Introduction
The Thanatos (the Greek god of death)-associated protein (THAP) domain is a sequence specific DNA binding domain that contains a C2-CH (Cys-Xaa2-4-Cys-Xaa35-50-Cys-Xaa2-His) zinc finger that is similar to the DNA domain of the P element transposase from Drosophila1. THAP containing proteins have been observed in the proteome of humans, pigs, cows, chickens, zebrafish, Drosophila, C. elegans, and Xenopus1. To date there are no known THAP domain proteins in plants, yeast, or bacteria. There are 12 indentified human THAP domain-containing proteins (THAP0 – 11). In all human THAP protein, the THAP domain is located at the N-terminus and is approximately 90 residues in length. While all of the human THAP containing proteins have a homologous N-terminus, there is extensive variation in both the predicted structure and length of the remaining protein1. Even though the exact function of these THAP proteins is not well defined, there is evidence they play a role in cell proliferation2, apoptosis3, cell cycle modulation4, chromatin modification5, and transcriptional regulation6. THAP containing proteins have also been implicated in a number of human disease states including heart disease4, neurological defects7 and several types of cancers6,8,9.
Human THAP4 is a 577 residue protein of unknown function that is proposed to bind DNA in a sequence specific manner similar to THAP110,11 and has been found to be upregulated in response to heat shock12. THAP4 is expressed in a relatively uniform manner in a broad range of tissues and appears to be upregulated in lymphoma cells and highly expressed in heart cells13. The C-terminal domain of THAP4 (residues 415-577), designated here as cTHAP4, is evolutionarily conserved and is observed in all know THAP4 orthologues. Several single domain proteins lacking a THAP domain are found in plants and bacteria, and show significant levels of homology to cTHAP4. It appears that cTHAP4 belongs to a large class of proteins that have yet to be fully functionally characterized.
On the basis of prior work, we predicted that cTHAP4 is composed of a heme-binding nitrobindin domain14, making THAP4 the only human THAP protein predicted to bind a cofactor. Nitrobindin, a recently characterized protein from Arabidopsis thaliana14, is structurally similar and exhibits nitric oxide (NO) binding properties that resemble the heme-binding nitrophorins. Nitrophorins utilize a heme moiety to store, transport, and release NO in a pH specific manner15. While the exact function of nitrobindin is not fully known, the similarities between the well characterized NPs implies a role in NO transport, sensing, or metabolism14. In order to better elucidate the possible function of THAP4, we solved the heme-bound structure of cTHAP4 to a resolution of 1.79 Å.
Material and Methods
Protein purification
The selenomethionine labeled protein was generated utilizing the standard Center for Eukaryotic Structural Genomics (CESG) pipeline protocols for cloning, protein expression, protein purification, and overall information management16-19. Incorporation of the hemin group was completed as previously described14.
Crystallization, Diffraction Data Collection, and Structure Solution
Crystals utilized for data collection were grown at 4° C by hanging drop vapor diffusion by mixing 2 μL of an 10 mg•ml-1 cTHAP4 solution (5 mM HEPES buffer, pH 5.0 containing NaCl (50 mM) and NaN3 (0.3 mM)) with 2 μL of reservoir solution (100 mM Triethanolamine buffer at pH 7.5 containing polypropylene glycol 400 (55%)).
X-ray diffraction data were collected at the Life Sciences Collaborative Access Team (LS-CAT) 21-ID-G beamline at the Advanced Photon Source, Argonne National Laboratories. Diffraction images were indexed, integrated, and scaled using HKL2000 20. The cTHAP4 selenium substructure was determined using Hyss 21. The results from Hyss indicated there were fourteen heavy atom sites per asymmetric unit corresponding to twelve selenomethionine and two iron sites. cTHAP4 was phased to 1.79 Å using a single wavelength (0.97857 Å) with autoSharp 22. The experimental electron density map was of excellent quality, and the bound heme moites were apparent in the density modified map. The structure was completed with alternate cycles of model building in Coot 23 and refinement in Phenix 24. All refinement steps were monitored using an Rfree value based on 5.0% of the independent reflections.
The final model of cTHAP4 was refined to a resolution of 1.79 Å with an Rcryst of 0.165 and an Rfree of 0.201. All pertinent information on data collection, refinement, phasing, and model statistics are summarized in Table I. Model quality was assessed using MolProbity25. cTHAP4 renderings were generated using PyMOL 26.
Table I.
Summary of crystal parameters, data collection, structure phasing and refinement statistics. Values in parentheses are for the highest resolution shell.
| Crystal parameters | |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 41.933, b = 74.694, c = 103.064 |
|
| |
| Data collection statistics | |
| Wavelength (Å) | 0.97857 |
| Resolution range (Å) | 35.12 – 1.79 (1.84 – 1.79) |
| No. of reflections (measured / unique) | 173378 / 29830 |
| Completeness (%) | 95.9 (76.2) |
| Rmerge* | 0.093 (0.457) |
| Redundancy | 5.8 (4.8) |
| Mean I / sigma (I) | 18.16 (2.92) |
|
| |
| Phasing statistics‡ | |
| Mean FOM (centris / acentric) | 0.306 / 0.116 |
| Phasing power (anomalous) | 0.963 |
| Cullis R-factor (anomalous) | 0.846 |
|
| |
| Refinement and model statistics | |
| Resolution range (Å) | 35.12 – 1.79 |
| No. of reflections (work / test) | 29830 / 1510 |
| Rcryst§ | 0.165 (0.192) |
| Rfree¶ | 0.201 (0.248) |
| RMSD bonds (Å) | 0.009 |
| RMSD angles (°) | 1.449 |
| B factor (protein / solvent / Heme) (Å2) | 23.43 / 23.14 / 24.94 |
| No. of protein atoms | 2636 |
| No. of waters | 273 |
| No. of auxiliary molecules | 2 Heme |
|
| |
| Ramachandran plot (%) | |
| Favorable region | 98.44 |
| Additional allowed region | 1.56 |
| Disallowed region | 0.00 |
|
| |
| PDB ID | 3IA8 |
Rmerge = Σh Σi | Ii (h) − <I(h)>| / ΣhΣi Ii(h), where Ii(h) is the intensity of an individual measurement of the reflection and <I(h)> is the mean intensity of the reflection.
Phasing in autoSharp
Rcryst = Σh ‖Fobs| − |Fcalc‖ / Σh |Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes,
Rfree was calculated as Rcryst using 5.0 % of randomly selected unique reflections that were omitted from the structure refinement.
Results and Discussion
Overall structure and structure quality
The cTHAP4 structure had well-defined electron density corresponding to residues 415-576. There was not sufficient electron density to definitively place residues 413, 414, and 577, and subsequently, these residues were not modeled in the cTHAP4 structure. Two molecules of cTHAP4 were observed per asymmetric unit and the structure belonged to the space group P212121.
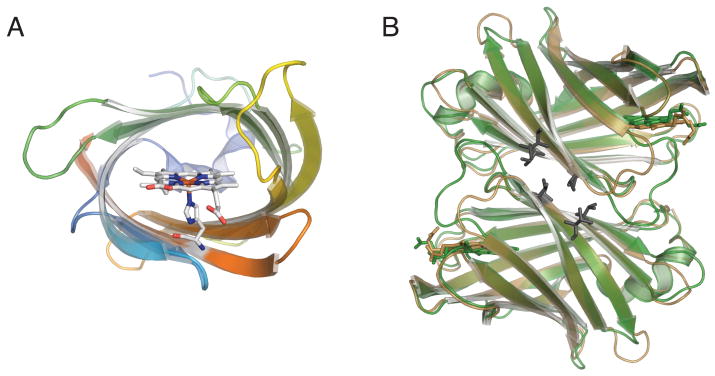
cTHAP4 is composed almost exclusively of β-strands. Ten antiparallel strands form a compact β-barrel, which is capped on one end by ten residue long 310 α-helix. The other end of the β-barrel is exposed to the solvent (Figure 1A). The overall fold of cTHAP4 is identical to the fold observed in heme-binding protein nitrobindin14, which is not surprising, given that cTHAP4 and nitrobindin share 59% sequence similarity. A large cavity composed of primarily hydrophobic residues is formed in the interior of the β-barrel. In the cTHAP4 structure, a heme molecule is bound in this hydrophobic cavity and is coordinated by His567 (Figure 1A). Similar to Nitrobindin, there is no distal histidine within bonding distance of the heme iron. However, His480 is positioned in such a way that the imizidazole ring in parallel to the heme plane. One of the two heme propionate groups forms a hydrogen bond with Thr444, while the other propionate group is solvent-exposed and does not hydrogen bond with any amino acid side chains. The heme pocket is relatively open when compared to the globins with the heme primarily solvent-exposed.
Figure 1.
A. Cartoon representation of cTHAP4 is shown going from blue at the N-terminus to red at the C-terminus. The bound heme is shown as sticks with carbon in white, nitrogen in blue, oxygen in red, and iron in orange. Looking down the barrel shows the heme coordinated by His567 and the extended hydrophobic cavity formed by the β-barrel. B. Structural alignment of the cTHAP4 dimer in green and the Nitrobindin dimer 14 (PDB ID 3EMM) in brown. Conserved hydrophobic residues that compose the dimer interface are shown as black sticks.
The two monomers of cTHAP4 in the asymmetric unit form an interface of approximately 1312 Å2 as calculated by the PISA server27, suggesting that cTHAP4 forms a dimer in the crystal lattice. The cTHAP4 dimer is identical to the dimer observed in the nitrobindin structure 14 (Figure 1B). A number of hydrophobic residues that are conserved between cTHAP4 and nitrobindin compose the dimer interface (Figure 1B). Nitrobindin likely forms a dimer in solution, further suggesting that the cTHAP4 dimmer is biologically relevant and perhaps plays a functional role.
cTHAP4 Homologues
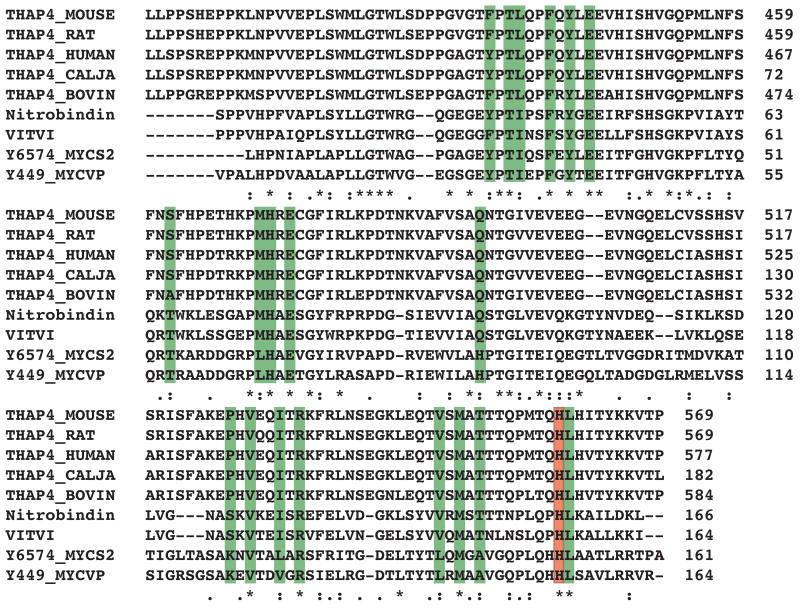
A BLAST search utilizing the cTHAP4 sequence revealed the closest cTHAP4 homologues to be THAP4 proteins from other mammalian species. In addition to the mammalian THAP4 proteins, a number of hypothetical proteins or proteins of unknown function from plant and bacteria were found to be 38% to 34% identical to cTHAP4. Of these additional proteins the only one that has been characterized is the NO binding nitrobindin. A sequence alignment of cTHAP4 and its closest homologues reveals that the heme-binding histidine (His567) and the residues within 4 Å of the bound heme are highly conserved (Figure 2). Given the conservation of the heme-binding histidine it is extremely likely that all THAP4 proteins coordinate a heme.
Figure 2.
Sequence alignment of THAP4 from Mus musculus (Q6P3Z3), THAP4 from Rattus norvegicus (Q642B6), THAP4 from Homo sapiens (Q8WY91), THAP4 from Callithrix jacchus (A6MKW1), THAP4 from Bos taurus (Q2TBI2), Nitrobindin from Arabidopsis thaliana (O64527), uncharacterized protein from Vitis vinifera (A5BBZ0), fatty acid-binding like protein from Mycobacterium smegmatis (A0R6J8), and fatty acid-binding like protein from Mycobacterium vanbaalenii (A1T297). Residues that are within 4 A of the bound heme of cTHAP4 are highlighted in green. The strictly conserved histidine that coordinates the heme is highlighted in red.
A number of cTHAP4 structural homologues were identified by Dali28. The top structure indentified was nitorbindin (PDB ID 3EMM Z = 32.8). cTAHP4 and nitrobindin are structurally nearly identical and have an overall Cα RMS deviation of 0.68 Å (Figure 1B). Two uncharacterized proteins from Mycobacterium tuberculosis (PDB ID 2FR2 Z = 24.3 and PDB ID 2FWV Z = 23.4) were close structural homologues to cTHAP4. Despite having a histidine in a structurally similar position as cTHAP4 both 2FR2 and 2FWV were solved without a bound heme 29. Several additional proteins were identified as possible structural homologues that were either hypothetical proteins or functionally annotated as fatty acid binding proteins.
Possible Function
Given the structural similarities between cTHAP4 and nitrobindin, and to a lesser extent the nitophorins15, it is probable that cTHAP4 binds NO and has similar NO binding properties to nitrobindin14. Perhaps cTHAP4 acts as a NO sensor, which utilizes the heme coordinated to His567 as a NO binding platform that can modulate the DNA binding activity of the N-terminal THAP domain. This would allow THAP4 to act as a NO dependent transcriptional regulator. While none of the THAP domain containing proteins are known to interact with NO, THAP7 and THAP11 have been shown to be transcriptional regulators 5,6. In addition to transcriptional regulation, THAP proteins has also been implicated in mediating cell death3,4. It is possible that THAP4 functions in a similar manner and is involved in NO regulated apoptosis30. While the function of THAP4 still remains unknown, the structure of cTHAP4 provides not only insight into a possible function, but also a direction for future research on the role of THAP4.
Acknowledgments
CESG has been funded by the following NIH/NIGMS grant numbers GM074901 (John L. Markley; 07/01/05-06/30/10) and GM064598 (John L. Markley; 01/01/02-08/31/05). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. GM/CA-CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). The authors would also like to thank all members of the Center for Eukaryotic Structural Genomics for various contributions.
References
- 1.Roussigne M, Kossida S, Lavigne AC, Clouaire T, Ecochard V, Glories A, Amalric F, Girard JP. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends in biochemical sciences. 2003;28(2):66–69. doi: 10.1016/S0968-0004(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 2.Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, Lazar V, Dessen P, Mantovani R, Aguilar L, Girard JP. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood. 2007;109:584–594. doi: 10.1182/blood-2006-03-012013. [DOI] [PubMed] [Google Scholar]
- 3.Roussigne M, Cayrol C, Clouaire T, Amalric F, Girard JP. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene. 2003;22:2432–2442. doi: 10.1038/sj.onc.1206271. [DOI] [PubMed] [Google Scholar]
- 4.Balakrishnan MP, Cilenti L, Mashak Z, Popat P, Alnemri ES, Zervos AS. THAP5 is a human cardiac-specific inhibitor of cell cycle that is cleaved by the proapoptotic Omi/HtrA2 protease during cell death. Am J Physiol-Heart C. 2009;297:H643–H653. doi: 10.1152/ajpheart.00234.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macfarlan T, Kutney S, Altman B, Montross R, Yu JJ, Chakravarti D. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. Journal of Biological Chemistry. 2005;280:7346–7358. doi: 10.1074/jbc.M411675200. [DOI] [PubMed] [Google Scholar]
- 6.Zhu C, Li C, Li Y, Zhan Y, Li Y, Xu C, Xu W, Sun HB, Yang X. Cell growth suppression by thanatos-associated protein 11(THAP11) is mediated by transcriptional downregulation of c-Myc. Cell death and differentiation. 2009;16:395–405. doi: 10.1038/cdd.2008.160. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs T, Gavarini S, Saunders-Pullman R, Raymond D, Ehrlich ME, Bressman SB, Ozelius LJ. Mutations in the THAP1 gene are responsible for DYT6 primary torsion dystonia. Nature genetics. 2009;41(3):286–288. doi: 10.1038/ng.304. [DOI] [PubMed] [Google Scholar]
- 8.Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, Lazar V, Dessen P, Mantovani R, Aguilar L, Girard JP. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood. 2007;109(2):584–594. doi: 10.1182/blood-2006-03-012013. [DOI] [PubMed] [Google Scholar]
- 9.De Souza Santos E, De Bessa SA, Netto MM, Nagai MA. Silencing of LRRC49 and THAP10 genes by bidirectional promoter hypermethylation is a frequent event in breast cancer. International journal of oncology. 2008;33(1):25–31. [PubMed] [Google Scholar]
- 10.Bessière D, Lacroix C, Campagne S, Ecochard V, Guillet V, Mourey L, Lopez F, Czaplicki J, Demange P, Milon A, Girard JP, Gervais V. Structure-function analysis of the THAP zinc finger of THAP1, a large C2CH DNA-binding module linked to Rb/E2F pathways. J Biol Chem. 2008;283(7):4352–4363. doi: 10.1074/jbc.M707537200. [DOI] [PubMed] [Google Scholar]
- 11.Campagne S, Saurel O, Gervais V, Milon A. Structural determinants of specific DNA-recognition by the THAP zinc finger. Nucleic acids research. 2010;38(10):3466–3476. doi: 10.1093/nar/gkq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gau B, Chu I, Huang M, Yang K, Chiou S, Fan Y, Chen M, Lin J, Chuang C, Huang S, Lee W. Transcripts of enriched germ cells responding to heat shock as potential markers for porcine semen. Theriogenology. 2008;69:758–766. doi: 10.1016/j.theriogenology.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 13.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bianchetti CM, Blouin GC, Bitto E, Olson JS, Phillips GN., Jr The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1. Proteins. 2009 doi: 10.1002/prot.22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montfort WR, Weichsel A, Andersen JF. Nitrophorins and related antihemostatic lipocalins from Rhodnius prolixus and other blood-sucking arthropods. Biochimica et biophysica acta. 2000;1482(1-2):110–118. doi: 10.1016/s0167-4838(00)00165-5. [DOI] [PubMed] [Google Scholar]
- 16.Jeon WB, Aceti DJ, Bingman CA, Vojtik FC, Olson AC, Ellefson JM, McCombs JE, Sreenath HK, Blommel PG, Seder KD, Burns BT, Geetha HV, Harms AC, Sabat G, Sussman MR, Fox BG, Phillips GN., Jr High-throughput purification and quality assurance of Arabidopsis thaliana proteins for eukaryotic structural genomics. Journal of structural and functional genomics. 2005;6(2-3):143–147. doi: 10.1007/s10969-005-1908-7. [DOI] [PubMed] [Google Scholar]
- 17.Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, Aceti DJ, Frederick RO, Phillips GN, Jr, Fox BG. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein expression and purification. 2005;40(2):256–267. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 18.Thao S, Zhao Q, Kimball T, Steffen E, Blommel PG, Riters M, Newman CS, Fox BG, Wrobel RL. Results from high-throughput DNA cloning of Arabidopsis thaliana target genes using site-specific recombination. Journal of structural and functional genomics. 2004;5(4):267–276. doi: 10.1007/s10969-004-7148-4. [DOI] [PubMed] [Google Scholar]
- 19.Zolnai Z, Lee PT, Li J, Chapman MR, Newman CS, Phillips GN, Jr, Rayment I, Ulrich EL, Volkman BF, Markley JL. Project management system for structural and functional proteomics: Sesame. Journal of structural and functional genomics. 2003;4(1):11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski ZM. Wladek Processing of x-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Part A. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta crystallographica. 2003;59(Pt 11):1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 22.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods in molecular biology (Clifton, NJ. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 23.Cowtan PEaK. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallographica Section D - Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta crystallographica. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 25.Lovell Simon C, D IW, Arendall W Bryan, III, de Bakker Paul IW, Word J Michael, Prisant Michael G, Richardson Jane S, Richardson David C. Structure validation by C-alpha geometry: phi, psi, and C-beta deviation. Proteins: Structure, Function, and Genetics. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific LLC; 2002. [Google Scholar]
- 27.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic acids research. 38(Suppl):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shepard W, Haouz A, Grana M, Buschiazzo A, Betton JM, Cole ST, Alzari PM. The crystal structure of Rv0813c from Mycobacterium tuberculosis reveals a new family of fatty acid-binding protein-like proteins in bacteria. Journal of bacteriology. 2007;189(5):1899–1904. doi: 10.1128/JB.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brune B, von Knethen A, Sandau KB. Nitric oxide (NO): an effector of apoptosis. Cell death and differentiation. 1999;6(10):969–975. doi: 10.1038/sj.cdd.4400582. [DOI] [PubMed] [Google Scholar]