Abstract
DNA methylation serves as the principal form of post-replicative epigenetic modification. It is intricately involved in gene regulation and silencing in eukaryotic cells, making significant contributions to cell phenotype. Much of it is mitotically inherited; some is passed on from one filial generation to the next. Establishment and maintenance of DNA methylation patterns in mammals is governed by three catalytically active DNA methyltransferases – DNMT3a, DNMT3b and DNMT1. While the first two are responsible mainly for de novo methylation, DNMT1 maintains the methylation patterns by preferentially catalyzing S-adenosyl methionine-dependant transfer of a methyl group to cytosine at hemimethylated CpG sites generated as a result of semi-conservative DNA replication. DNMT1 contains numerous regulatory domains that fine-tune associated catalytic activities, deregulation of which is observed in several diseases including cancer. In this minireview, we analyze the regulatory mechanisms of various sub-domains of DNMT1 protein and briefly discuss its pathophysiological and pharmacological implications. A better understanding of DNMT1 function and structure will likely reveal new applications in the treatment of associated diseases.
Keywords: DNMT1, catalytic regulation, epigenetics, disease implications
Introduction
Somatic cells within an organism contain identical genetic material but remarkably exhibit a variety of phenotypes, giving rise to different cell types, tissues, and organs. The basis of these differences lies in the epigenetic code, the bulk of which is encoded as a methyl mark on a specific DNA sequence: the CpG dinucleotide. It is estimated that about half of all CpG dinucleotides in the genome are methylated; the variation in methylation pattern determines cell type [1]. CpG methylation occurs on the 5’ carbon of cytosine in both strands of complementary DNA, altering chromatin structure and regulating specific gene expression patterns. Establishment and maintenance of methylation patterns are mediated by a family of enzymes called DNA methyltransferases (DNMTs). The human genome contains three active enzymes: DNMT1, DNMT3a, and DNMT3b.
Primary establishment of methylation patterns on the genome following embryonic fertilization requires DNMT3a and DNMT3b enzymes which have high affinity for non-methylated DNA [2, 3]. Knock out of these enzymes proved to be fatal at the embryonic stage, suggesting that any overlapping functions of other variants of DNMT are not sufficient in establishing de novo methy-lation patterns [4],[5]. DNMT1, the most abundant form of DNMT in mammalian cells, is highly expressed during the S phase and faithfully maintains the DNA methylation marks in somatic cells after each replication cycle [6]. DNMT1 is mostly localized to nuclei of somatic cells during G1 and G2 phases and is actively targeted to replication foci during the S-phase [7, 8]. DNMT1's preference for hemimethylated CpG sites compared to unmethylated CpG sites is thought to form the basis of high-fidelity maintenance of the epigenetic code during replication [9].
However this long-established model of DNA methylation patterns being maintained by DNMT1 has recently been put into question. A new model is emerging that suggests maintenance of DNA methylation relies not only on the recognition of hemimethylated DNA by DNMT1 but also on the localization of the DNMT3a and DNMT3b enzymes to specific chromatin regions that contain methylated DNA [9, 10]. According to this model, DNMT3a and DNMT3b complete the methylation process and correct errors that are left by the DNMT1 enzyme thereby assuring high-fidelity replication of DNA methylation patterns. Significant cooperation between the DNMTs is required to maintain DNA methylation patterns during the post-replication phase [11]. Moreover, interaction of DNMT1 with the multidomain UHRF1 (or Np95) protein has been recently proposed as the mechanism of hemimethylated site recognition [12, 13]. The SRA domain of UHRF1 exhibits binding affinity for hemimethylated CpG sites and loads DNMT1 to these sites [14, 15]. By specifically recognizing hemi-methylated CpG dinucleotides through its SRA domain and by recruiting DNMT1, UHRF1 acts as a fidelity factor. Null mutants of UHRF1 in murine embryonic stem cells and embryos displayed global hypomethylation, showing its close association with DNMT1 [16]. In addition, UHRF1 binds to histone H3 tails on trimethylated lysine 9 via the Tudor domain [17]. The interactions of UHRF1 with transcriptionrepressive histone and DNA modifications implicate its role as a mediator of gene silencing and epigenetic control. Though mammalian (murine and human) DNMT1 has been cloned and characterized, the structure and exact functions of its regulatory domains have yet to be elucidated [18, 19]. The present paper will review the regulation and catalytic mechanisms of DNMT1 as well as the consequences of its deregulation.
Regulatory domains of DNMT1
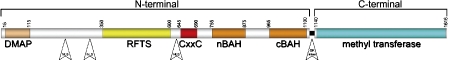
Human DNMT1 contains 1616 amino acid residues; the first three quarters contain the regulatory domains; the DNA methyltransferaseassociated protein 1 (DMAP1)-binding domain, the Replication Foci Targeting Sequence (RFTS) domain, the CxxC zinc binding domain and two Bromo-Adjacent Homology (BAH) domains (Figure 1). The remaining C terminal region (residues 1140–1616) corresponds to the catalytic methyltransferase domain. Thirteen alternating repeats of Gly-Lys residues links the regulatory with the catalytic domain. The DMAP binding domain [residues 12-105] of DNMT1 binds DMAP which serves as a co-repressor of transcription by interacting with histone deacetylase 2 (HDAC2) [20]. The function of the RFTS domain is unclear, but has been implicated in the proper targeting of DNMT1 to the replication foci during the S phase of the cell cycle [21] and in homodimerization of DNMT1 which may aid in the discrimination of hemimethylated DNA [22]. We recently solved the high resolution crystal structure of the replication foci targeting sequence (RFTS) [residues 350-600] domain revealing a novel fold with an extended and conserved electronegative surface (PDB code: 3EPZPDB code: 3EPZ, F.S. et al., manuscript in preparation). The CxxC domain [residues 621-698] was shown to contribute to catalytic activity by interacting with unmethylated CpG DNA substrates [23, 24]. Proteolytic cleavage between the N and C termini resulted in a higher de novo methylation rate suggesting that N-terminal sequences may function as an inhibitor of endogenous de novo methylation activity of the C-terminal methyltransferase domain [23]. It was proposed that DNMT1 may have evolved from a fusion of a replication foci localization domain with the prokaryote methyltransferase [25]. In the absence of the regulatory domains, the isolated catalytic domain is inactive [26], suggesting that the regulatory domains may either relieve auto-inhibition or that they contribute to the catalytic machinery. In contrast, the N -terminal regulatory region of murine DNMT1 fused with prokaryotic DNA methyltransferase yielded an enzyme with a 2.5 fold preference for hemimethylated DNA [27]. Since preference is not observed in the simpler prokaryotic methyltransferase, this suggests that the nucleotide specificity of DNMT1 resides within the C-terminal domain whereas preference for hemimethylated sites is attributed by the N-terminal domain.
Figure 1.
Domain architecture of DNA Methyltransferase 1. N-terminal domain contains: DMAP1 binding site (15– 115), PCNA binding site (160–240), RFTS (350–600), CxxC (645–690), BAH domains-1, 2 (BAH1 755-875) & BAH2 (965–1100), NLS (191–211, 259–378 and 630–757). Gly-Lys dipeptide linker separates the N-terminal and a catalytic C-terminal domain. C-terminal domain (1140–1616) contains 10 conserved amino acid motifs, shared with many prokaryotic 5-methyl-cytosine methyltransferases. The catalytic center and coenzyme binding site reside within this domain.
DNMT1 contains many motifs, including the PCNA-binding motif, residues 162-171, which allows DNMT1 to be recruited to replication foci during replication and repair [28]. While this interaction has been shown to increase the enzyme efficiency by two-fold, recruitment to the replication foci is not necessary to maintain post-replicative global methylation levels [29, 30]. In addition, the N-terminal region of DNMT1 contains several nuclear localization signals sites (NLS) [residues 191-211, 259-378, 630-757] that could be important for nucleocytoplasmic shuttling [31].
Evidence of intramolecular interactions between the N-terminal CXXC region and the catalytic domain was shown, leading to allosteric activation of the enzyme after binding to methylated DNA [26]. An interaction between the N-terminal region encompassing the RFTS and the catalytic domains was also reported [32]. These observations indicate that physical interactions between the N- and C-termini may be important for the regulation of enzymatic activity.
Catalytic domain of DNMT1
The catalytic domain of eukaryotic DNMT1 contains 10 highly conserved motifs with prokaryotic methyl transferases, some of which have been characterized in detail using structural biology [33]. While prokaryotic orthologs lack regulatory N-terminal domains, all DNMTs follow a similar catalytic pathway and share at least some specificity for CpG sites [34].
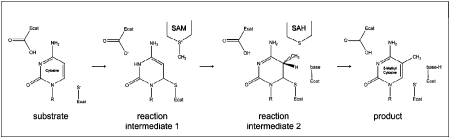
Within its concave binding pocket for duplex DNA (Figure 2) the methyltransferase catalyzes the addition of a methyl group onto the 5’ position of cytosine through a complex, multi-step process. The catalytic site contains a conserved motif I FxFxG where the S-Adenosyl-L-Methionine (AdoMet) cofactor and methyl donor binds [35]. A general catalytic mechanism for prokaryotic DNA methytransferases has been described according to which the enzyme extracts the base from its double helical form (“base-flipping”) and inserts it into its catalytic pocket [36], which is in close proximity to AdoMet. DNMT1 catalyzes methyl transfer according to the Michael addition reaction pathway as shown in Figure 3 [35]. A conserved cysteine residue initiates the reaction through a nucleophilic attack on C6, forming a covalently bonded intermediate between the cytosine and the enzyme. Early steps include the formation of [Enzyme-Substrate] complexes and [Enzymesubstrate-co-factor] complexes that accumulate in rapid equilibrium with each other (this step precisely involves base-flipping/base restacking). This is followed by deprotonation of the endocyclic N3, initiating the formation of the covalent bond between the methyl group of the donor and C5 of cytosine, and releases S-adenosyl-L-homocysteine (AdoHcy). The proton on C5 is then abstracted by a basic residue in the catalytic site and the methyl transfer reaction is completed through a beta-elimination step. The methylated cytosine is then released from the catalytic site and returned into its double helical conformation [35]. Proton transfer is the rate-limiting step of catalysis [37]. The overall catalytic rate depends on the formation, accumulation and concentration of the reaction intermediates as well as on the factors controlling the proton transfer during the methyltransfer process.
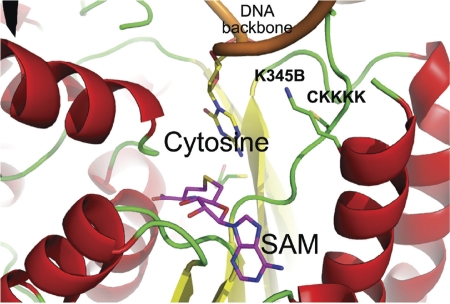
Figure 2.
Model of the catalytic pocket of DNMT1. Shown is a ribbon representation of a model of the methyltransferase domain of human DNMT1, generated by the FFAS server (http://ffas.ljcrf.edu) using the homologous prophage Cp-933R methyl trans-ferase structure (PDB 3G7U). The concave catalytic site of the enzyme extracts the cytosine base out of its double helical form and brings it into close proximity with SAM. After the methyl transfer, both 5-methyl cytosine and SAH are released from the catalytic site. The enzyme subsequently binds to another SAM co-factor to regenerate the catalytic site.
Figure 3.
Reaction mechanism. Methyl transfer by DNMT1 through Michael addition reaction using S-Adenosyl me-thionine (SAM) as methyl donor. Ecat and base represent catalytic or basic residues in the enzyme's active site. S-adenosyl homocysteine (SAH) is the cofactor product.
Isoforms of DNMT1
Several isoforms of DNMT1 have been purified and characterized from human cells. DNMT1-b is a splice isoform of the primary DNMT1 gene transcript detectable in all human tissue cell lines [38]. DNMT1-b contains a 48 nucleotide exon sequence from intron 4 of the DNMT1 gene between the exons 4 and 5 [39]. It has been demonstrated to be a fully functional methyltransferase with a similar binding affinity for AdoMet to DNMT1 [38]. While the DNMT1-b protein is expressed at only 2-5% of the cellular level of DNMT1 in healthy cell lines, transcript abundance has been shown to be up to six folds higher in certain oncogenic tissues [39]. The DNMT1-b transcript is detectable in higher primates but its role in carcinogenesis and development, however, remains unclear [38].
DNMT1 lacking the first 118 residues of the N-terminal domain (therefore lacking the DMAP domain) was found to be uniquely expressed in oocytes (DNMT1o) [40]. DNMT1o is actively retained in the cytoplasm, which prevents binding to its DNA substrate in the nucleus and thereby contributes to the erasure of gamete-specific epigenetic information (epigenetic reprogramming) during early mammalian development [31, 41, 42]. Embryos derived from DNMT1o-deficient oocytes, showed a loss of allelespecific expression and methylation at certain imprinted loci. Transient nuclear localization of DNMT1o in 8-cell embryos suggests that this variant of DNMT1 provides maintenance methyltransferase activity at imprinted loci during the fourth embryonic S phase [41, 43, 44], however this effect was later challenged by other studies (Refs to be inserted). These findings indicate that DNMT1o is cytoplasmic during early development; its presence in the embryos of mouse [42], pig [45] and cow [46] further suggests that DNMT1o is conserved in mammals. Another truncated isoform of DNMT1 was found specifically expressed in differentiated myotubes and in testis suggesting an active role of this isoform in DNA methylation during muscle differentiation and gametogenesis [47, 48]. A different somatic-type cytoplasmic isoform of DNMT1 was identified in post-mitotic neurons in mouse brain [48] and was recently shown to relocalize to the nucleus after traumatic injury [49]. However the reasons for this atypical cytoplasmic localization or the re-translocation of DNMT1 to the nucleus remain unclear. The transcriptional activation of specific genes by passive demethylation of their promotors in the nucleus appears likely to be the mechanism behind cytoplasmic localization of DNMT1o.
Deregulation of DNMT1
DNA methylation and its maintenance play significant roles in embryonic and germ cell development through parental imprinting and X-chromosome inactivation, and in somatic cell development and differentiation through genome stability and regulation of gene expression. The DNA methylation epigenetic code in mammalian organisms is almost completely erased at two instances; in germ cells and in pre-implantation embryos, and re-established to silence imprinted genes. Genomic imprinting refers to the parental-dependent expression of a small subset of monoallelic genes via differential methylation and silencing of one parental allele [50]. Improper imprinting in humans resulting from deficiency, dysregulation or mutations of DNMT1 and DNMT3a/b have been associated with several developmental syndromes which are characterized by severe growth and behavioral defects [51], [52], [53], [54]. For example, Beckwith-Wiedemann syndrome (BWS) is a natal overgrowth disorder associated with an increased risk of childhood cancer [55]. Prader–Willi syndrome (PWS) is a rare genetic disorder in which seven genes (or some subset thereof) on chromosome 15 (q 11-13) are deleted or unexpressed (chromosome 15q partial deletion) on the paternal chromosome. Conversely, the sister syndrome Angelman syndrome presents maternally deleted and paternally imprinted genes in the same genetic region.
DNMT1 was found to be either downregulated or upregulated during cellular differentiation. In many instances, for example in muscle cells [47], F9 mouse tumor cells [56], PC12 neuronal cells [57] and in skin cells [58] DNMT1 expression and/or activity levels are decreased in cells when differentiation is induced experimentally. DNMT1 expression is upregulated in certain malignant blood cells [59], [60]. The role of DNMT1 in cell differentiation appears to be unclear and very complex and is dependent on the cell-type, the nature of stimuli and on the cell cycle phases [61]. It is believed that in selfrenewing tissues, the proliferative capacity of cells is maintained through active repression (by hypermethylation) of genes involved in differentiation thereby delaying premature terminal differentiation [58].
Altered levels of DNMT1 expression have frequently been associated with several types of tumors. Increased DNA methylation following overexpression of DNMT1 led to oncogenic transformation of human cells [62]. Furthermore, recent studies revealed that elevated levels of DNMT1 have been detected in tumor and cancer cells and may be associated to the progression of disease symptoms in pancreatic carcinoma [63]. It was proposed that carcinogenesis may be due to silencing of tumor suppressor genes by hypermethylation [64]. In contrast, many tumor types show evidence of ge-nomic hypomethylation. Mice mutants expressing DNMT1 at only 10% of the physiological levels displayed chromosomal instability and developed highly aggressive T-cell lymphomas [65]. More drastically, a DNMT1-knockout murine embryonic stem cell showed a three-fold reduction in CG methylation, increased global gene expression, elevated mitotic recombination, chromosome deletions and expression of previously silent genes and integrated retroviral DNA; these mutant embryonic stem cells, however, did not show obvious growth or morphological abnormalities [66–68]. In contrast, DNMT1-knockout murine embryos show genomic instability and undergo p53 mediated cell cycle arrest in G1 phase and apoptosis soon after gastrulation [66, 68]. This difference in viability may be attributed to the lack of p53 activity in embryonic stem cells [69]. In general cancer cells are characterized by global hypomethylation (including oncogenes) and hypermethylation of tumour suppressor genes. The underlying mechanisms of this dysregulation remains largely unknown while some evidence points out to a close coupling between the DNA methylation and histone acetylation machineries.
DNMT1 was shown to contribute to the cellular response to DNA damage. A rapid and transient recruitment of DNMT1 to DNA double strand breaks and its interaction with multiple components of the DNA damage response machinery including PCNA and the ATR effector kinase CHK1 was reported [47]. DNMT1's exact role in this process remains unclear.
DNA demethylation
The concept of DNA demethylation being catalyzed by an enzyme was suggested several years ago, but remained poorly supported due to lack of candidate genes [70]. However, the recent discovery of several lysine and arginine demethylases has generated renewed interest in the active DNA demethylase hypothesis. Interestingly, a mammalian methyl DNA binding protein (MBD2) and de novo DNA methyltransferase DNMT3A and DNMT3B were shown experimentally to possess detectable DNA demethylase activity [12, 71].
Pharmacological inhibitors of DNMT1
DNMT1, a key maintenance methyltransferase in mammals, is highly expressed in cancer cells and is associated with cellular transformation. Being regulated by multiple oncogenic pathways and affecting cancer cell phenotype, DNMT1 constitutes a potential anti-cancer target. Methyltransferase inhibitors that competitively bind to the catalytic site of DNMT such as 5-aza-2'-deoxycytidine have been used in clinical trials to treat leukemia group B patients [72], [73]. Other potent cytidine analogs including 1-(beta-D-ribofuranosyl)-2(1H)-pyrimidinone (Zebularine) was shown to inhibit DNMT by forming a stable covalent complex with the enzyme when incorporated into DNA thereby hindering the methylation and decreasing the dissociation, resulting in decreased enzymatic turnover [74]. Subsequently other non-nucleoside DNMT inhibitors in form of maleimide derivatives, were designed and synthesized [75]. DNMT1 inhibition was also tested experimentally in cancer cells by alternate pathways using HDAC inhibitors that indirectly promote ubiquitin-dependent proteasomal degradation of DNMT1 [76]. A similar mechanism of degradation of unbound (non-chromatin bound) DNMT1 via the proteasomal pathway was described in mouse embryonic cells treated by 5-aza-2'-deoxycytidine [77]. Although transient inhibition of DNMT1 in cancer cells may lead to demethylation of silenced tumor suppressor genes [78], a complete inactivation of DNMT1 in human colorectal carcinoma cell lines caused genomic instability, defective chromatin as well as mitotic arrest [79]. While it is still unknown why increased or decreased levels of DNMT1 causes oncogenesis, further investigation may lead to potent treatments for cancer patients in the future.
Almost 30 years after the discovery of the first small molecule inhibitor, the 5-aza-2'-deoxycytidine, little has been achieved in terms of specific and safe inhibition of DNMTs. Few plausible reasons for this lag could be: (i) lack of knowledge about the protein structure (ii) insufficient insights into the molecular mechanisms of regulation (iii) plethora of interacting and regulatory protein factors and (iv) high risk of potential DNA mutagenesis and carconigenicity. In addition to small molecule inhibitors, siRNAs would be an interesting alternative approach to knock down DNMT1 effects.
Concluding remarks
This review represents an appreciation of the vital role played by DNMT1 in the maintenance of the epigenetic code and the regulation of associated biological functions. However, further studies probing the function and structure of DNMT1 may shed light on the underlying mechanisms and facilitate development of efficient therapeutic measures in the treatment of associated diseases.
Abbreviations
- DNMT1
DNA methyltransferase 1
- RFTS
Replication Foci Targeting Sequence domain (residues 351-600 of DNMT1)
- DMAP1
DNA methyltransferase-associated protein 1
- PCNA
proliferating cell nuclear antigen
- BAH
Bromo-Adjacent Homology (BAH) domain
- RING
Really Interesting New Gene
- SAM
S-Adenosyl methionine
- CpG
Cytosine-phosphate-Guanine
- UHRF1
Ubiquitin-like, containing PHD and RING finger domains, 1
- SRA
SET and RING-associated domain
- CxxC
C is cysteine and x is any amino acid
- NLS
nuclear localization signal.
References
- [1].Razin A, Webb C, Szyf M, Yisraeli J, Rosenthal A, Naveh-Many T, Sciaky-Gallili N, Cedar H. Variations in DNA methylation during mouse cell differentiation in vivo and in vitro. Proc Natl Acad Sci U S A. 1984;81:2275–2279. doi: 10.1073/pnas.81.8.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- [3].Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- [4].Dodge JE, Okano M, Dick F, Tsujimoto N, Chen T, Wang S, Ueda Y, Dyson N, Li E. Inactiva-tion of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- [5].Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- [6].Detich N, Ramchandani S, Szyf M. A conserved 3'-untranslated element mediates growth regulation of DNA methyltransferase 1 and inhibits its transforming activity. J Biol Chem. 2001;276:24881–24890. doi: 10.1074/jbc.M103056200. [DOI] [PubMed] [Google Scholar]
- [7].Easwaran HP, Leonhardt H, Cardoso MC. Cell cycle markers for live cell analyses. Cell Cycle. 2005;4:453–455. doi: 10.4161/cc.4.3.1525. [DOI] [PubMed] [Google Scholar]
- [8].Easwaran HP, Schermelleh L, Leonhardt H, Cardoso MC. Replication-independent chroma-tin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1181–1186. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- [10].Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kim GD, Ni J, Kelesoglu N, Roberts RJ, Prad-han S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. Embo J. 2002;21:4183–4195. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ooi SK, Bestor TH. Cytosine methylation: remaining faithful. Curr Biol. 2008;18:R174–176. doi: 10.1016/j.cub.2007.12.045. [DOI] [PubMed] [Google Scholar]
- [14].Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a baseflipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- [15].Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- [16].Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- [17].Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 38:1796–1804. doi: 10.1093/nar/gkp1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- [19].Yen RW, Vertino PM, Nelkin BD, Yu JJ, el-Deiry W, Cumaraswamy A, Lennon GG, Trask BJ, Celano P, Baylin SB. Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res. 1992;20:2287–2291. doi: 10.1093/nar/20.9.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- [21].Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- [22].Fellinger K, Rothbauer U, Felle M, Langst G, Leonhardt H. Dimerization of DNA methyltransferase 1 is mediated by its regulatory domain. J Cell Biochem. 2009;106:521–528. doi: 10.1002/jcb.22071. [DOI] [PubMed] [Google Scholar]
- [23].Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. Embo J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pradhan M, Esteve PO, Chin HG, Samaranayke M, Kim GD, Pradhan S. CXXC domain of human DNMT1 is essential for enzymatic activity. Biochemistry. 2008;47:10000–10009. doi: 10.1021/bi8011725. [DOI] [PubMed] [Google Scholar]
- [25].Margot JB, Aguirre-Arteta AM, Di Giacco BV, Pradhan S, Roberts RJ, Cardoso MC, Leon-hardt H. Structure and function of the mouse DNA methyltransferase gene: Dnmt1 shows a tripartite structure. J Mol Biol. 2000;297:293–300. doi: 10.1006/jmbi.2000.3588. [DOI] [PubMed] [Google Scholar]
- [26].Fatemi M, Hermann A, Pradhan S, Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J Mol Biol. 2001;309:1189–1199. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- [27].Pradhan S, Roberts RJ. Hybrid mouseprokaryotic DNA (cytosine-5) methyltransferases retain the specificity of the parental C-terminal domain. Embo J. 2000;19:2103–2114. doi: 10.1093/emboj/19.9.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- [29].Schermelleh L, Haemmer A, Spada F, Rosing N, Meilinger D, Rothbauer U, Cardoso MC, Leonhardt H. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–4312. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Langst G, Leonhardt H. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176:565–571. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cardoso MC, Leonhardt H. DNA methyltransferase is actively retained in the cytoplasm during early development. J Cell Biol. 1999;147:25–32. doi: 10.1083/jcb.147.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Margot JB, Ehrenhofer-Murray AE, Leonhardt H. Interactions within the mammalian DNA methyltransferase family. BMC Mol Biol. 2003;4:7. doi: 10.1186/1471-2199-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- [34].Kumar S, Cheng X, Klimasauskas S, Mi S, Pos-fai J, Roberts RJ, Wilson GG. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994;22:1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci U S A. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Svedruzic ZM, Reich NO. DNA cytosine C5 methyltransferase Dnmt1: catalysis-dependent release of allosteric inhibition. Biochemistry. 2005;44:9472–9485. doi: 10.1021/bi050295z. [DOI] [PubMed] [Google Scholar]
- [38].Hsu DW, Lin MJ, Lee TL, Wen SC, Chen X, Shen CK. Two major forms of DNA (cytosine-5) methyltransferase in human somatic tissues. Proc Natl Acad Sci U S A. 1999;96:9751–9756. doi: 10.1073/pnas.96.17.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bonfils C, Beaulieu N, Chan E, Cotton-Montpetit J, MacLeod AR. Characterization of the human DNA methyltransferase splice variant Dnmt1b. J Biol Chem. 2000;275:10754–10760. doi: 10.1074/jbc.275.15.10754. [DOI] [PubMed] [Google Scholar]
- [40].Mertineit C, Yoder JA, Taketo T, Laird DW, Tra-sler JM, Bestor TH. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125:889–897. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- [41].Grohmann M, Spada F, Schermelleh L, Alenina N, Bader M, Cardoso MC, Leonhardt H. Restricted mobility of Dnmt1 in preimplantation embryos: implications for epigenetic reprogramming. BMC Dev Biol. 5:18. doi: 10.1186/1471-213X-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ratnam S, Mertineit C, Ding F, Howell CY, Clarke HJ, Bestor TH, Chaillet JR, Trasler JM. Dynamics of Dnmt1 methyltransferase expression and intracellular localization during oogenesis and preimplantation development. Dev Biol. 2002;245:304–314. doi: 10.1006/dbio.2002.0628. [DOI] [PubMed] [Google Scholar]
- [43].Doherty AS, Bartolomei MS, Schultz RM. Regulation of stage-specific nuclear translocation of Dnmt1o during preimplantation mouse development. Dev Biol. 2002;242:255–266. doi: 10.1006/dbio.2001.0534. [DOI] [PubMed] [Google Scholar]
- [44].Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- [45].Jeong YS, Oh KB, Park JS, Kim JS, Kang YK. Cytoplasmic localization of oocyte-specific variant of porcine DNA methyltransferase-1 during early development. Dev Dyn. 2009;238:1666–1673. doi: 10.1002/dvdy.21975. [DOI] [PubMed] [Google Scholar]
- [46].Lodde V, Modina SC, Franciosi F, Zuccari E, Tessaro I, Luciano AM. Localization of DNA methyltransf erase-1 du ring ooc yte differentiation, in vitro maturation and early embryonic development in cow. Eur J Histochem. 2009;53:199–207. doi: 10.4081/ejh.2009.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Aguirre-Arteta AM, Grunewald I, Cardoso MC, Leonhardt H. Expression of an alternative Dnmt1 isoform during muscle differentiation. Cell Growth Differ. 2000;11:551–559. [PubMed] [Google Scholar]
- [48].Inano K, Suetake I, Ueda T, Miyake Y, Nakamura M, Okada M, Tajima S. Maintenance-type DNA methyltransferase is highly expressed in post-mitotic neurons and localized in the cytoplasmic compartment. J Biochem. 2000;128:315–321. doi: 10.1093/oxfordjournals.jbchem.a022755. [DOI] [PubMed] [Google Scholar]
- [49].Lundberg J, Karimi M, von Gertten C, Holmin S, Ekstrom TJ, Sandberg-Nordqvist AC. Traumatic brain injury induces relocalization of DNA-methyltransferase 1. Neurosci Lett. 2009;457:8–11. doi: 10.1016/j.neulet.2009.03.105. [DOI] [PubMed] [Google Scholar]
- [50].Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- [51].Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol. 2007;211:261–268. doi: 10.1002/path.2116. [DOI] [PubMed] [Google Scholar]
- [52].Turek-Plewa J, Jagodzinski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647. [PubMed] [Google Scholar]
- [53].Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- [54].El-Osta A, Wolffe AP. DNA methylation and histone deacetylation in the control of gene expression: basic biochemistry to human development and disease. Gene Expr. 2000;9:63–75. doi: 10.3727/000000001783992731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bestor TH. Imprinting errors and developmental asymmetry. Philos Trans R Soc Lond B Biol Sci. 2003;358:1411–1415. doi: 10.1098/rstb.2003.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Teubner B, Schulz WA. Regulation of DNA methyltransferase during differentiation of F9 mouse embryonal carcinoma cells. J Cell Physiol. 1995;165:284–290. doi: 10.1002/jcp.1041650209. [DOI] [PubMed] [Google Scholar]
- [57].Deng J, Szyf M. Downregulation of DNA (cytosine-5-)methyltransferase is a late event in NGF-induced PC12 cell differentiation. Brain Res Mol Brain Res. 1999;71:23–31. doi: 10.1016/s0169-328x(99)00147-3. [DOI] [PubMed] [Google Scholar]
- [58].Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].DePaoli-Roach A, Roach PJ, Zucker KE, Smith SS. Selective phosphorylation of human DNA methyltransferase by protein kinase C. FEBS Lett. 1986;197:149–153. doi: 10.1016/0014-5793(86)80316-7. [DOI] [PubMed] [Google Scholar]
- [60].Nakajima K, Kusafuka T, Takeda T, Fujitani Y, Nakae K, Hirano T. Identification of a novel interleukin-6 response element containing an Ets-binding site and a CRE-like site in the junB promoter. Mol Cell Biol. 1993;13:3027–3041. doi: 10.1128/mcb.13.5.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Szyf M, Detich N. Regulation of the DNA methylation machinery and its role in cellular transformation. Prog Nucleic Acid Res Mol Biol. 2001;69:47–79. doi: 10.1016/s0079-6603(01)69044-5. [DOI] [PubMed] [Google Scholar]
- [62].Wu J, Issa JP, Herman J, Bassett DE, Jr., Nelkin BD, Baylin SB. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc Natl Acad Sci U S A. 1993;90:8891–8895. doi: 10.1073/pnas.90.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Peng DF, Kanai Y, Sawada M, Ushijima S, Hira-oka N, Kosuge T, Hirohashi S. Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci. 2005;96:403–408. doi: 10.1111/j.1349-7006.2005.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- [65].Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- [66].Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- [67].Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- [68].Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- [69].Sabapathy K, Klemm M, Jaenisch R, Wagner EF. Regulation of ES cell differentiation by functional and conformational modulation of p53. Embo J. 1997;16:6217–6229. doi: 10.1093/emboj/16.20.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Reik W, Dean W, Walter J. Epigenetic repro-gramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- [71].Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol Carcinog. 2002;33:163–171. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- [72].Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, Stone RM, Nelson D, Powell BL, DeCastro CM, Ellerton J, Larson RA, Schiffer CA, Holland JF. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- [73].Szyf M. The DNA methylation machinery as a target for anticancer therapy. Pharmacol Ther. 1996;70:1–37. doi: 10.1016/0163-7258(96)00002-2. [DOI] [PubMed] [Google Scholar]
- [74].Champion C, Guianvarc'h D, Senamaud-Beaufort C, Jurkowska RZ, Jeltsch A, Ponger L, Arimondo PB, Guieysse-Peugeot AL. Mechanistic insights on the inhibition of c5 DNA methyltransferases by zebularine. PLoS One. 5:e12388. doi: 10.1371/journal.pone.0012388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Suzuki T, Tanaka R, Hamada S, Nakagawa H, Miyata N. Design, synthesis, inhibitory activity, and binding mode study of novel DNA methyltransferase 1 inhibitors. Bioorg Med Chem Lett. 20:1124–1127. doi: 10.1016/j.bmcl.2009.12.016. [DOI] [PubMed] [Google Scholar]
- [76].Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Cancer Res. 2008;6:873–883. doi: 10.1158/1541-7786.MCR-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B. Targeting of 5-aza-2'-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 38:4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- [79].Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human. [DOI] [PubMed]