for further functional characterization of loci of importance. In addition to being excellent model systems, many insects that harbor bacterial symbionts are also human disease vectors or agricultural pests. Given that many symbioses are indispensable for host physiology, understanding these relationships are also of applied interest as they can lead to the development of more efficient insect control strategies. To this end, it has been possible to cultivate several insect commensal symbionts in vitro, and genetically modify and reintroduce the recombinant symbionts back into their native host. These ‘paratransgenic’ transformation systems are a promising alternative to those that involve direct modification of the host germ-line. Expression of anti-pathogen products by genetically modified insect symbionts may prevent the transmission of harmful agents to their eukaryotic hosts and thus provide a novel means of disease control [5–8].
Insect symbiotic systems
Symbiotic associations have been described in insects that rely on nutritionally restricted diets, such as vertebrate blood (i.e. tsetse flies and triatome bugs), plant phloem (i.e. aphids and psyliids) or wood (i.e. termites and carpenter ants). Presumably these microbes supplement their hosts with nutrients that are limited or absent in their diet or that they cannot produce on their own. Such mutualistic symbioses involve bacteria (typically referred to as the primary symbionts) that are vertically transmitted and that are indispensable for their hosts to survive in unique or resource-limited environments. Phylogenetic reconstruction studies have indicated that these host-microbe relationships are often ancient, and presumably neither partner can live in the absence of the other. During the long co-evolutionary history with their hosts, primary symbiont genomes have undergone drastic size reductions, losing genes and pathways that are no longer necessary in the unique metabolic niches the hosts provide [9]. Among the well-characterized primary symbionts are Buchnera from aphids [10], Carsonella from sharpshooters [11], Blochmannia from ants [12], SOPE from rice weevils [13, 14] and Wigglesworthia from tsetse (Box 1) [15, 16].
Box 1. Tsetse and its associated microbiome.
Three endosymbionts with distinct phylogenetic and genomic characteristics have been described from tsetse from laboratory colonies (Figure 1). These symbioses reflect varying levels of host integration, and as such have a different impact on tsetse’s physiology [71, 72]. While the microbiome is highly restricted in laboratory maintained flies, analysis of tsetse in natural field populations indicates the presence of additional microbes, which are presumably environmentally acquired. The acquisition routes, persistence and the role(s) of these microbes on host physiology remain unknown [73–75]. Below are biological highlights of tsetse’s heritable endosymbionts.
Wigglesworthia glossinidia
Obligate mutualist from the family Enterobacteriaceae [16, 76].
Phylogenetic analyses indicate that tsetse and Wigglesworthia have evolved concordantly for approximately 50–80 million years [77].
Highly streamlined genome (700 Kb in size) exhibits exceptional A–T bias (82%) and contains no transposons or phage-related elements [15, 78].
Chromosome encodes several vitamin biosynthesis pathways that likely supplement tsetse’s vertebrate blood-specific diet [15]. Female tsetse that lack Wigglesworthia are sterile [44].
Resides intracellularly in tsetse’s bacteriome in the midgut and extracellularly in the female milk gland lumen [59]. The milk gland population is essential for transmission to progeny, while the bacteriome population is essential for fecundity [44].
Must be present during larval maturation in order for tsetse’s immune system to develop and function properly during adulthood (see text).
Sodalis glossinidius
Facultative commensal from the family Enterobacteriaceae [19].
Recent association with tsetse as Sodalis from different species are closely related [79]. Several other insects, including stinkbugs [80], hippoboscid flies [81], grain weevils [13, 82] and slender pigeon lice [83] harbor endosymbionts that are closely related to Sodalis.
Detected in all colony flies, but not present in all field populations of tsetse. Found both inter- and intracellularly in the midgut, muscle, fat body, milk gland and salivary glands of tsetse [84].
Genome size (4.2 Mb) and chromosomal synteny indicates a close relationship to free-living enteric bacteria, including Salmonella and Yersinia [18, 85].
Sodalis’ genome has a low protein coding capacity (49%) and a high number of pseudogenes (972). Both of these characteristics are indicative of a transition from a free-living to endosymbiotic life style [85, 86].
Sodalis encodes three symbiosis islands (SSI) that share significant synteny and sequence homology with Type III secretion systems (TTSS) identified from pathogenic bacteria [85]. These SSIs presumably facilitate Sodalis’ intracellular lifestyle [87–89].
Sodalis may impact tsetse vector competence and longevity [90, 91].
Modifications associated with Sodalis’ Outer Membrane Protein A may facilitate host tolerance to this symbiont [92].
Field studies indicate a positive correlation between specific Sodalis genotypes and trypanosome infections in tsetse. However, flies that lack Sodalis are still competent for trypanosome infections [93–95].
Wolbachia
Rickettsia-like bacterium found in many insect taxa, including tsetse [96].
Infections with Wolbachia are restricted to tsetse’s reproductive tissues.
Certain tsetse species harbor Wolbachia infections, and the field infection prevalence and the Wolbachia strains infecting different species in the different populations investigated varies [97].
Many insects also harbor commensal microbes (referred to as secondary symbionts). These symbiotic associations apparently originated more recently and may be transient in nature. In some cases they are found to be dispensable for their hosts as not every individual in a population carries these microbes. While primary symbionts are vertically transmitted from mother to progeny with high fidelity, secondary symbionts can be acquired through multiple means; i.e. vertically, horizontally or from the environment. Examples of secondary symbionts include Hamiltonella defensa in sap-feeding insects including aphids, psyllids and whiteflies [17] and Sodalis glossinidius in tsetse (Box 1) [18, 19].
In addition to microbes with commensal and mutualistic associations, many insects also harbor maternally heritable parasitic microbes. The most extensively studied of these belong to the genus Wolbachia [20, 21]. A number of reproductive abnormalities are associated with Wolbachia infections, the most prominent being cytoplasmic incompatibility (CI). CI, which results in early embryo death, occurs when a Wolbachia-uninfected female mates with an infected male. Since infected females can successfully mate with males infected with the same strain of Wolbachia or with uninfected males, the Wolbachia infected female has a reproductive advantage, and the associated female genotypes can spread quickly through a population. CI has been proposed as a means of spreading desirable traits into populations, including genotypes or other maternally transmitted symbionts conferring disease resistance [22, 23]. In addition to CI, certain Wolbachia strains have been found to reduce host lifespan. These strains are also of interest from an applied perspective, as older adults typically contribute more to disease transmission while younger individuals contribute to reproductive output [24]. Thus, skewing the age of the population towards younger individuals without reducing the reproductive output can provide an evolutionarily sustainable means of disease control. Finally, it has recently been observed that infections with certain strains of Wolbachia confer pathogen resistance traits to their hosts, presumably through host immune induction (as described below and reviewed in [25]).
While insects with single diets of limited nutritional value have established associations with heritable primary and secondary symbionts, insects with multiple diets can harbor a wide range of environmentally acquired microbes. Both laboratory lines and field populations of Anopheline mosquitoes host a robust gut-associated microbiota that is principally comprised of Gram-negative (Gram−) members of the family Enterobacteriaceae. For example, 16 bacterial species from 14 genera were identified from field populations of the predominant African malaria vectors Anopheles gambiae and Anopheles funestus [26]. Furthermore, laboratory populations of An. gambiae and Anopheles stephensi (the primary vector of human malaria in Asia) also harbor a wide variety of bacteria, the most abundant of which come from the genera Asaia, Enterobacter, Mycobacterium, Sphingomonas, Serratia and Chryseobacterium [27–29]. The dominant symbiont Asaia (an α-proteobacterium belonging to the family Acetobacteriaceae) has been observed in all of the developmental stages of wild and laboratory reared colonies of Anopheline as well as Aedes mosquitoes [30]. A study on the genetic variation present in the Asaia symbiont indicates the presence of various strains in four different mosquito species analyzed [31]. This symbiont has been detected in the midgut, salivary glands and reproductive organs of adult mosquitoes [30]. Fluorescent in situ hybridization (FISH) analysis on the reproductive tract of female An. gambiae showed a concentration of Asaia at the very periphery of the eggs, suggesting that transmission of Asaia from mother to offspring is likely mediated by a mechanism of egg-smearing [30].
Insect pathogen associations
In addition to carrying beneficial microbes that enhance their overall physiological functions, some insects also carry and transmit microbes that are pathogenic to their mammalian or plant hosts (Table 1) [32]. Most of these insects (disease vectors of humans) utilize blood as a food source and typically acquire pathogens while feeding on infected hosts and then pass the disease agents on to other hosts during the course of subsequent meals. Once acquired by the insect host, viruses need to replicate, and protozoan pathogens and worms interact with different insect tissues and undergo several differentiation events before they can be transmitted to a subsequent host. This process of pathogen development in an insect can be significantly long and is known as the Extrinsic Incubation Period. Insects that transmit disease to humans include tsetse flies (Diptera: Glossinidae), triatome bugs (Hemiptera: Triatominae) and sand flies (Diptera: Psychodidae), which transmit kinetoplastid parasites that cause Human African trypanosomiasis (HAT), Chagas disease and leishmaniasis, respectively. In addition, lice (order Phthiraptera) can vector Epidemic typhus and Trench fever, and fleas (order Siphonaptera) transmit bacterial pathogens including the causative agent of Bubonic plague. Finally, mosquitoes are prolific disease vectors that inhabit a broad geographic range and host a diverse array of pathogens including dengue, West Nile and Chickungunya viruses, filarial worms and Plasmodium parasites.
Table 1.
Insect model systems that harbor immunomodulatory bacterial symbionts
| Insect host | Symbiotic bacteria (genera) | Pathogen/parasite | Refs. |
|---|---|---|---|
| Tsetse (Glossina spp.) | Wigglesworthia, Sodalis, Wolbachia | Trypanosoma brucei spp. | 15,16, 18, 19, 44, 51, 58, 59, 76,77 |
| ‘Kissing’ bug (Triatomine spp.) | Rhodococcus, Serratia, Nocardia, Gordonia | Trypanosoma cruzi | 101, 102, 103, 104 |
| Mosquito (Aedes, Anopheles, Culex spp.) | Asaia, Wolbachia, Serratia | Protozoan parasites, filarial worms, viruses | 27,28,29, 30,31 |
| Pea aphid (Acyrthosiphon pisum) | Buchnera, Serratia, Hamiltonella | N/Aa (agricultural pest) | 10,17, 48, 49 |
| Grain weevil (Sitophilus oryzae) | SOPE (Sitophilus oryzae primary endosymbiont), Wolbachia | N/A (agricultural pest) | 13,14, 53, 54 |
| Carpenter ant (Camponotus ligniperdus) | Blochmannia, Wolbachia | N/A (agricultural pest) | 12 |
N/A, not applicable
Interestingly, many vector insects exhibit an innate resistance to the pathogens they transmit. Both laboratory and field studies indicate that only a small proportion of insects that acquire pathogens actually allow for these organisms to establish successful infections for subsequent transmission to the next host. In fact, the majority of insects are capable of eliminating pathogens in their midgut shortly after acquisition in the bloodmeal. An insect’s genetic ability to transmit pathogens is measured in terms of its vector competence. An important component of vector competence is the proficiency of the host insect’s immune responses.
Insect vector competence and a role for the microbiome
Multiple immunity pathways are implicated in vector competence, which results in insect host resistance to pathogen infection (for mosquito responses to malarial parasites, see [33]). An initial study that investigated gene expression responses of An. gambiae to microbial and malaria challenges by using cDNA microarrays constructed from an EST-clone collection noted that the response to malaria parasites partially overlapped with the response to Gram positive (+) and Gram− bacteria [34]. Using a similar array approach the mosquito response to the filarial worm has also been found to involve the induction of a large number of genes functioning in the innate immune pathways shown to respond to microbial challenge, although their role in parasite transmission remains to be confirmed [35].
In the case of tsetse flies, trypanosome infections induced tsetse’s innate immune responses that are typically involved in clearance of Gram− bacteria; i.e. the Immune deficient (Imd) pathway [36]. Trypanosome infection prevalence increased in flies when the expression of the Imd pathway regulator relish or the downstream expressed antimicrobial peptide (AMP) effector (attacin) were downregulated by an RNA interference (RNAi)-based reverse genetic analysis before subjecting flies to parasite infections [36, 37]. In addition, the AMPs Diptericin and Attacin displayed trypanocidal activity both in vitro and in vivo in the tsetse’s midgut [38].
To understand the role of Aedes aegypti mosquito immune responses to dengue virus pathogen transmission, Xi et al. used high-throughput analysis of gene expression and an RNAi approach, reporting that another innate immune pathway (Toll pathway) regulates viral resistance in mosquitoes [39]. Interestingly, the same study showed that regulation of genes in this immune pathway was also stimulated by natural gut microbiota. Furthermore, when mosquitoes were reared aseptically (in the absence of their endogenous bacterial flora), dengue virus was present in midguts at 2-fold higher titers compared to wild type mosquitoes, implicating once again the microbial fauna in influencing levels of immune resistance.
Early studies on mosquitoes had noted a positive correlation between midgut microbiota and inhibition of Plasmodium sporozoite development. This phenomenon was demonstrated independently in two different mosquito vectors of Plasmodium. In one set of experiments An. stephensi adults were offered a P. falciparum gametocyte-enriched blood meal that also contained one of four distinct non-native Gram− bacteria or two distinct Gram+ species. All of the Gram− bacteria tested were found to partially or completely inhibit oocyst development within the mosquito host. Conversely, the presence of neither Gram+ bacteria resulted in an inhibitory phenotype [40]. A similar experiment was performed using aseptic An. albimanus adults and several bacteria that are found naturally in both wild type laboratory and field-captured populations. In this situation, the number of mosquitoes infected with oocysts, and oocyst density, was significantly lower in mosquitoes that received each of the bacteria separately as compared to controls that received no bacteria [41, 42]. Although no physiological mechanism was proposed at the time, this study indicated that gut microbes have the potential to reduce the capacity of mosquitoes to vector Plasmodium. Recently aseptic and septic adult An. gambiae were fed with Plasmodium gametocytes and subsequently monitored parasite infection status in each group [27]. Aseptic mosquitoes displayed an increased susceptibility to Plasmodium infection. This study also found that co-feeding mosquitoes bacteria and P. falciparum gametocytes resulted in lower than normal infection levels. While ookinete number was the same in the midgut lumen of both mosquito lines, significantly more oocysts were found in the antibiotic treated aseptic mosquitoes. These results indicated that bacteria affect parasite viability prior to the oocyst formation. This may occur while the ookinete is in the midgut or while invading midgut epithelial cells. Global transcription profiling of septic and aseptic mosquitoes identified a significant subset of immune genes in the septic mosquitoes that were presumably upregulated by the host's microbial flora. These immunity genes included several anti-Plasmodium factors [27, 43]. Both expression and infection analyses suggest that the observed anti-Plasmodium effect is caused by the mosquito’s antimicrobial immune response, possibly through activation of basal immunity [33].
The commensal and obligate microbes of tsetse (Sodalis and Wigglesworthia, respectively) have also been implicated in trypanosome transmission in tsetse. There has been a correlation observed with the presence of the commensal symbiont Sodalis and in particular specific Sodalis genotypes and the presence of trypanosome infections in several natural tsetse populations. Also in the absence of the obligate symbiont Wigglesworthia in the laboratory, flies have been found to be highly susceptible to parasite infections, as discussed below [44].
Microbiota may influence their hosts’ vectorial competence by means of direct interaction with parasites. This may occur through inhibitory bioactivity of secreted enzymes or toxins. Alternatively, microbiota may constrain pathogen development indirectly by inducing activity of the host immune system that in turn can clear the pathogenic microbes. Recent studies in several insect systems indicate that both direct and indirect microbiota-induced phenotypes can affect an insect host’s capacity to transmit pathogens (discussed in Box 2). In fact, both heritable symbionts and environmentally acquired commensal microfauna have been shown to influence this function (reviewed in [45, 46]). Below we discuss examples of symbiont produced anti-pathogen products, symbiont induced host anti-pathogen products and host immune priming by resident gut fauna as well as by Wolbachia.
Box 2. Manipulating the microbiome to modulate insect host vector competence.
Endosymbionts are being investigated for their ability to decrease host vector competence. One promising symbiont-based strategy currently under development is called ‘paratransgenesis’. This procedure involves isolating and genetically modifying symbiotic bacteria to express an anti-pathogen molecule. The recombinant symbionts are then reintroduced into their host, where they subsequently increase host resistance to pathogens. Three disease vector systems where this approach has been applied are described below.
Tsetse: Tsetse’s commensal symbiont, Sodalis, has been genetically modified in vitro to express a marker gene, and then returned to fertile females where they are subsequently passed on to future generations (reviewed in [5]). Sodalis exhibits a natural resistance to several trypanocidal molecules, including tsetse antimicrobial peptides. Expression of these molecules by Sodalis can result in parasite resistance in tsetse’s midgut [98–100].
Triatome bugs: Triatomines that transmit the causative agent of Chagas disease (Trypanosoma cruzi) harbor genetically modifiable, gut-associated symbionts that are horizontally transmitted via copharagy [101]. These symbionts can be cultured and genetically modified [101–103]. Host recolonization with genetically modified symbionts that express anti-parasitic molecules subsequently blocks parasite transmission [104].
Mosquito: Anopheline mosquitoes form stable associations with bacteria from the genus Asaia that may be used for paratransgenesis [30]. Stable infections can be established in multiple epidemiologically relevant tissues when female mosquitoes were reconstituted with recombinant bacteria [28, 105].
Research is currently under way to improve the efficiency of paratransgenesis by optimizing several variables. These include (i) screening for other commensal symbionts that are stably associated with insect disease vectors, (ii) identifying novel anti-pathogen effector molecules that can be expressed in symbionts, (iii) engineering expression constructs that encode efficacious promoters and secretion signals, and (iv) establishing a mechanism to drive paratransgenic bacteria into field-based insect populations.
Endosymbiont produced anti-pathogen products
Aphids harbor a beneficial facultative endosymbiont, Candidatus Hamiltonella defensa that can increase host survival following attack by parasitoid wasps [17]. Interestingly, different strains of H. defensa vary in the degree of protection they confer upon their aphid host. This symbiont-mediated protective phenotype depends on the presence of a lysogenic bacteriophage (APSE) present in certain symbiont host strains [47]. Aphids that host H. defensa infected with APSE are significantly more resistant to parasitoid wasps than are their counterparts that host APSE-uninfected symbionts [48]. In laboratory studies, phage loss occurs repeatedly in H. defensa-infected aphid clonal lines. In each instance this phenomenon results in increased host susceptibility to parasitism [47]. APSE-mediated protection results when eukaryote-targeted toxins are expressed from the bacteriophage genome [48, 49]. Thus, mobile genetic elements can code for ecologically important traits including defense against parasitoids. This characteristic can endow endosymbionts with direct benefits that extend to their animal hosts.
Symbiont induced host anti-pathogen products
How the beneficial microbiome can evade their host’s immune responses is actively being investigated, a phenomenon referred to as host tolerance. One suggested mechanism leading to host tolerance involves symbiont suppression of host immune responses as a form of self-protection. An alternative possible mechanism is the absence of molecular signals on symbiont cell surfaces that are typically recognized by insect hosts as foreign following exposure to microbes. One molecular signal that mediates host recognition of bacteria is the peptidoglycan (PGN) structure present in bacterial membranes. A family of host PGN recognition proteins (PGRPs) have been identified with PGN binding activity and include pattern recognition receptors (PRRs), which serve as the initial component of signal transduction pathways that result in immune activation following exposure [7]. For example in insects, the Imd pathway has been shown to respond to the presence of Gram− bacterial PGN and results in the production of a battery of antimicrobial peptides. Given that the symbiotic microbes also have PGN as part of their cell structures, it has been of interest if and how these microbes can evade host recognition and/or subsequent immune damage.
In Drosophila, one of the PGRPs, PGRP-LB, has been shown to exhibit amidase catalytic activity to degrade PGN [50]. In this way, metabolically costly insect immune pathways are not activated in response to environmental microbes present at low concentrations. In the presence of a persistent infection, however, the immune responses are activated when the level of the endogenous PGRP-LB is exhausted. Interestingly in tsetse, PGRP-LB is expressed at high levels in the Wigglesworthia symbiont-containing midgut bacteriome organ [51]. Furthermore, the level of pgrp-lb expression increases as a function of host age and Wigglesworthia density [52]. This positive correlation between Wigglesworthia density and host pgrp-lb expression levels was also demonstrated in natural field populations [51]. In the weevil, Sitophilus zeamais, which also has mutualistic symbionts, pgrp-lb expression was also detected in the bacteriome organ and found to be upregulated in the nymphal phase during a time when the symbionts are released from host cells [53, 54]. Thus, symbiont density regulation through the action of catalytic PGRPs may be a general mechanism that insects use to downregulate their immune responses, which could otherwise be damaging to their symbiotic fauna. In fact when pgrp-lb levels were reduced through the use of RNAi, the density of Wigglesworthia was significantly reduced. This decrease in Wigglesworthia numbers probably resulted from the activation of the Imd pathway, and induced synthesis of AMPs in tsetse as symbiont numbers were restored when both PGRP-LB and Imd pathway functions were downregulated [51]. Thus, host PGRP-LB expression appears to be essential to protect Wigglesworthia from damage by its host’s immune responses.
In addition to protecting the indispensable symbiosis with Wigglesworthia, PGRP-LB also plays a role in parasite transmission in tsetse, as flies with diminished pgrp-lb levels were found to exhibit increased susceptibility to trypanosomes [51]. It remains to be seen, however, if tsetse PGRP-LB may have an anti-protozoal activity. One PGRP in Drosophila and all PGRPs in humans have direct antibacterial properties [55]. Thus PGRP-LB may have a dual function in tsetse. First, PGRP-LB expressed in response to Wigglesworthia may protect tsetse’s mutualistic symbiosis by negatively modulating the activity of the Imd pathway, which when induced can be harmful to this bacterium. Secondly, high PGRP-LB levels benefits tsetse by preventing the establishment of trypanosome infections, which in turn negatively impact host reproductive fitness and decrease fecundity by increasing the larval development period [56].
Immune priming by resident gut fauna
Recent studies indicate that An. gambiae immune response to malaria infection involves an intimate association between the presence of microbiota in the gut and hemocytes in the hemolymph [57]. Hemocytes, which are functionally homologous to mammalian macrophages, are an essential component of insect cellular immunity. Recent research demonstrated that when mosquitoes are pre-exposed to infection with Plasmodium ookinetes they subsequently exhibit enhanced immunity to a second parasite challenge [57]. During establishment of the initial infection, malaria ookinetes irreversibly compromise physical barriers that otherwise prevent bacteria normally found only in the gut lumen from directly interacting with host epithelial cells. When this atypical encounter occurs, hemocytes that are attached to the basal surface of host epithelial cells, as well as those circulating freely in the hemolymph, differentiate into a subtype (granulocytes) that enhances host immunity to the second challenge with malaria parasites. This ‘priming’ of the host immune system was only effective when endogenous midgut bacteria were present in the system and did not occur in aseptic mosquitoes.
Tsetse flies rely on their symbiotic microbiota for proper immune function development [58]. During tsetse’s unique viviparous mode of reproduction, the symbionts are vertically transmitted from mother to developing intrauterine larvae via milk gland secretions (Figure 1). In adult tsetse two distinct populations of obligate Wigglesworthia are present. The first population is intracellular within tsetse’s bacteriome organ, while the second population is extracellular in milk in the milk gland organ [44, 59]. Treatment of pregnant females with the antibiotic ampicillin results in the clearance of the second population, extracellular in the milk, while the intracellular bacteriome-associated Wigglesworthia are left undisturbed. Thus, immature progeny of ampicillin-treated females (GmmWgm−) lack Wigglesworthia throughout intrauterine development and later as adults [44]. Adult GmmWgm− exhibit a highly immuno-compromised phenotype compared to their wild type counterparts. In fact, challenge of GmmWgm− individuals with Escherichia coli results in bacterial sepsis and death while wild type flies eliminate the same infection and survive. Furthermore, when challenged with trypanosomes, gut infections are established in the majority of GmmWgm− flies, while wild type flies are highly resistant and can clear parasite infections effectively [44, 60]. Interestingly, Wigglesworthia does not directly enhance immunity in wild type individuals, as elimination of Wigglesworthia from adult flies via antibiotic tetracycline treatment does not result in an immune compromised phenotype. Instead, this obligate must be present during the larval immature stages in order for tsetse’s immune system to develop and function properly during adulthood [58]. Gene expression studies indicate that adult GmmWgm− exhibit highly depleted humoral and cellular immune responses. This condition is characterized by reduced expression of genes that encode AMPs (cecropin and attacin), hemocyte-mediated processes (thioester-containing proteins 2 and 4 and prophenoloxidase) and signal-mediating molecules (inducible nitric oxide synthase) following challenge with bacteria. Furthermore, GmmWgm− adults house a reduced population of sessile and circulating hemocytes. This phenotype may result from a significant decrease in larval expression of serpent and lozenge, both of which are transcription factors regulating the process of early hemocyte differentiation [58]. The specific physiological mechanism(s) underlying Wigglesworthia-induced maturation of tsetse immune system development, and the implications this has on host vector competence, remains to be determined.
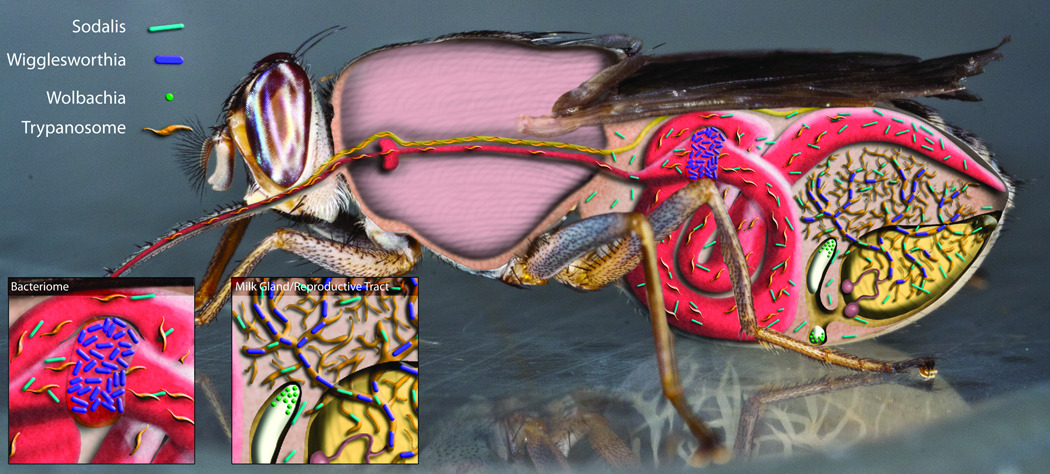
Figure 1.
Tsetse female with its symbionts Sodalis, Wigglesworthia and Wolbachia and the parasite African trypanosome. The cartoon shows the major organs where the symbionts are located. During its transmission in the fly, the trypanosome resides in the midgut and then in the salivary glands of the fly. In the midgut the trypanosome is in close proximity to Wigglesworthia housed in the bacteriome organ and Sodalis found in the midgut. Thus anti-pathogenic products expressed by Sodalis or induced by Wigglesworthia can have an adverse effect on trypanosome transmission (shown by inset labeled bacteriome). The symbionts Sodalis and Wigglesworthia are maternally transmitted to the intrauterine progeny in the milk while Wolbachia is transovarially transmitted (shown by inset labeled milk gland/reproductive tract). Contributed by Geoffrey Attardo.
Immune priming by Wolbachia
Until recently, it was thought that Wolbachia infections of insects were largely parasitic and had invaded host populations by manipulating the reproduction of their hosts to increase their transmission through the female germline. However, new studies are suggesting that Wolbachia infections in Drosophila can confer resistance to viruses and therefore act as mutualists [25]. Initial work with Drosophila showed that certain Wolbachia infections reduced virus proliferation of Drosophila C, cricket paralysis, Nora and Flock House viruses and delayed mortality in flies [61–63]. Subsequently, a similar pathogen blocking phenomenon was shown to affect A. aegypti mosquitoes. Transinfection of A. aegypti with the life-shortening Wolbachia strain wMelPop resulted in reduced transmission of medically important pathogens, including dengue and Chikungunya viruses, Plasmodium gallinaceum (cause of avian malaria) and Brugia pahangi (rodent filarial nematodes) [61, 62, 64–66]. Replication of dengue virus following either ingestion or intrathoracic injection was almost completely inhibited in Wolbachia infected mosquitoes. Similarly, Wolbachia infections reduced the number of mosquitoes infected with P. gallinaceum by about 50% and lowered oocyte numbers significantly [66]. Wolbachia infections in A. aegypti also significantly reduced the prevalence and mean number of infective third stage filarial nematodes by more than two-fold compared to uninfected controls [64]. This inhibition of nematode development has been hypothesized to result at least partially from a significant Wolbachia-induced up-regulation of immune genes that encode antimicrobial peptides (cecropin and defensin), thio-ester containing proteins (TEP) and C-type lectins, as shown by microarray analysis [66]. Wolbachia infections naturally associated with Culex quinquefasciatus have also been found to influence West Nile virus transmission. Wolbachia-infected Culex mosquitoes were found to produce lower virus titers and had 2 to 3-fold lower rates of virus transmission compared to mosquitoes lacking Wolbachia [67]. Stable infection of an An. gambiae cell line with wMelPop caused increased expression of malaria-related immune genes, indicating that host resistance to parasite infection may be regulated by symbiont-induced immunity. While it has not been possible to establish stable infection of An. gambiae mosquitoes with wMelPop, transient somatic infections can be established by intrathoracic inoculation. This procedure induces the expression of several host immune-related genes, including LRIM1 and TEP1 that have been shown to influence the development of malaria parasites. Plasmodium infection intensity in mosquitoes that received this treatment was thereupon significantly reduced in comparison to uninfected cohorts [68]. In contrast, in a different study where gene expression of Wolbachia infected mosquito cells were analyzed by Affymetrix GeneChip microarray, a significant down-regulation of many immune, stress and detoxification-related transcripts were noted [69]. It will remain to be seen if successful Wolbachia infections can be established in Anopheline mosquitoes where they confer a pathogen resistance phenotype. Thus in several different insect systems in the laboratory and in natural populations, infections with specific Wolbachia strains apparently can confer resistance to the pathogens typically transmitted by the same hosts. This pathogen blocking process appears to involve induced expression of host immune responses against the pathogens. It is important to understand the fitness cost of such Wolbachia infections on their hosts, in particular on fecundity for field applications. Ability to harness Wolbachia conferred pathogen blocking coupled with CI outcomes provides a novel approach for disease control where Wolbachia infected, pathogen resistant insects can be spread via CI and replace the susceptible populations [70].
Concluding remarks
In this paper, we reviewed major trends on symbiotic microbes that are associated with different insects, including those that are disease vectors. Evidence to date indicates that these beneficial microorganisms are indispensable for nutrient provisioning and for proper host immune system maturation during development. We also discussed vector control strategies that use insect symbionts to ‘artificially’ boost their host’s immune system are currently under development. The goal of these methods is to reduce the dissemination of pathogenic microbes. In Box 3 several future research directions are suggested that will enhance the ability to exploit insect symbiotic bacteria for the purpose of reducing disease transmission.
Box 3. Future Directions.
This review summarizes our current knowledge on the relationship between insect symbionts and host disease vector competence. Below we highlight future research avenues that will contribute to the development of microbiome-based strategies for the control of insect-transmitted diseases.
Characterize the microbiome of natural vector populations, with special emphasis placed on evaluating differences between infected and pathogen resistant insects. This research could lead to the identification of symbiotic microbes that naturally confer resistance to pathogens.
Identify Wolbachia endosymbiont strains that are capable of modifying host immune competence and/or fitness as a novel means of vector control.
Ecological studies must be performed to better understand host-symbiont infection dynamics as a function of space and time and varying environmental conditions. This information is essential for the success of downstream field applications.
Large-cage studies, followed by open field releases, are necessary to access the ecological stability of modified insects in natural habitats. While technical aspects associated with these methodologies are advancing, these programs now need to address societal challenges such as regulatory approval and ethical acceptance. One such study is ongoing in Australia for control of the dengue vectors (http://www.eliminatedengue.org/en/HOME.aspx).
Despite challenges, these self-sustainable and cost-effective programs can provide unprecedented opportunities for control of tropical disease, for which limited funds are available.
Acknowledgements
We are grateful to our laboratory members for their contributions to the work presented on the tsetse system. We thank Geoffrey Attardo for the artwork provided in this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Round JL, et al. Coordination of tolerogenic immune responses by the commensal microbiota. J Autoimmun. 2010;34:J220. doi: 10.1016/j.jaut.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazmanian SK, et al. Capsular polysaccharides of symbiotic bacteria modulate immune responses during experimental colitis. J Pediatr Gastroenterol Nutr. 2008;46 Suppl 1:E11. doi: 10.1097/01.mpg.0000313824.70971.a7. [DOI] [PubMed] [Google Scholar]
- 4.Mazmanian SK, et al. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 5.Rio RV, et al. Strategies of the home-team: symbioses exploited for vector-borne disease control. Trends Microbiol. 2004;12:325. doi: 10.1016/j.tim.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Beard CB, et al. Bacterial symbiosis and paratransgenic control of vector-borne Chagas disease. International journal for parasitology. 2001;31:621. doi: 10.1016/s0020-7519(01)00165-5. [DOI] [PubMed] [Google Scholar]
- 7.Gross R, et al. Immunity and symbiosis. Molecular microbiology. 2009;73:751. doi: 10.1111/j.1365-2958.2009.06820.x. [DOI] [PubMed] [Google Scholar]
- 8.Bextine B, et al. Delivery of a genetically marked Alcaligenes sp. to the glassy-winged sharpshooter for use in a paratransgenic control strategy. Curr Microbiol. 2004;48:327. doi: 10.1007/s00284-003-4178-2. [DOI] [PubMed] [Google Scholar]
- 9.Wernegreen JJ, et al. Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet. 2002;3:850. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
- 10.Shigenobu S, et al. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 11.Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 12.Degnan PH, et al. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 2005;15:1023. doi: 10.1101/gr.3771305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heddi A, et al. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci U S A. 1999;96:6814. doi: 10.1073/pnas.96.12.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil R, et al. Massive presence of insertion sequences in the genome of SOPE, the primary endosymbiont of the rice weevil Sitophilus oryzae. Int Microbiol. 2008;11:41. [PubMed] [Google Scholar]
- 15.Akman L, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32:402. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- 16.Aksoy S. Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int J Syst Bacteriol. 1995;45:848. doi: 10.1099/00207713-45-4-848. [DOI] [PubMed] [Google Scholar]
- 17.Degnan PH, et al. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci U S A. 2009;106:9063. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akman L, et al. Genome size determination and coding capacity of Sodalis glossinidius, an enteric symbiont of tsetse flies, as revealed by hybridization to Escherichia coli gene arrays. J Bacteriol. 2001;183:4517. doi: 10.1128/JB.183.15.4517-4525.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dale C, Maudlin I. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. International Journal of Systematic Bacteriology. 1999;49(Pt 1):267. doi: 10.1099/00207713-49-1-267. [DOI] [PubMed] [Google Scholar]
- 20.Werren JH. Biology of Wolbachia. Annu Rev Entomol. 1997;42:587. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- 21.Werren JH, et al. Evolution and Phylogeny of Wolbachia - Reproductive Parasites of Arthropods. Proceedings of the Royal Society of London Series B-Biological Sciences. 1995;261:55. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- 22.Sinkins S, O'Neill S. In: Insect Transgenesis. Handler A, James A, editors. New York: CRC Press; 2000. pp. 271–287. [Google Scholar]
- 23.Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat Rev Genet. 2006;7:427. doi: 10.1038/nrg1870. [DOI] [PubMed] [Google Scholar]
- 24.Cook PE, et al. Modifying insect population age structure to control vector-borne disease. Adv Exp Med Biol. 2008;627:126. doi: 10.1007/978-0-387-78225-6_11. [DOI] [PubMed] [Google Scholar]
- 25.Cook PE, McGraw EA. Wolbachia pipientis: an expanding bag of tricks to explore for disease control. Trends in parasitology. 2010;26:373. doi: 10.1016/j.pt.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Lindh JM, et al. 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl Environ Microbiol. 2005;71:7217. doi: 10.1128/AEM.71.11.7217-7223.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong Y, et al. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Favia G, et al. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9047. doi: 10.1073/pnas.0610451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zouache K, et al. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol Ecol. 2010 doi: 10.1111/j.1574-6941.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- 30.Damiani C, et al. Mosquito-bacteria symbiosis: the case of Anopheles gambiae and Asaia. Microb Ecol. 2010;60:644. doi: 10.1007/s00248-010-9704-8. [DOI] [PubMed] [Google Scholar]
- 31.Chouaia B, et al. Molecular evidence for multiple infections as revealed by typing of Asaia bacterial symbionts of four mosquito species. Applied and environmental microbiology. 2010;76:7444. doi: 10.1128/AEM.01747-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill CA, et al. Arthropod-borne diseases: vector control in the genomics era. Nat Rev Microbiol. 2005;3:262. doi: 10.1038/nrmicro1101. [DOI] [PubMed] [Google Scholar]
- 33.Cirimotich CM, et al. Mosquito immune defenses against Plasmodium infection. Dev Comp Immunol. 2010;34:387. doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimopoulos G, et al. Genome expression analysis of Anopheles gambiae: responses to injury, bacterial challenge, and malaria infection. Proc Natl Acad Sci U S A. 2002;99:8814. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erickson SM, et al. Mosquito infection responses to developing filarial worms. PLoS neglected tropical diseases. 2009;3:e529. doi: 10.1371/journal.pntd.0000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hao Z, et al. Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12648. doi: 10.1073/pnas.221363798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu C, Aksoy S. Innate immune responses regulate trypanosome parasite infection of the tsetse fly Glossina morsitans morsitans. Molecular microbiology. 2006;60:1194. doi: 10.1111/j.1365-2958.2006.05180.x. [DOI] [PubMed] [Google Scholar]
- 38.Hu Y, Aksoy S. An antimicrobial peptide with trypanocidal activity characterized from Glossina morsitans morsitans. Insect Biochem. Mol. Biol. 2005;35:105. doi: 10.1016/j.ibmb.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Xi Z, et al. The Aedes aegypti toll pathway controls dengue virus infection. PLoS pathogens. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pumpuni CB, et al. Plasmodium falciparum: inhibition of sporogonic development in Anopheles stephensi by gram-negative bacteria. Exp Parasitol. 1993;77:195. doi: 10.1006/expr.1993.1076. [DOI] [PubMed] [Google Scholar]
- 41.Pumpuni CB, et al. Bacterial population dynamics in three anopheline species: the impact on Plasmodium sporogonic development. Am J Trop Med Hyg. 1996;54:214. doi: 10.4269/ajtmh.1996.54.214. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez-Ceron L, et al. Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J Med Entomol. 2003;40:371. doi: 10.1603/0022-2585-40.3.371. [DOI] [PubMed] [Google Scholar]
- 43.Dong Y, et al. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pais R, et al. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Applied and environmental microbiology. 2008;74:5965. doi: 10.1128/AEM.00741-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azambuja P, et al. Gut microbiota and parasite transmission by insect vectors. Trends in parasitology. 2005;21:568. doi: 10.1016/j.pt.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 46.Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49:71. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 47.Oliver KM, et al. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Degnan PH, Moran NA. Evolutionary genetics of a defensive facultative symbiont of insects: exchange of toxin-encoding bacteriophage. Mol Ecol. 2008;17:916. doi: 10.1111/j.1365-294X.2007.03616.x. [DOI] [PubMed] [Google Scholar]
- 49.Degnan PH, Moran NA. Diverse phage-encoded toxins in a protective insect endosymbiont. Applied and environmental microbiology. 2008;74:6782. doi: 10.1128/AEM.01285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, et al. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12133. doi: 10.1073/pnas.0901226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rio RV, et al. Dynamics of multiple symbiont density regulation during host development: tsetse fly and its microbial flora. Proc Biol Sci. 2006;273:805. doi: 10.1098/rspb.2005.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anselme C, et al. Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Applied Environmental Microbiology. 2006;72:6766. doi: 10.1128/AEM.00942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anselme C, et al. Identification of the weevil immune genes and their expression in the bacteriome tissue. BMC Biol. 2008;6:43. doi: 10.1186/1741-7007-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dziarski R, Gupta D. Mammalian PGRPs: novel antibacterial proteins. Cell Microbiol. 2006;8:1059. doi: 10.1111/j.1462-5822.2006.00726.x. [DOI] [PubMed] [Google Scholar]
- 56.Hu C, et al. Infections with immunogenic trypanosomes reduce tsetse reproductive fitness: potential impact of different parasite strains on vector population structure. PLoS Negl Trop Dis. 2008;2:e192. doi: 10.1371/journal.pntd.0000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodrigues J, et al. Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science. 2010;329:1353. doi: 10.1126/science.1190689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss B, et al. Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 2011 doi: 10.1371/journal.pbio.1000619. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Attardo GM, et al. Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J Insect Physiol. 2008;54:1236. doi: 10.1016/j.jinsphys.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aksoy S, et al. Interactions between tsetse and trypanosomes with implications for the control of trypanosomiasis. Advances in parasitology. 2003;53:1. doi: 10.1016/s0065-308x(03)53002-0. [DOI] [PubMed] [Google Scholar]
- 61.Hedges LM, et al. Wolbachia and virus protection in insects. Science. 2008;322:702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- 62.Teixeira L, et al. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osborne SE, et al. Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS pathogens. 2009;5:e1000656. doi: 10.1371/journal.ppat.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kambris Z, et al. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science. 2009;326:134. doi: 10.1126/science.1177531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bian G, et al. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS pathogens. 2010;6:e1000833. doi: 10.1371/journal.ppat.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreira LA, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139:1268. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 67.Glaser RL, Meola MA. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PloS one. 2010;5:e11977. doi: 10.1371/journal.pone.0011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kambris Z, et al. Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS pathogens. 2010;6 doi: 10.1371/journal.ppat.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hughes GL, et al. Wolbachia infections in Anopheles gambiae cells: transcriptomic characterization of a novel host-symbiont interaction. PLoS Pathog. 2011;7:e1001296. doi: 10.1371/journal.ppat.1001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Enserink M. Infectious diseases. Australia to test 'mosquito vaccine' against human disease. Science. 2010;330:1460. doi: 10.1126/science.330.6010.1460. [DOI] [PubMed] [Google Scholar]
- 71.Aksoy S. Tsetse--A haven for microorganisms. Parasitology today (Personal ed. 2000;16:114. doi: 10.1016/s0169-4758(99)01606-3. [DOI] [PubMed] [Google Scholar]
- 72.Rio RV, Aksoy S. Interactions among multiple genomes: tsetse, its symbionts and trypanosomes. Insect Biochem and Molecular Biology. 2005;691 doi: 10.1016/j.ibmb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 73.Lindh JM, Lehane MJ. The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie Van Leeuwenhoek. 2011 doi: 10.1007/s10482-010-9546-x. [DOI] [PubMed] [Google Scholar]
- 74.Geiger A, et al. Serratia glossinae sp. nov., isolated from the midgut of the tsetse fly Glossina palpalis gambiensis. Int J Syst Evol Microbiol. 2010;60:1261. doi: 10.1099/ijs.0.013441-0. [DOI] [PubMed] [Google Scholar]
- 75.Geiger A, et al. First isolation of Enterobacter, Enterococcus, and Acinetobacter spp. as inhabitants of the tsetse fly (Glossina palpalis palpalis) midgut. Infect Genet Evol. 2009;9:1364. doi: 10.1016/j.meegid.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 76.Aksoy S, et al. Mycetome endosymbionts of tsetse flies constitute a distinct lineage related to Enterobacteriaceae. Insect molecular biology. 1995;4:15. doi: 10.1111/j.1365-2583.1995.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 77.Chen X, et al. Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J Mol Evol. 1999;48:49. doi: 10.1007/pl00006444. [DOI] [PubMed] [Google Scholar]
- 78.Akman L, Aksoy S. A novel application of gene arrays: Escherichia coli array provides insight into the biology of the obligate endosymbiont of tsetse flies. Proc. Natl. Acad. Sci. USA. 2001;98:7546. doi: 10.1073/pnas.131057498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weiss BL, et al. Inter-specific transfer of bacterial endosymbionts between tsetse species: infection establishment and effect on host fitness. Appl Environ Microbiol. 2006;72:7013. doi: 10.1128/AEM.01507-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaiwa N, et al. Primary gut symbiont and secondary, Sodalis-allied symbiont of the Scutellerid stinkbug Cantao ocellatus. Applied and environmental microbiology. 2010;76:3486. doi: 10.1128/AEM.00421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Novakova E, Hypsa V. A new Sodalis lineage from bloodsucking fly Craterina melbae (Diptera, Hippoboscoidea) originated independently of the tsetse flies symbiont Sodalis glossinidius. FEMS Microbiol Lett. 2007;269:131. doi: 10.1111/j.1574-6968.2006.00620.x. [DOI] [PubMed] [Google Scholar]
- 82.Heddi A, et al. Molecular Characterization of the Principal Symbiotic Bacteria of the Weevil Sitophilus oryzae: A Peculiar G+C Contents of an Endocytobiotic DNA. J Molecular Evolution. 1998;47:52. doi: 10.1007/pl00006362. [DOI] [PubMed] [Google Scholar]
- 83.Fukatsu T, et al. Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Applied and environmental microbiology. 2007;73:6660. doi: 10.1128/AEM.01131-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng Q, Aksoy S. Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect molecular biology. 1999;8:125. doi: 10.1046/j.1365-2583.1999.810125.x. [DOI] [PubMed] [Google Scholar]
- 85.Toh H, et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Belda E, et al. Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC genomics. 2010;11:449. doi: 10.1186/1471-2164-11-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dale C, Moran NA. Molecular Interactions between Bacterial Symbionts and Their Hosts. Cell. 2006;126:453. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 88.Dale C, et al. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Natl Acad Sci USA. 2002;99:12397. doi: 10.1073/pnas.182213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dale C, et al. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci U S A. 2001;98:1883. doi: 10.1073/pnas.021450998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dale C, Welburn SC. The endosymbionts of tsetse flies: manipulating host-parasite interactions. International journal for parasitology. 2001;31:628. doi: 10.1016/s0020-7519(01)00151-5. [DOI] [PubMed] [Google Scholar]
- 91.Welburn SC, Maudlin I. Tsetse-trypanosome interactions: rites of passage. Parasitology today (Personal ed. 1999;15:399. doi: 10.1016/s0169-4758(99)01512-4. [DOI] [PubMed] [Google Scholar]
- 92.Weiss BL, et al. An insect symbiosis is influenced by bacterium-specific polymorphisms in outer-membrane protein A. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15088. doi: 10.1073/pnas.0805666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Geiger A, et al. Vector competence of Glossina palpalis gambiensis for Trypanosoma brucei s.l. and genetic diversity of the symbiont Sodalis glossinidius. Molecular biology and evolution. 2007;24:102. doi: 10.1093/molbev/msl135. [DOI] [PubMed] [Google Scholar]
- 94.Farikou O, et al. Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes--an epidemiological approach in two historical human African trypanosomiasis foci in Cameroon. Infect Genet Evol. 2010;10:115. doi: 10.1016/j.meegid.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 95.Geiger A, et al. Vector Competence of Glossina palpalis gambiensis for Trypanosoma brucei s.l. and Genetic Diversity of the Symbiont Sodalis glossinidius. Mol Biol Evol. 2006 doi: 10.1093/molbev/msl135. [DOI] [PubMed] [Google Scholar]
- 96.O'Neill SL, Gooding RH, Aksoy S. Phylogenetically distant symbiotic microorganisms reside in Glossina midgut and ovary tissues. Medical and Veterinary Entomology. 1993;7:377. doi: 10.1111/j.1365-2915.1993.tb00709.x. [DOI] [PubMed] [Google Scholar]
- 97.Cheng Q. Tissue distribution and prevalence of Wolbachia infections in tsetse flies, Glossina spp. Med Vet Entomol. 2000;14:44. doi: 10.1046/j.1365-2915.2000.00202.x. [DOI] [PubMed] [Google Scholar]
- 98.Hu Y, Aksoy S. An antimicrobial peptide with trypanocidal activity characterized from Glossina morsitans morsitans. Insect biochemistry and molecular biology. 2005;35:105. doi: 10.1016/j.ibmb.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 99.Haines LR, et al. Cationic antimicrobial peptide killing of African trypanosomes and Sodalis glossinidius, a bacterial symbiont of the insect vector of sleeping sickness. Vector Borne Zoonotic Dis. 2003;3:175. doi: 10.1089/153036603322662165. [DOI] [PubMed] [Google Scholar]
- 100.Haines LR, et al. Killing of trypanosomatid parasites by a modified bovine host defense peptide, BMAP-18. PLoS neglected tropical diseases. 2009;3:e373. doi: 10.1371/journal.pntd.0000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Durvasala R, et al. In: Insect Symbiosis. Bourtzis K, Thomas AM, editors. New York: CRC Press; 2003. [Google Scholar]
- 102.Durvasula RV, et al. Genetic transformation of a Corynebacterial symbiont from the Chagas disease vector Triatoma infestans. Experimental parasitology. 2008;119:94. doi: 10.1016/j.exppara.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sundaram RK. Expression of a functional single-chain antibody via Corynebacterium pseudodiphtheriticum. Eur J Clin Microbiol Infect Dis. 2008;27:617. doi: 10.1007/s10096-008-0483-9. [DOI] [PubMed] [Google Scholar]
- 104.Durvasula R, et al. Expression of a functional antibody fragment in the gut of Rhodnius prolixus via transgenic bacterial symbiont Rhodococcus rhodnii. Med Vet Entomology. 1999;13:115. doi: 10.1046/j.1365-2915.1999.00175.x. [DOI] [PubMed] [Google Scholar]
- 105.Favia G, et al. Bacteria of the genus Asaia: a potential paratransgenic weapon against malaria. Adv Exp Med Biol. 2008;627:49. doi: 10.1007/978-0-387-78225-6_4. [DOI] [PubMed] [Google Scholar]