Comparative DNA sequence analysis of orthologous centromeres from three rice species revealed the presence of seven conserved genes. Surprisingly, all seven genes are under strong purifying selection despite being harbored in a region that is free of detectable chromosomal exchanges (crossing-over), a phenomenon suggestive of strong functional constraints on these genes.
Abstract
Recombination is strongly suppressed in centromeric regions. In chromosomal regions with suppressed recombination, deleterious mutations can easily accumulate and cause degeneration of genes and genomes. Surprisingly, the centromere of chromosome8 (Cen8) of rice (Oryza sativa) contains several transcribed genes. However, it remains unclear as to what selective forces drive the evolution and existence of transcribed genes in Cen8. Sequencing of orthologous Cen8 regions from two additional Oryza species, Oryza glaberrima and Oryza brachyantha, which diverged from O. sativa 1 and 10 million years ago, respectively, revealed a set of seven transcribed Cen8 genes conserved across all three species. Chromatin immunoprecipitation analysis with the centromere-specific histone CENH3 confirmed that the sequenced orthologous regions are part of the functional centromere. All seven Cen8 genes have undergone purifying selection, representing a striking phenomenon of active gene survival within a recombination-free zone over a long evolutionary time. The coding sequences of the Cen8 genes showed sequence divergence and mutation rates that were significantly reduced from those of genes located on the chromosome arms. This suggests that Oryza has a mechanism to maintain the fidelity and functionality of Cen8 genes, even when embedded in a sea of repetitive sequences and transposable elements.
INTRODUCTION
Recombination via chromosomal crossing-over plays a significant role in gene and genome evolution (Gaut et al., 2007; Li et al., 2007). In chromosomal regions with suppressed or reduced recombination from crossing-over, deleterious mutations can easily accumulate due to inefficient natural selection caused by Hill-Robertson Inference (Haddrill et al., 2007; Comeron et al., 2008; Betancourt et al., 2009; Charlesworth et al., 2009). Such interference is thought to be a major factor leading to genetic degeneration of genes and genomes. Suppression or reduction of recombination resulting from structural rearrangements along chromosomes can also prevent gene flow and hinder the introgression of alleles, thereby contributing to speciation and/or persistence as demonstrated by many empirical studies (Noor et al., 2001; Rieseberg, 2001; Ortíz-Barrientos et al., 2002; Navarro and Barton, 2003; Butlin, 2005; Stump et al., 2005).
Recent studies have demonstrated the presence of active genes in recombination-suppressed chromosomal domains of mammals (Mudge and Jackson, 2005), Drosophila melanogaster (Hoskins et al., 2002), and plants (Haupt et al., 2001; Yan et al., 2005, 2008), which raises questions as to how the fidelity and function of such genes are maintained in an environment presumed to be void of recombination. Maintenance of structure and function of genes in the human Y chromosome has been shown to occur by intrachromatid gene conversion mediated via homologous recombination between opposing arms of large palindromic sequences (Lange et al., 2009). In Drosophila, regional and ancient recombination events in heterochromatin have been postulated to maintain heterochromatic genes (Schulze et al., 2006). Polymorphism and divergence data from chromosome 4 of Drosophila, which was initially believed to be recombination suppressed, revealed the presence of extremely low levels of recombination; this reduced level was sufficient to maintain normal gene density and gene functionality (Arguello et al., 2010).
Centromeres are defined by the presence of a centromere-specific histone variant CENH3 (CID in Drosophila, CENP-A in humans) (Allshire and Karpen, 2008). Unlike other chromosomal domains, such as pericentromeric heterochromatin, in which self- or low-frequency recombination events have been observed, centromeres are thought to be completely devoid of crossover recombination (Beadle, 1932; Lambie and Roeder, 1986; Jackson et al., 1996; Anderson et al., 2003; Shi et al., 2010). Interestingly, sequence and transcriptome analysis of the centromere of rice (Oryza sativa) chromosome8 (Cen8) revealed the presence of 16 transcribed genes (Nagaki et al., 2004; Yan et al., 2005); the first set of genes found in the functional domain of a eukaryotic centromere. An intriguing question emerged from this discovery: What selective forces are driving the evolution and existence of transcribed genes in genomic regions devoid of crossover recombination?
To address this question, we sequenced Cen8 in two additional Oryza species, Oryza glaberrima and Oryza brachyantha, which have diverged from rice for ~0.5 to 1 and ~10 to 15 million years (MY), respectively (Ammiraju et al., 2008). The centromeres of O. brachyantha chromosomes were previously demonstrated to contain completely different sets of repetitive DNA sequences compared with cultivated rice (Lee et al., 2005; Gao et al., 2009). Here, we demonstrate the persistence of orthologous transcribed Cen8 genes in these three Oryza species. The conserved Cen8 genes showed strong functional constraints in both O. glaberrima and O. brachyantha, representing a striking phenomenon of active gene survival in a recombination-free zone over a 10 to 15 MY evolutionary time span.
RESULTS
Identification of Active Genes in Cen8 of O. glaberrima and O. brachyantha
We sequenced and assembled Cen8 sequences of O. glaberrima (1.3 Mb) and O. brachyantha (1.1 Mb) corresponding to ~1 Mb of the O. sativa ssp japonica var Nipponbare (referred to hereafter simply as O. sativa) Cen8 (Nip-Cen8), including the ~750-kb CENH3 binding domain (Yan et al., 2008). The crossover-suppressed domain in Cen8 is 2312 kb and encompasses this 750-kb CENH3 binding domain (Yan et al., 2005). The pseudomolecules of both species included centromere-specific satellite repeats of unknown size.
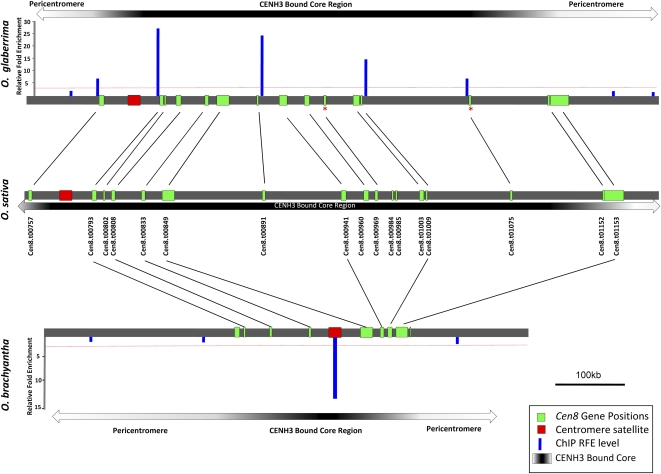
The 1-Mb sequence of O. sativa contains 17 transcribed genes based on expression evidence and gene ontology (Yan et al., 2005) (see Supplemental Table 1 online; Cen8.t00802 was not described in Yan et al., 2005). To determine the extent of gene conservation in orthologous Cen8 regions of O. glaberrima and O. brachyantha, we manually annotated these regions and identified 15 of the 17 (88%) orthologous genes in O. glaberrima (13 with intact open reading frames and two pseudogenes [Cen8.t00969 and Cen8.t01075]) and 7 of the 17 (41.2%) orthologous genes in O. brachyantha (Figure 1, Table 1). Two orthologous genes in O. glaberrima (Cen8.t00969 and Cen8.t01075) appeared to be pseudogenes as each had single base pair deletions that resulted in frame-shift mutations in their protein coding sequences (Figure 1). The order and orientation of all orthologous Cen8 genes were conserved across the three Oryza species (Figure 1).
Figure 1.
ChIP Analysis and Orthologous Cen8 Gene Alignment in O. sativa, O. glaberrima, and O. brachyantha.
O. sativa Refseq and gene annotation from the Rice Genome Annotation database were used as references, and O. glaberrima and O. brachyantha orthologous genes are matched by straight lines. Two O. glaberrima orthologous pseudogenes are marked with a red asterisk and contain single frame-shift point mutations. The size of the centromere satellite (CentO in O. sativa and O. glaberrima and CentO-F in O. brachyantha) was not drawn to scale as the full sequence could not be determined. The CENH3 binding functional domains are shown as gray shaded bars. The black to white gradient illustrates the decrease in CENH3 binding from the satellite core of the centromere to more sparsely bound regions approaching the pericentromere. The red dashed lines indicate the ChIP significance level at a P value of 0.01.
Table 1.
Sequence Divergence of Orthologous Cen8 Active Genes
| OS_CDS | TIGR Gene ID | Comparison | Identity | Length | Ka | Ks | Ka/Ks | LRT P Value |
| Cen8.t00757 | Os08g21660 | OG-OS | 99.9 | 978 | 0.0000 | 0.0034 | 0.0010 | 0.1261 |
| Cen8.t00793 | Os08g21700 | OG-OS | 99.7 | 576 | 0 | 0.0106 | 0.0000 | 0.0365 |
| OB-OS | 89.9 | 555 | 0.0827 | 0.1908 | 0.4332 | 0.0044** | ||
| OG-OB | 89.5 | 555 | 0.0830 | 0.2062 | 0.4026 | 0.0016** | ||
| Cen8.t00802 | Os08g21720 | OG-OS | 98.6 | 285 | 0.0207 | 0.0000 | ∞ | 0.0928 |
| Cen8.t00808 | Os08g21760 | OG-OS | 99.8 | 483 | 0.0026 | 0.0000 | ∞ | 0.4997 |
| OB-OS | 92.8 | 483 | 0.0203 | 0.2949 | 0.0689 | 0.0000*** | ||
| OG-OB | 93.5 | 483 | 0.0116 | 0.2818 | 0.0412 | 0.0000*** | ||
| Cen8.t00833 | Os08g21840 | OG-OS | 99.5 | 885 | 0.0035 | 0.0069 | 0.5010 | 0.4949 |
| OB-OS | 90.5 | 873 | 0.0464 | 0.2621 | 0.1770 | 0.0000*** | ||
| OG-OB | 92.1 | 873 | 0.0286 | 0.2420 | 0.1182 | 0.0000*** | ||
| Cen8.t00849 | NA | OG-OS | 98.4 | 567 | 0.0142 | 0.0223 | 0.6384 | 0.5475 |
| OB-OS | 93.3 | 570 | 0.0222 | 0.2223 | 0.0997 | 0.0000*** | ||
| OG-OB | 93.5 | 567 | 0.0198 | 0.2255 | 0.0878 | 0.0000*** | ||
| Cen8.t00891 | Os08g22060 | OG-OS | 100 | 102 | 0.0000 | 0.0000 | ∞ | 0.9980 |
| Cen8.t00941 | Os08g22149 | OG-OS | 99.7 | 705 | 0.0020 | 0.0050 | 0.4015 | 0.5288 |
| OB-OS | 91.0 | 696 | 0.0538 | 0.2215 | 0.2431 | 0.0000*** | ||
| OG-OB | 90.7 | 696 | 0.0560 | 0.2170 | 0.2442 | 0.0000*** | ||
| Cen8.t00960 | Os08g22200 | OG-OS | 99.7 | 882 | 0.0049 | 0.0000 | ∞ | 0.1515 |
| Cen8.t01003 | Os08g22354 | OG-OS | 99.8 | 1980 | 0.0014 | 0.0037 | 0.3891 | 0.3603 |
| OB-OS | 93.5 | 1968 | 0.0233 | 0.2235 | 0.1043 | 0.0000*** | ||
| OG-OB | 93.6 | 1968 | 0.0234 | 0.2170 | 0.1077 | 0.0000*** | ||
| Cen8.t01009 | NA | OG-OS | 99.0 | 294 | 0.0096 | 0.0121 | 0.7918 | 0.7152 |
| Cen8.t01152 | Os08g22852 | OG-OS | 99.8 | 564 | 0.0000 | 0.0046 | 0.0000 | 0.1639 |
| Cen8.t01153 | Os08g22864 | OG-OS | 99.7 | 3174 | 0.0020 | 0.0048 | 0.4106 | 0.2465 |
| OB-OS | 95.0 | 3108 | 0.0211 | 0.1487 | 0.1416 | 0.0000*** | ||
| OG-OB | 95.0 | 3108 | 0.0206 | 0.1511 | 0.1361 | 0.0000*** |
NA, not available; OB, O. brachyantha; OG, O. glaberrima; OS, O. sativa; TIGR, The Institute for Genomic Research. ∞, infinite when there is no synonymous substitution and therefore the denominator (Ks) as 0; **, significant level as P < 0.01; ***, significant level as P < 0.001.
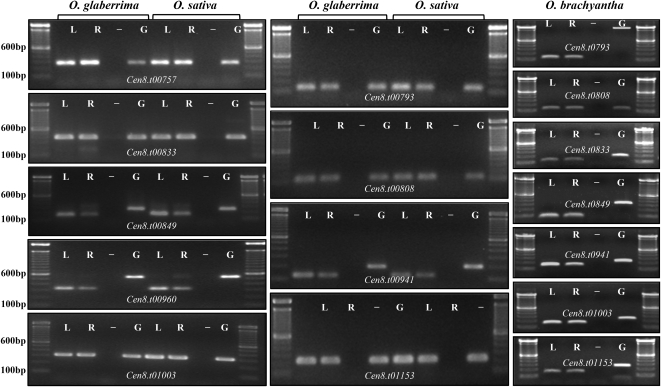
Nine Cen8 genes conserved between O. sativa and O. glaberrima and seven conserved O. brachyantha Cen8 genes were tested for transcriptional activity using cDNA synthesized from both leaf and root tissues. Genomic DNA and an RT negative control (produced using cDNA synthesis reaction without addition of reverse transcriptase) were also included in the analysis as controls. Each of the nine primer sets in O. glaberrima/O. sativa and the seven sets in O. brachyantha faithfully amplified products of predicted size from both tissues, except Cen8.t00793 in O. brachyantha, which produced a much larger genomic product (Figure 2; for primers, see Supplemental Table 2 online). Furthermore, transcripts were also detected for all seven O. brachyantha Cen8 genes based on Illumina-based transcriptome profiling data obtained from leaf and root tissues (see Supplemental Table 3 online). Overall, by combining RT-PCR results, transcriptome analysis, and an EST search analysis, our data showed that 12 of the 15 orthologous O. glaberrima Cen8 genes and all seven O. brachyantha Cen8 genes were transcribed (see Supplemental Table 4 online).
Figure 2.
RT-PCR Results of Cen8 Genes from O. sativa, O. glaberrima, and O. brachyantha.
L, leaf; R, root; (−), negative control; G, genomic DNA.
Confirmation of the Centromeric Position of the Orthologous Cen8 Sequences from O. glaberrima and O. brachyantha
We conducted chromatin immunoprecipitation (ChIP) experiments to confirm the centromeric location of the Cen8 sequences produced from O. glaberrima and O. brachyantha. PCR primers were designed from regions near the seven conserved Cen8 genes in both species (see Supplemental Table 5 online). ChIP was performed using an antibody against rice CENH3 (Nagaki et al., 2004). Here, we surveyed the Cen8 landscape in O. glaberrima and O. brachyantha for evidence of CENH3 binding. Eight O. glaberrima and four O. brachyantha primers were designed to interrogate this region spanning ~1 Mb of Cen8 sequence. In O. glaberrima, 13 of the 15 conserved Cen8 genes are localized within a region where eight primer sets showed significant CENH3 enrichment using ChIP-PCR (Figure 1). The remaining two genes, Cen8.t01152 and Cen8.t01153, are 100 kb away from one of the ChIP-PCR markers (Figure 1). In O. brachyantha, significant CENH3 enrichment of the centromere specific CentO-F satellite array was confirmed (Figure 1). DNA sequences outside of the seven Cen8 genes were not enriched by ChIP-PCR. However, active genes are associated with H3 (rather than CENH3) nucleosomes (Yan et al., 2008); thus, the Cen8 genes cannot be used for testing CENH3 enrichment. Since the O. brachyantha Cen8 genes are only 500 kb away from the CentO-F satellite, we predict that these genes are either within the CENH3 domain or immediately outside of the CENH3 domain, depending on the size of the CentO-F satellite array in this centromere. Based on the fact that the crossover suppressed domains are severalfold larger than the CENH3 domains in all rice centromeres (Yan et al., 2008), we presumed that all O. brachyantha Cen8 genes are located within the crossover-suppressed domain in this centromere.
Purifying Selection of Conserved Orthologous Cen8 Genes in Three Oryza Species
Annotation and expression data revealed the presence of seven conserved and transcribed orthologous genes between two independently domesticated rice species and their distant wild relative O. brachyantha. To determine if any of these genes were under functional constraints, the ratios of nonsynonymous substitution rate (Ka) and synonymous substitution rate (Ks) for all seven Cen8 orthologs were calculated (Table 1). The results showed that all Ka/Ks (ω) ratios between either O. sativa and O. brachyantha (OS versus OB) or O. glaberrima and O. brachyantha (OG versus OB) were <0.5 (average 0.1811 for OS versus OB, and 0.1625 for OG versus OB) (Table 1), suggesting that all seven genes are under strong functional constraint with purifying selection. Likelihood ratio tests (LRTs) for all seven Ka/Ks values deviated significantly from neutrality (ω = 1).
Ka/Ks ratios for the seven genes between O. sativa and O. glaberrima as well as the six remaining genes shared by these two species indicated that 10 genes were under purifying selection. The remaining three genes (Cen8.t00802, Cen8.t00808, and Cen8.t00960) had Ka/Ks ratios > 1 (Table 1). However, LRT tests for all 13 comparisons between O. sativa and O. glaberrima did not deviate significantly from 1, suggesting that the short divergence time between these two species limits the statistical power of this analysis.
Low Molecular Evolution Rates of Cen8 Coding Sequences versus Noncentromeric Genes
To investigate the pattern of molecular evolution of Cen8 genes versus genes located in recombining regions, we analyzed orthologous gene sets derived from sequenced short arms of chromosome 3 (Chr3S) of O. sativa, O. glaberrima, and O. brachyantha. Comparisons between O. sativa and O. glaberrima were made using 1515 orthologous gene pairs (see Supplemental Figure 1 online). These Chr3S genes were scattered along the entire chromosome arm excluding the recombination free centromeric region. Both the mean Ks and Ka values for all 1515 pairs were significantly higher than that of the 13 pairs of Cen8 genes (P = 7.7E-11 for Ks and P = 0.00083 for Ka, one-sided t test; Table 2), and tests of mutation rate in the coding region (/bp/MY) yielded similar results (P = 1.35E-5; Table 2). For O. brachyantha, we selected a total of 268 and 230 Chr3S genes that are orthologous to O. sativa and O. glaberrima genes, respectively (see Supplemental Figures 2 and 3 online). The average Ks of orthologous genes pairs between O. sativa and O. brachyantha and between O. glaberrima and O. brachyantha (Supplemental Figures 2 and 3 online) was significantly higher (P = 0.00182, 3.62E-5, respectively, one-sided t test) than that for the seven pairs of Cen8 genes (Table 2). However, the Ka of Cen8 genes and Chr3S genes were not statistically different (Table 2). The mutation rate across the entire Chr3S of O. brachyantha revealed nearly significant higher mutation rates than those found for all conserved active Cen8 genes across the three Oryza species (P = 0.169 and 0.059) (Table 2).
Table 2.
Statistical Tests of Synonymous (Ks) and Nonsynonymous (Ka) Substitution Rates and Mutation Rates between Cen8 Genes and Chr3S Noncentromeric Genes
| Test | Gene | OS versus OG | OS versus OB | OG versus OB |
| Ks | Chr3S genes | 0.0287 (1515) | 0.3000 (268) | 0.3245 (238) |
| Cen8 genes | 0.0056 (13) | 0.2234 (7) | 0.2219 (7) | |
| Welch t test P value (one tail) | 7.716E-11*** | 0.00182** | 3.62E-5*** | |
| Ka | Chr3S genes | 0.0113 (1515) | 0.0436 (268) | 0.0505 (238) |
| Cen8 genes | 0.0043 (13) | 0.0385 (7) | 0.0347 (7) | |
| Welch t test P value (one tail) | 0.000828*** | 0.3000 | 0.0796 | |
| Mutation rate (/bp/MY) | Chr3S gene | 0.0066 (1515) | 0.00466 (268) | 0.00486 (238) |
| Cen8 genes | 0.00250 (13) | 0.00393 (7) | 0.00381 (7) | |
| Welch t test P value (one tail) | 1.3553E-5*** | 0.169 | 0.0594 |
OB, O. brachyantha; OG, O. glaberrima; OS, O. sativa. **, significant level as P < 0.01; ***, significant level as P < 0.001. The numbers in parentheses indicate the number of genes used for each test.
Functional Constraints of Cen8 Genes Suggested by O. glaberrima and O. brachyantha Cen8 Polymorphism Patterns
To obtain supporting evidence for purifying selection of the conserved Cen8 genes, we performed a population genetic analysis using two data sets. First, we obtained O. glaberrima Cen8 region polymorphisms using whole-genome single nucleotide polymorphism (SNP) data from eight accessions of O. glaberrima. In a 651,584-bp region of O. glaberrima Cen8, we found a total of 388 SNPs giving a polymorphism rate of 0.595/kb, which is equal to half of the average polymorphism rate across the whole genome (1.118/kb). Of the 388 SNPs in Cen8, 358 SNPs were located in intergenic regions, and only 30 were present in Cen8 genes. Of the 30 SNPs, only five were located in exons, including one nonsynonymous substitution and four synonymous substitutions (see Supplemental Table 6 online). Statistical tests indicated that both an excess of SNPs in Cen8 genes and synonymous substitutions in coding regions significantly deviated from neutral expectations (see Supplemental Tables 7 and 8 online).
Second, we amplified and sequenced a complete orthologous Cen8 gene (OB_t00833 [3028 bp, excluding gaps]) and part of the OB_t01153 (1022 bp) gene, from 15 O. brachyantha accessions. Sequence analysis from pooling two sequence regions revealed the presence of 35 polymorphic sites (31 in OB_t00833 and four in OB_t01153), 30 of which were in noncoding sequences (27 in OB_t00833 and three in OB_t01153) and five of which were in coding sequences (four in OB_t00833 and one in OB_t01153): four as synonymous substitutions and one as a replacement mutation (see Supplemental Figure 4 online). Having both an excess of polymorphic sites in noncoding regions and an excess of synonymous substitutions in coding sequences significantly deviated from neutral expectations (χ2 test, P = 0.048 and 0.029, respectively; see Supplemental Tables 7 and 8 online), thus supporting our observation that the conserved OB_t00833 and OB_t01153 coding sequences are under strong functional constraints.
We conducted additional population genetic analyses using the polymorphisms identified from the eight O. glaberrima accessions and 15 O. brachyantha accessions to infer the evolutionary pattern of Cen8 genes in O. glaberrima and O. brachyantha. Each of the three recombination and gene conversion tests we performed failed to reject the null hypothesis of no recombination (see Supplemental Tables 9 to 11 online), indicating that no recombination or conversion within these centromeric genes could be detected. A negative Tajima’s D value in O. glaberrima significantly deviated from neutrality, indicating O. glaberrima Cen8 is likely undergoing purifying selection (Table 3). Tajima’s D values inferred from either silent polymorphic sites or total polymorphic sites in O. brachyantha were positive but did not deviate from neutral expectations (P = 0.84) (Table 3).
Table 3.
Average Sequence Diversity of O. brachyantha Cen8 Genes and O. glaberrima Cen8 Region
| Statistic | OB_t00833 and OB_t01153 | OG_Cen8 |
| Sample size | 16 | 8 |
| Length (bp) | 4,427 | 651,484 |
| Segregating sites (S) | 35 | 388 |
| θw per kb | 3.17 | 0.23 |
| θπ per kb | 2.66 | 0.15 |
| Tajima’s D | 0.82494, P = 0.839 | −1.84077, P = 0.00001** |
OB, O. brachyantha; OG, O. glaberrima. **, significant level as P < 0.01.
Pfam Analysis of Seven Conserved Cen8 Genes
The presence of seven highly conserved genes within a functional centromere, spanning the Oryza phylogeny, may indicate that these genes are biologically essential. Pfam analysis (http://pfam.sanger.ac.uk/) of these genes revealed that five of the seven (except Cen8.t00793 and Cen8.t00849) belong to gene families assigned to physiological and/or cellular functions (see Supplemental Table 12 online). In particular, Cen8.t01003 (Os08g22354) was found to contain nine conserved domains: four RNA/DNA binding sites, four RRM dimerization sites, and poly-adenylate binding protein, providing scaffolds to which proteins can bind and mediate processes such as export, translation, transcript turnover, and regulation of development at the transcriptional level (Bandziulis et al., 1989; Birney et al., 1993; Mangus et al., 2003).
DISCUSSION
Centromeres in higher eukaryotes are embedded within highly heterochromatic pericentromeric chromatin. In most plant and animal species described, centromeres contain satellite repeats and transposable elements (Henikoff et al., 2001; Jiang et al., 2003) and are nonrecombinogenic. These attributes have hindered centromere research, especially at the sequence level. In fact, virtually all whole-genome shotgun assemblies have completely ignored centromeres as they are difficult to recognize and assemble. The genus Oryza provides a unique model for centromere research for three reasons. First, the centromeres of several rice chromosomes have been fully or partially sequenced (Zhang et al., 2004; Yan et al., 2008; Wu et al., 2009). Second, transcribed genes located within CENH3-associated chromatin domains in rice provide a platform to study the evolution of genes located in recombination suppressed chromosomal domains. Third, a set of BAC-based physical maps representing 13 Oryza species and all 10 Oryza genome types (six diploids and four polyploids) has been developed, thereby providing unprecedented access to virtually any region of the collective Oryza genome for interrogation, including centromeres (Ammiraju et al., 2006; Kim et al., 2008).
We sequenced and compared the Cen8 regions of O. glaberrima and O. brachyantha that span the CENH3 binding domains and contain centromere-specific satellite repeats. We demonstrated that O. glaberrima shared 12 active Cen8 genes with O. sativa-Cen8, whereas only six orthologous Cen8 genes were shared between the two subspecies O. sativa ssp japonica and O. sativa ssp indica (Wu et al., 2009). A significant finding was that seven active genes were conserved in O. brachyantha, which diverged roughly 10 to 15 MY ago from O. sativa and O. glaberrima. This observation raises important questions regarding gene loss, gene gain, and gene mutation in the three Oryza centromeres. It is unknown if the ancestral state of Cen8 more closely resembled O. brachyantha Cen8 or O. sativa Cen8. If the structure of O. sativa Cen8 reflects the ancestral state, then both O. glaberrima and O. brachyantha have undergone gene loss (2 to 10 genes, respectively) and pseudogenization in Cen8. Alternatively, if O. brachyantha Cen8 is a closer to reflection of the ancestral state, then O. glaberrima Cen8 has acquired additional expressed genes, and O. sativa Cen8 is still acquiring genes. The latter scenario is highly unlikely because centromeres are thought to have evolved from noncentromeric regions via neocentromere formation and accumulation of repetitive DNA (Nagaki et al., 2004). In addition, an analysis of sequences flanking the O. glaberrima Cen8 genes did not reveal the presence of helitron or MULE sequences, which could be used to explain such gene acquisition.
The most surprising discovery was that all seven genes were not only transcribed but appear to be under strong purifying selection based on two lines of evidence (Ka/Ks ratios and a population genetics analysis of centromeric genes). Such syntenic conservation and purifying selection implies that the fidelity of centromeric genes can be preserved without crossover recombination, even when embedded in a sea of highly dynamic and constantly evolving transposable elements and tandem satellite repeats.
It is generally understood that low recombination rates will reduce sequence diversity due to rapid elimination or fixation of mutations (Nachman, 2002). We tested the synonymous substitution rate, which is assumed to be neutral, between three Oryza species. The significantly smaller Ks rates for centromeric genes compared with noncentromeric genes are consistent with the observation of low intraspecific polymorphisms detected in centromeric regions from various organisms, including yeast, maize (Zea mays), Drosophila, and rice (Aguade et al., 1989; Begun and Aquadro, 1992; Gerton et al., 2000; Gore et al., 2009; Schacherer et al., 2009). It is known that recombination can facilitate chromosomal rearrangements, gene copy number changes, and even the generation of single-nucleotide mutations (Lercher and Hurst, 2002; Hellmann et al., 2003; Jelesko et al., 2004; Schuermann et al., 2005). Therefore, given the lack of crossover recombination in centromeres, a reduced mutation rate could also contribute to low sequence divergence rates of centromeric genes.
Speciation and species differentiation may be enhanced by the suppression of recombination. Both modeling and recent empirical studies suggest that recombination can reduce speciation events (Noor et al., 2001; Ortíz-Barrientos et al., 2002), while suppression of recombination can allow species to diverge by preventing gene flow between individuals. For example, in Drosophila, the hybrid incompatibility genes Lhr, Zhr, and OdsH are associated with speciation, and all map to recombinationally suppressed pericentric and heterochromatic regions that showed reduced or undetectable levels of recombination (Sawamura et al., 1993; Brideau et al., 2006; Bayes and Malik, 2009). In plants, several sets of genes involved in speciation and reproductive isolation have been localized to highly heterochromatic regions where recombination is suppressed. For example, the “A” locus for gametophytic apomixis, a phenomenon that results in asexual reproduction, was identified in a region completely devoid of recombination (Ozias-Akins and van Dijk, 2007). Moreover, self-incompatibility genes (S-locus) were found to be recombinationally suppressed due to their subcentromeric location in Petunia (Coleman and Kao, 1992; Entani et al., 1999) and Antirrhinum (Ma et al., 2003; Yang et al., 2007), the presence of repetitive DNA in Nicotiana (Matton et al., 1995), and conserved linkage in Prunus (Ikeda et al., 2005). It is believed that S-locus genes experienced strong balancing selection that resulted in high local population polymorphisms but low population differentiation (Ruggiero et al., 2008). The low levels of sequence divergence found between Cen8 genes in our three species comparisons, along with the population genetic data analysis from the O. brachyantha accessions, suggests that the conserved genes found within these functional centromeres have undergone molecular evolutionary events similar to those observed in S-locus genes. Functional assays of the conserved Cen8 genes may provide new evidence that centromeres serve as islands of speciation (Noor and Bennett, 2009).
METHODS
Sequencing of BAC Tiles from Cen8 Centromeric Regions of Oryza glaberrima and Oryza brachyantha
We used the rice (Oryza sativa) Cen8 sequence (~1 Mb DNA), which includes the 750-kb CENH3 binding domain (Yan et al., 2008), as a reference sequence to identify Cen8 genomic regions of O. glaberrima and O. brachyantha. Minimum tiling paths of overlapping BAC clones spanning Cen8 in the two species were developed, including 12 (O. glaberrima) and 9 (O. brachyantha) BACs. Each BAC was shotgun Sanger sequenced and finished using previously described methods (Project, 2005). The O. glaberrima and O. brachyantha BAC sequences were assembled into 1.3- and 1.0-Mb pseudomolecule sequences, respectively. Each individual BAC ID and GenBank accession number is listed in Supplemental Table 13 online.
ChIP Analysis to Determine Functional Centromeres
Nuclei were isolated from young leaf tissue of O. glaberrima and O. brachyantha, and ChIP was performed using antibodies against the centromere histone H3 (CENH3) of rice (Nagaki et al., 2004; Lee et al., 2005). As a negative control, a mock ChIP experiment was also performed, in tandem, by replacing the anti-CENH3 antibody with normal rabbit serum. Centromere sequences bound to CENH3 were identified with quantitative PCR using primers that spanned and flanked the proposed set of centromeric genes. Primers were designed to amplify products between 119 and 325 bp. Quantitative real-time PCR was conducted to determine the enrichment of centromere sequences within the ChIP samples compared with the mock. The quantitative real-time PCR was performed in triplicate using a DyNAmo HS SYBR Green qPCR kit (Finnzymes) using the following cycling parameters: 94°C for 15 min, 45 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. The relative fold enrichment was calculated for each primer pair using a noncentromere primer set (NonCenControl 1) as a reference. For each primer pair, Δcycle threshold (Ct) for mock was calculated as ΔCt(mock) = Ct(centromere primer) – Ct(NonCenControl 1), and ΔCt for ChIP was calculated as ΔCt(ChIP) = Ct(centromere primer) – Ct(noncentromere primer). Lastly, the relative fold enrichment (2−ΔΔCt) was calculated, where ΔΔCt = ΔCt(ChIP) – ΔCt(mock). An enrichment cutoff line was placed based on P values assigned using a one-tailed Student’s t test at a significance level of α = 0.01.
Identification and Expression Analysis of Orthologous Cen8 Genes in O. sativa, O. glaberrima, and O. brachyantha
Coding sequences (CDSs) of all annotated genes in the O. sativa Cen8 region were used as queries to search for orthologous genes in the O. glaberrima and O. brachyantha Cen8 pseudomolecules using MEGABLAST (parameters: O. glaberrima e-value <1e-3, >95% sequence identity over the entire CDS; O. brachyantha e-value <1e-1, >90% sequence identity). Active genes were classified as those having RT-PCR and/or ESTs expression evidence. Gene ontology annotations were described in the Rice Genome Annotation database (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/). Expression profiles of all Cen8 genes in O. sativa were obtained from RT-PCR experiments (Yan et al., 2005) and/or the most recent whole-genome UniGene rice EST/mRNA data set (http://www.ncbi.nlm.nih.gov/UniGene/UGOrg.cgi?TAXID=4530). Evidence for expression of O. glaberrima Cen8 genes was obtained using RT-PCR from leaf and/or root tissue. Evidence for expression of O. brachyantha Cen8 genes was obtained by RT-PCR and analysis of an Illumina sequence-based genome-wide transcriptome data set derived from root and shoot cDNA, which were kindly provided by M. Chen at the Chinese Academy of Sciences, Beijing, China. Gene expression levels were expressed as reads (number of Illumina reads that mapped onto CDS without mismatch) and RPKM (number of reads per kilobase per million reads).
A total of nine non–transposable element related O. glaberrima Cen8 genes and seven O. brachyantha Cen8 genes found within Cen8 in O. sativa were selected for transcriptional analysis via RT-PCR. Primer pairs were designed from conserved regions within the gene exons such that the same primers could be used to perform RT-PCR in both O. sativa and O. glaberrima. However, since the priming sites were not conserved between O. glaberrima and O. brachyantha, the primers used for O. brachyantha were designed de novo. Total mRNA was isolated from leaf and root tissues sampled from O. sativa, O. glaberrima, and O. brachyantha plants grown in the Biotron facilities at the University of Wisconsin-Madison under normal rice growth conditions. First-stand cDNA synthesis was completed using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). One mRNA sample from each species was taken through an amended cDNA synthesis procedure in which the addition of the reverse transcriptase enzyme was omitted. These samples represent the negative controls used in subsequent PCR to confirm the absence of contaminating genomic DNA. PCR from the synthesized cDNAs was performed under the following cycling conditions: 95°C for 5 min followed by 33 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s, ending with a final 4-min extension at 72°C.
Sequence Divergence Analysis and Estimation of Mutation Rate
Genomic, CDS, and protein sequences of orthologous Cen8 genes from the three Oryza species were compared to reveal the structure, origin, and evolution of the orthologous genes. Ka/Ks ratios (ω) using the maximum likelihood algorithm were computed using PAML (Yang, 2007). The significance of ω that deviated from neutrality (ω = 1.0) was tested using LRT. Protein sequences of homologous gene pairs were aligned using MUSCLE (Edgar, 2004), and codon-based DNA sequences were aligned using the aligned protein sequences as guides with the Pal2nal script (Suyama et al., 2006). Codeml with fixed (ω = 1) and free omega (ω = estimated) models was used to test whether any homologous gene pairs were under selective constraint (Yang, 2007). We further calculated the mutation rate in coding regions (/base pair/MY) using the total number of substitutions (synonymous and nonsynonymous) divided by the product of CDS length (bp) and double species divergent time (Gillespie, 2004), 1 MY for O. sativa and O. glaberrima and 10 MY for O. sativa and O. brachyantha, and O. glaberrima and O. brachyantha (Ammiraju et al., 2008, 2010).
Population Polymorphism Analysis of O. glaberrima and O. brachyantha Cen8 Genes
SNP data of O. glaberrima Cen8 genes were generated from eight accessions (see Supplemental Table 14 online) using Illumina Solexa resequencing (R.A. Wing, Y. Yu, and C. Fan, unpublished data). By mapping the resequencing reads to the O. glaberrima Cen8 pseudomolecule, we obtained sequence polymorphisms for O. glaberrima Cen8 genes and intergenic regions. The number of SNPs and the rate of SNPs per kilobase was further calculated and compared. For O. brachyantha, one O. brachyantha Cen8 gene, OB_t00833 (3.9 kb) and one gene fragment OB_t01153 (partial sequence is 1022 bp), were PCR amplified and sequenced from 15 O. brachyantha accessions collected from several African countries (see Supplemental Table 15 online). OB_t00833 is located immediately adjacent to the centromeric satellite domain (CentO in O. sativa and O. glaberrima, and CentO-F in O. brachyantha; Figure 1), and OB_t01153 resides in the left boundary of the satellite domain, where crossover recombination is presumed to be totally suppressed. Sequence alignment of population polymorphism data allowed us to calculate several population genetic parameters, including polymorphism frequency spectra, and Tajima’s D (Tajima, 1989) as implemented in DnaSPv5 (Librado and Rozas, 2009) using nucleotide diversity (θπ) and Watterson’s sequence variation (θw) (Watterson, 1975). Assessment of significant deviation from neutrality was simulated using a coalescence approach. Since OB_t01153 has fewer polymorphic sites, it cannot be used for statistical analysis if we analyzed it individually; therefore, we pooled the two sequence data sets together to perform population genetic and recombination analyses. Using the combined polymorphism data, we further performed three powerful recombination detection methods (Piganeau et al., 2004), which are the most efficient and sensitive methods for detecting recombination and gene conversion events: (1) maxichi, maximum χ2 recombination test using Maynard Smith’s method (Smith, 1992); (2) LDr2, tests the correlation between the measure of linkage disequilibrium, r2 (Hill and Robertson, 1966), and the distance between sites; and (3) geneconv, detects gene conversion events using Sawyer's method (Sawyer, 1989).
Pfam Analysis of Cen8 Genes
Protein sequences of seven conserved Cen8 genes were used to find matching protein family at http://pfam.sanger.ac.uk/. The search was performed using HMM (hidden Markov model) model and E-value of 1.0 as threshold.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AC240787, AC237093, AC223444, AC240789, AC237092, AC223443, AC240788, AC223442, AC240786, AC223445, AC237091, AC223441, AC223438, AC240777, AC223440, AC249775, AC223439, AC240778, AC237085, AC237086, and AC240776.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Boxplot of Ks and Ka Values between O. sativa and O. glaberrima.
Supplemental Figure 2. Boxplot of Ks and Ka Values between O. sativa and O. brachyantha.
Supplemental Figure 3. Boxplot of Ks and Ka Values between O. glaberrima and O. brachyantha.
Supplemental Figure 4. Gene Structure and Polymorphism Distribution in One O. brachyantha Cen8 Gene (OB_t0083) and One Gene Fragment (OB_t01153).
Supplemental Table 1. List of O. sativa Cen8 Genes.
Supplemental Table 2. Primers Used for RT-PCR.
Supplemental Table 3. Expression Profile of Seven Active O. brachyantha Cen8 Genes.
Supplemental Table 4. Expression of O. glaberrima Cen8 Genes.
Supplemental Table 5. ChIP Analysis.
Supplemental Table 6. Summary of O. glaberrima SNP Data.
Supplemental Table 7. Statistical Test of Polymorphism Substitution Pattern in OB_t00833.
Supplemental Table 8. Statistical Test of Polymorphism Distribution in O. brachyantha Cen8 Genes.
Supplemental Table 9. Maximum χ2 Recombination Test.
Supplemental Table 10. Relationship between LD and Distance (LDr2 Test).
Supplemental Table 11. Sawyer’s Gene Conversion Detection (Geneconv Test).
Supplemental Table 12. Pfam Search Results for Seven Conserved Cen8 Genes.
Supplemental Table 13. Summary of BACs and GenBank Accessions Used for Cen8 Sequencing in O. glaberrima and O. brachyantha.
Supplemental Table 14. Sampling of O. glaberrima Accessions.
Supplemental Table 15. O. brachyantha Population Accession Sampling.
Supplementary Material
Acknowledgments
We thank Mingsheng Chen at the Institute of Genetics and Development, Chinese Academy of Sciences, Beijing, for providing O. brachyantha transcripts and the Arabidopsis Genome Initiative staff for shotgun library construction, sequence generation, and finishing. This study was supported by National Science Foundation Grant 0603927 to J.J. and R.A.W.
AUTHOR CONTRIBUTIONS
C.F., J.J., and R.A.W. designed the research. C.F. and J.G.W. performed the research. C.F., J.G.W., J.Z., and C.D.H. analyzed data. C.F., J.G.W., C.D.H., J.J., and R.A.W. wrote the article. J.J. and R.A.W. are joint senior authors who contributed equally.
References
- Aguade M., Miyashita N., Langley C.H. (1989). Reduced variation in the yellow-achaete-scute region in natural populations of Drosophila melanogaster. Genetics 122: 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R.C., Karpen G.H. (2008). Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat. Rev. Genet. 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammiraju J., et al. (2010). Spatio-temporal patterns of genome evolution in allotetraploid species of the genus Oryza. Plant J. 63: 430–442 [DOI] [PubMed] [Google Scholar]
- Ammiraju J.S., et al. (2008). Dynamic evolution of oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell 20: 3191–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammiraju J.S., et al. (2006). The Oryza bacterial artificial chromosome library resource: Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 16: 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L.K., Doyle G.G., Brigham B., Carter J., Hooker K.D., Lai A., Rice M., Stack S.M. (2003). High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165: 849–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello J.R., Zhang Y., Kado T., Fan C., Zhao R., Innan H., Wang W., Long M. (2010). Recombination yet inefficient selection along the Drosophila melanogaster subgroup’s fourth chromosome. Mol. Biol. Evol. 27: 848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandziulis R.J., Swanson M.S., Dreyfuss G. (1989). RNA-binding proteins as developmental regulators. Genes Dev. 3: 431–437 [DOI] [PubMed] [Google Scholar]
- Bayes J.J., Malik H.S. (2009). Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326: 1538–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G.W. (1932). A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA 18: 160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D.J., Aquadro C.F. (1992). Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520 [DOI] [PubMed] [Google Scholar]
- Betancourt A.J., Welch J.J., Charlesworth B. (2009). Reduced effectiveness of selection caused by a lack of recombination. Curr. Biol. 19: 655–660 [DOI] [PubMed] [Google Scholar]
- Birney E., Kumar S., Krainer A.R. (1993). Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21: 5803–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau N.J., Flores H.A., Wang J., Maheshwari S., Wang X., Barbash D.A. (2006). Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295 [DOI] [PubMed] [Google Scholar]
- Butlin R.K. (2005). Recombination and speciation. Mol. Ecol. 14: 2621–2635 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Betancourt A.J., Kaiser V.B., Gordo I. (2009). Genetic recombination and molecular evolution. Cold Spring Harb. Symp. Quant. Biol. 74: 177–186 [DOI] [PubMed] [Google Scholar]
- Coleman C.E., Kao T. (1992). The flanking regions of two Petunia inflata S alleles are heterogeneous and contain repetitive sequences. Plant Mol. Biol. 18: 725–737 [DOI] [PubMed] [Google Scholar]
- Comeron J.M., Williford A., Kliman R.M. (2008). The Hill-Robertson effect: Evolutionary consequences of weak selection and linkage in finite populations. Heredity 100: 19–31 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entani T., Iwano M., Shiba H., Takayama S., Fukui K., Isogai A. (1999). Centromeric localization of an S-RNase gene in Petunia hybrida Vilm. Theor. Appl. Genet. 99: 391–397 [DOI] [PubMed] [Google Scholar]
- Gao D., et al. (2009). A lineage-specific centromere retrotransposon in Oryza brachyantha. Plant J. 60: 820–831 [DOI] [PubMed] [Google Scholar]
- Gaut B.S., Wright S.I., Rizzon C., Dvorak J., Anderson L.K. (2007). Recombination: An underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 8: 77–84 [DOI] [PubMed] [Google Scholar]
- Gerton J.L., DeRisi J., Shroff R., Lichten M., Brown P.O., Petes T.D. (2000). Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J.H. (2004). Population Genetics: A Concise Guide. (Baltimore and London: The Johns Hopkins University Press; ). [Google Scholar]
- Gore M.A., Chia J.M., Elshire R.J., Sun Q., Ersoz E.S., Hurwitz B.L., Peiffer J.A., McMullen M.D., Grills G.S., Ross-Ibarra J., Ware D.H., Buckler E.S. (2009). A first-generation haplotype map of maize. Science 326: 1115–1117 [DOI] [PubMed] [Google Scholar]
- Haddrill P.R., Halligan D.L., Tomaras D., Charlesworth B. (2007). Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol. 8: R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt W., Fischer T.C., Winderl S., Fransz P., Torres-Ruiz R.A. (2001). The centromere1 (CEN1) region of Arabidopsis thaliana: Architecture and functional impact of chromatin. Plant J. 27: 285–296 [DOI] [PubMed] [Google Scholar]
- Hellmann I., Ebersberger I., Ptak S.E., Pääbo S., Przeworski M. (2003). A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72: 1527–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Hill W.G., Robertson A. (1966). The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294 [PubMed] [Google Scholar]
- Hoskins R., et al. (2002). Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 3: RESEARCH0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Ushijima K., Yamane H., Tao R., Hauck N.R., Sebolt A.M., Iezzoni A.F. (2005). Linkage and physical distances between the S-haplotype S-RNase and SFB genes in sweet cherry. Sex. Plant Reprod. 17: 261–313 [Google Scholar]
- Jackson M.S., See C.G., Mulligan L.M., Lauffart B.F. (1996). A 9.75-Mb map across the centromere of human chromosome 10. Genomics 33: 258–270 [DOI] [PubMed] [Google Scholar]
- Jelesko J.G., Carter K., Thompson W., Kinoshita Y., Gruissem W. (2004). Meiotic recombination between paralogous RBCSB genes on sister chromatids of Arabidopsis thaliana. Genetics 166: 947–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Birchler J.A., Parrott W.A., Dawe R.K. (2003). A molecular view of plant centromeres. Trends Plant Sci. 8: 570–575 [DOI] [PubMed] [Google Scholar]
- Kim H., et al. (2008). Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza. Genome Biol. 9: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie E.J., Roeder G.S. (1986). Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics 114: 769–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J., Skaletsky H., van Daalen S.K., Embry S.L., Korver C.M., Brown L.G., Oates R.D., Silber S., Repping S., Page D.C. (2009). Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell 138: 855–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.R., Zhang W., Langdon T., Jin W., Yan H., Cheng Z., Jiang J. (2005). Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 102: 11793–11798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher M.J., Hurst L.D. (2002). Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18: 337–340 [DOI] [PubMed] [Google Scholar]
- Li J., Hsia A.P., Schnable P.S. (2007). Recent advances in plant recombination. Curr. Opin. Plant Biol. 10: 131–135 [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Ma W.-S., Zhou J.-L., Lai Z., Zhang Y.-S., Xue Y.-B. (2003). The self-incompatibilty S locus of Antirrhinum resides in a pericentromeric region. Acta Bot. Sin. 45: 47–52 [Google Scholar]
- Mangus D.A., Evans M.C., Jacobson A. (2003). Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matton D.P., Mau S.L., Okamoto S., Clarke A.E., Newbigin E. (1995). The S-locus of Nicotiana alata: Genomic organization and sequence analysis of two S-RNase alleles. Plant Mol. Biol. 28: 847–858 [DOI] [PubMed] [Google Scholar]
- Mudge J.M., Jackson M.S. (2005). Evolutionary implications of pericentromeric gene expression in humans. Cytogenet. Genome Res. 108: 47–57 [DOI] [PubMed] [Google Scholar]
- Nachman M.W. (2002). Variation in recombination rate across the genome: Evidence and implications. Curr. Opin. Genet. Dev. 12: 657–663 [DOI] [PubMed] [Google Scholar]
- Nagaki K., Cheng Z., Ouyang S., Talbert P.B., Kim M., Jones K.M., Henikoff S., Buell C.R., Jiang J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36: 138–145 [DOI] [PubMed] [Google Scholar]
- Navarro A., Barton N.H. (2003). Chromosomal speciation and molecular divergence—Accelerated evolution in rearranged chromosomes. Science 300: 321–324 [DOI] [PubMed] [Google Scholar]
- Noor M.A., Bennett S.M. (2009). Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity 103: 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M.A., Grams K.L., Bertucci L.A., Reiland J. (2001). Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortíz-Barrientos D., Reiland J., Hey J., Noor M.A. (2002). Recombination and the divergence of hybridizing species. Genetica 116: 167–178 [PubMed] [Google Scholar]
- Ozias-Akins P., van Dijk P.J. (2007). Mendelian genetics of apomixis in plants. Annu. Rev. Genet. 41: 509–537 [DOI] [PubMed] [Google Scholar]
- Piganeau G., Gardner M., Eyre-Walker A. (2004). A broad survey of recombination in animal mitochondria. Mol. Biol. Evol. 21: 2319–2325 [DOI] [PubMed] [Google Scholar]
- Project, I.R.G.S.; International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H. (2001). Chromosomal rearrangements and speciation. Trends Ecol. Evol. (Amst.) 16: 351–358 [DOI] [PubMed] [Google Scholar]
- Ruggiero M.V., Jacquemin B., Castric V., Vekemans X. (2008). Hitch-hiking to a locus under balancing selection: High sequence diversity and low population subdivision at the S-locus genomic region in Arabidopsis halleri. Genet. Res. (Camb.) 90: 37–46 [DOI] [PubMed] [Google Scholar]
- Sawamura K., Yamamoto M.T., Watanabe T.K. (1993). Hybrid lethal systems in the Drosophila melanogaster species complex. II. The Zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics 133: 307–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. (1989). Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6: 526–538 [DOI] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J.A., Ruderfer D.M., Kruglyak L. (2009). Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458: 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann D., Molinier J., Fritsch O., Hohn B. (2005). The dual nature of homologous recombination in plants. Trends Genet. 21: 172–181 [DOI] [PubMed] [Google Scholar]
- Schulze S.R., McAllister B.F., Sinclair D.A., Fitzpatrick K.A., Marchetti M., Pimpinelli S., Honda B.M. (2006). Heterochromatic genes in Drosophila: A comparative analysis of two genes. Genetics 173: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wolf S.E., Burke J.M., Presting G.G., Ross-Ibarra J., Dawe R.K. (2010). Widespread gene conversion in centromere cores. PLoS Biol. 8: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.M. (1992). Analyzing the mosaic structure of genes. J. Mol. Evol. 34: 126–129 [DOI] [PubMed] [Google Scholar]
- Stump A.D., Fitzpatrick M.C., Lobo N.F., Traoré S., Sagnon N., Costantini C., Collins F.H., Besansky N.J. (2005). Centromere-proximal differentiation and speciation in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 102: 15930–15935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server issue): W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G.A. (1975). On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276 [DOI] [PubMed] [Google Scholar]
- Wu J., et al. (2009). Comparative analysis of complete orthologous centromeres from two subspecies of rice reveals rapid variation of centromere organization and structure. Plant J. 60: 805–819 [DOI] [PubMed] [Google Scholar]
- Yan H., Jin W., Nagaki K., Tian S., Ouyang S., Buell C.R., Talbert P.B., Henikoff S., Jiang J. (2005). Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 17: 3227–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Talbert P.B., Lee H.R., Jett J., Henikoff S., Chen F., Jiang J. (2008). Intergenic locations of rice centromeric chromatin. PLoS Biol. 6: e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Zhang D., Li Q., Cheng Z., Xue Y. (2007). Heterochromatic and genetic features are consistent with recombination suppression of the self-incompatibility locus in Antirrhinum. Plant J. 51: 140–151 [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Huang Y., Zhang L., Li Y., Lu T., Lu Y., Feng Q., Zhao Q., Cheng Z., Xue Y., Wing R.A., Han B. (2004). Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 32: 2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.