Abstract
For the first time, the possible binding site of nanoparticles on protein was revealed by crosslinking chemistry coupled with mass spectrometry. The peptides locating very close to the polyacrylic acid (PAA)-coated Fe3O4 nanoparticles (NPs) during interaction with human serum albumin (HSA) were crosslinked to the surface of NPs. Following protease digestion, the attached peptides were cleaved off the particle surface, and identified by matrix-assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS). The peptides were found to be part of the so-called drug binding site 2 of HSA; and the competitive binding to HSA between the corresponding drug, ibuprofen, and the NPs was observed. Our results demonstrated that crosslinking chemistry coupled with MS was a quick and simple method for locating the possible binding sites of NPs on protein. Information on NP-protein binding interface will benefit study of how the interactions are governed by the physicochemical properties of NPs, for guiding the design of functional bio-nano constructs. It can also help to predict the biological consequence of protein adsorption on NPs, for obtaining more knowledge on nanotoxicity.
Keywords: nanoparticle-protein interaction, crosslinking chemistry coupled with mass spectrometry, drug binding site, human serum albumin
Introduction
Finding out the possible binding sites of nanoparticles on proteins can benefit both the study of nanotoxicity and the preparation of functional bio-nanomaterials, helping to promote safer and more effective implementation of nanotechnology. The layer of adsorbed proteins on the surface of nanomaterials entering the bio-systems is believed to serve as the bio-signature of nanomaterials and impact their uptake,1 distribution, and excretion.2–4 The adsorption may also alter protein conformation and function,5,6 causing adverse health effects. By exploring where the NPs would bind on proteins, we can predict the biological consequence of protein adsorption and thus the potential toxicity. On the other hand, nano-bio hybrid materials are prepared by attaching proteins to nanomaterials for energy production,7 sensing,8 separation,9 biomedical imaging,10 drug delivery,11 etc. The locations of nanomaterials on proteins could be strategically controlled to prevent any interference to protein function, to allow efficient energy12 or electron transfer13 between the proteins and the supporting materials, or to permit natural molecule recognition.14,15 Tactical coupling of nanomaterials to proteins calls for more knowledge about the dependence of binding epitope on nanomaterial properties.
Few attempts have explored the binding interface of nanoparticles and proteins at the amino acid level, employing hydrogen/deuterium (H/D) exchange-mass spectrometry (MS)16 and nuclear magnetic resonance (NMR).17 These methods are technically demanding and require delicate treatment of proteins.18,19 Alternatively, chemical crosslinking in combination with MS has been used to reveal the topology of protein complexes.20–23 The reaction is rapid and a large variety of crosslinking reagents have been developed to target a wide range of functional groups. This method could be a facile solution to identification of the binding interface of the nanoparticle (NP) - protein complex. The judgement is based on two considerations: (1) the involvement of NPs could simplify peptide identification by providing a solid support for easy isolation and clean-up of the crosslinked peptides before MS analysis; (2) most of the functional bio-nanomaterials are prepared by covalent attachment to gain long-term stability.24 Hence, this proof-of-principle study was devoted to test the hypothesis of whether the crosslinking chemistry was suitable for exploration of the possible binding sites of the polyacrylic acid (PAA)-coated Fe3O4 nanoparticles (NPs) on human serum albumin (HSA). Three peptides were consistently identified to be close to the surface of NPs during protein adsorption, and they belong to the drug binding site 2 of HSA. Competition for this binding site between ibuprofen and NPs was observed to confirm the identification. These results demonstrated that crosslinking chemistry coupled with MS could be a quick approach to reveal peptides near the NPs when protein adsorption occurs, which will be beneficial to the study of interaction between nanoparticles and proteins for gaining more understanding on nanotoxicity and imposing better guidance to the design of biofunctional nanostructures.
Experimental Section
Synthesis and characterization of PAA-Fe3O4 NPs
The reagents for NPs synthesis were obtained from Sigma-Aldrich Corporation (St. Louis, MO, USA). A NaOH/diethylene glycol (DEG) stock solution was prepared by dissolving 50 mmol of NaOH in 20 mL of DEG. In a typical synthesis, a mixture of PAA (4 mmol, Mw = 1.8 kDa), FeCl3 (2 mmol), and DEG (15 mL) was heated up to 220 °C in a nitrogen atmosphere with stirring. A NaOH/DEG stock solution (4.5 mL) was then injected rapidly into the above hot mixture. The NPs with an average diameter around 8 nm were obtained by continuously heating the mixture for 12 hrs at 220 °C. An excess amount of PAA and other reagents in the reaction mixture were removed by centrifugation-assisted washing several times with a mixture of deionized water and ethanol. Finally, the PAA-Fe3O4 NPs were suspended in 10 mL of water to form the stock solutions.
Characterization of the obtained NPs was carried out and the results were displayed in Figure S1 in the Supporting Information. A Philips Tecnai 12 transmission electron microscope (TEM) was used to investigate the morphology of NPs. Inductively coupled plasma atomic emission spectroscopy (ICP-AES) tests were performed on a Perkin-Elmer Optima 2000 DV optical emission spectrometer to measure the concentration of Fe element in NPs. With the size measured under TEM and the element content obtained from ICP-AES, the molar concentration of the NP stock solutions was obtained. Hydrodynamic size was measured with a ZetaPALS system (Brookhaven, Holtsville, NY) at 25°C, which was equipped with a 660-nm laser and a build-in precision Peltier temperature controller.
Incubation, cross-linking and protein digestion
Trypsin from porcine pancreas (Type IX-S, lyophilized powder, 13,000–20,000 BAEE units/mg) and albumin from human serum (lyophilized powder, ≥97%) were attained from Sigma-Aldrich Corporation (St. Louis, MO, USA). The 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) (≥ 98%) was purchased from Fisher Scientific (Fairlawn, NJ). HSA (7.5 μg) was incubated with the PAA-Fe3O4 NPs (1.37 × 10−10 mol; the molar ratio of protein to NPs was about 1: 1) in the deionized water (Milli-Q water purification system, Billerica, MA) overnight with a total volume of 20 μL. Eighty microliters of 2-(N-morpholino)ethanesulfonic acid (MES) buffer (50 mM, pH 6.2) and the freshly prepared EDC solution (1μL 100 mg/mL) were added into the above mixture to allow crosslinking for 4 hrs at room temperature. We particularly skipped the denaturing step during the tryptic digestion, aiming to remove only the peptides far away from the NP surface but keep those belonging to the binding site on the NPs. Excess EDC and salts were removed with an Amicon 30 kDa filter (Millipore, Billerica, MA) by spinning it at 14 krcf for 10 min. The sample mixture was recovered by spinning the filter reversely in another clean centrifuge tube at 1 krcf for 2 min. Afterwards, 0.15 μg of trypsin was incubated with the recovered NP-HSA mixture in NH4HCO3 buffer (50 mM, pH 8.0) with a final reaction volume of 100 μL at 37 ºC overnight. Another centrifugation (16.1 krcf × 30 min) in 1.5-mL centrifuge tube helped to precipitate and remove the NPs. The supernatant containing the free peptides was collected, dried in a Savant SpeedVac® concentrator (Bridgepath Scientific, MD) and re-dissolved in 0.1% trifluoroacetic acid (TFA). After removing the NH4HCO3 with a C18 ziptip (Millipore), the peptides were recovered into 50% acetonitrile (ACN, HPLC grade, 99.9%, Fisher Scientific) containing 0.05% TFA and ready for matrix-assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS) or MALDI-quadrupole (Q)-TOF MS/MS. Supernatant analysis was repeated 10 times.
Peptide removal from the NPs after crosslinking and trypsin digestion
After trypsin digestion, the NPs recovered by centrifugation were washed sequentially by 0.09% sodium dodecyl sulfate (SDS) and deionized water to remove all the possibly adsorbed peptides. During the washes, the NPs were retained by the Amicon 30 kDa filter while the filter was spun at 14 krcf for 10 min to allow the adsorbed but not crosslinked peptides to go through. Then the NPs were treated with 10 μL of a mixture solution containing 40% dimethylamine (DMA, from Sigma-Aldrich), concentrated hydrochloride (12 N, Fisher Scientific) and acetonitrile at a volume ratio of 7:1:24 (final pH of 10) for 4 hrs at room temperature. Amicon 30 kDa filters were used to remove the NPs from the above mixtures at 14 krcf for 10 min. The solution containing the released peptides was collected and subject for MS analysis. Peptide removal by DMA after crosslinking was repeated 5 times.
MALDI-TOF-MS and MALDI-Q-TOF-MS/MS
MALDI-TOF-MS experiment was carried out on a Voyager-DE STR MALDI-TOF mass spectrometer (Applied Biosystems, Framingham, MA, USA) operating in positive reflective mode. The spectrometer is equipped with a pulsed nitrogen laser operated at 337 nm with 3-ns pulses. MS spectra were acquired as an average of 100 laser shots. Mass resolution is greater than 10,000. All the chemicals used for MS analysis are highly pure. The HPLC-grade alpha-Cyano-4-hydroxycinnamic acid (CHCA) from Sigma-Aldrich is > 99% pure, and the biochemistry-grade trifluoroacetic acid (TFA) from Acros Organics has a purity of 99.5. Peptide samples were cleaned up by using the C18 ziptips. For sample spotting, in brief, 0.5 μl of the saturated CHCA in 50% ACN and 0.05% TFA, and 0.5 μL of the clean peptide solution were sequentially spotted on MALDI plate. Adequate time was allowed for solvent evaporation. The sequences of the concerned peptides of m/z 961, m/z 1640, and m/z 2045 were verified by the Applied Biosystems Q-STAR XL oMALDI MS/MS (Carlsbad, CA) and the data were analyzed using Mascot.
Drug inhibition on NP-HSA interaction
Ibuprofen (> 98%) and fusidic acid were from Sigma-Aldrich. The drug was dissolved in 50% ethanol (HPLC grade, Fisher Scientific) to make a series of stock solutions with concentrations from 4.84 × 10−5 M to 2.42 × 10−3 M. Two microliters of the drug stock solution at different concentrations were mixed with 160 μg of HSA in a total volume of 18 μL (final HSA concentration was 1.19 × 10−4 M) and incubated at room temperature for 30 min. Then, 9.4 × 10−12 mol NPs were introduced into the HSA-drug mixture. The final mixture contained 5% ethanol with a final volume of 20 μL. After overnight incubation, these samples were analyzed by capillary electrophoresis in the Beckman Coulter P/ACETM MDQ system. A 50-cm fused-silica capillary (75 μm id, 365 μm od; Polymicro Technologies, Phoenix, AZ, USA) with an effective length of 40 cm was sequentially rinsed at 30 psi with 0.1 M NaOH (2 min), deionized water (1 min), and the running buffer (6 min) prior to injection. All CE separations were done at 25 kV at room temperature. A borate buffer (10 mM, pH 8.3) was used as the separation buffer. The areas of the complex and the free NPs peaks were calculated by the 32 KaratTM Version 8.0 accompanied with the Beckman system. Normalization was performed by dividing the complex peak areas measured at the presence of ibuprofen with that obtained without ibuprofen.
Results and Discussion
HSA is the most abundant protein in human serum with a well-studied crystal structure,25 and it can bind to various types of NPs, including iron oxides.2 Superparamagnetic Fe3O4 NPs have demonstrated great potential for biomedical applications, but their health impacts are not well understood so as the mechanisms of their transportation within circulation systems and delivery to tissues or cells.11,26,27 Study the binding of the Fe3O4 NPs to HSA and their locations on HSA may shed some lights on these mechanisms, since HSA has been known to be the carrier for a wide range of xenobiotic chemicals crossing the membranous structures.28 The polyacrylic acid (PAA)-coated Fe3O4 NPs prepared as reported by Ge et. al.29 possess high water solubility that can be attributed to the strong coordination of carboxylate groups with surface iron cations and the multiple anchor points for every single polymer chain. It has been demonstrated that a large amount of carboxylate groups remained on the surface of nanocrystals after synthesis and extensive washing.29 The average size of these NPs was 8.02 ± 1.26 nm, with a hydration size in water of 40.0 ± 3.9 nm and a zeta potential of −44.03 ± 0.08 mV (Supporting Information Figure S1b). These NPs interact with various proteins in aqueous solutions (Supporting Information Figure S1c), including HSA. The dissociation constant (KD) of the NP-HSA complex in water was 5.1 × 10−6 ± 1.8 × 10−7 M and that in 1 × PBS was 6.3 × 10−5 ± 5.3 × 10−7 M, measured by our reported method.30
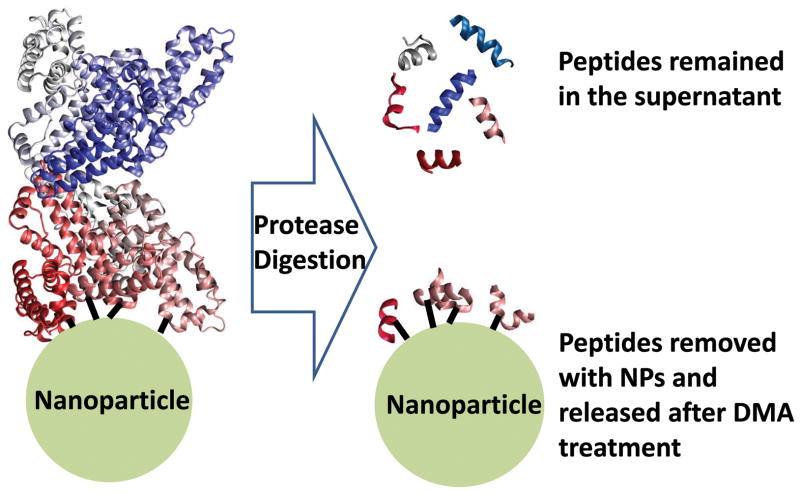
Chemical crosslinking was performed after the stable HSA adsorption on NPs (at a 1:1 protein to NPs ratio) was obtained from overnight incubation. Water was chosen as the incubation environment in the present study to enhance the total amount of the NP-bound HSA and facilitate MS analysis. The carboxyl groups on the PAA coating were coupled to the free amines on HSA by a zero-length, amine-reactive crosslinker, EDC.31 Then, we digested the protein by trypsin, leaving the crosslinked peptides on particle surface and those far away from the binding site of NPs in supernatant (Figure 1). The crosslinked peptides were isolated together with the NPs by centrifugation; cleaned thoroughly with several rounds of sequential wash to remove the passively adsorbed fragments; and finally cleaved off the particle surface by the base-assisted hydrolysis. Both the cleaved peptides, i.e. interfacial peptides, and those remained in the supernatant, i.e. peripheral peptides, were analyzed by MALDI-TOF-MS.
Figure 1.
Identification of peptides associated with nanoparticle-protein interaction by crosslinking and MS.
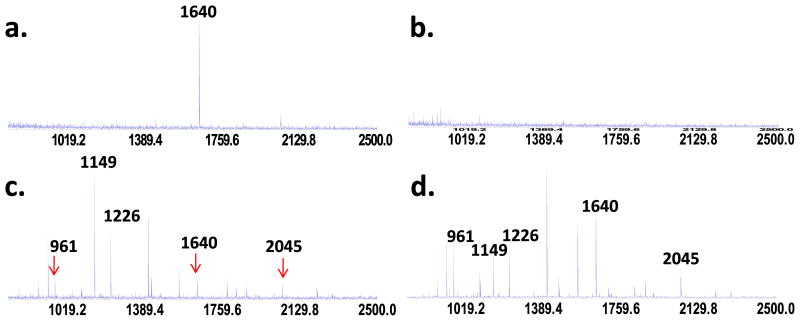
The typical MS result on peptides crosslinked to the NPs and then released by hydrolysis was shown in Figure 2a. Only one peptide with an m/z value of 1640 was found in this sample. Its sequence was confirmed to be (K) KVPQVSTPTLVEVSR (414–428, sequence numbers adopted from PDB ID: 2VUF) by MALDI-Q-TOF-MS/MS (Supporting Information Figure S2). No peptide was recovered from the NPs if no crosslinking reaction took place (Figure 2b), meaning that the non-specifically adsorbed peptides were completely washed off. Agreeing with these results, supernatant analysis revealed that the peak intensity of m/z 1640 dropped significantly in the crosslinking sample (Figure 2c), but not in the not-crosslinked sample (Figure 2d). A total of 13 peptides were identified under our digestion and analysis conditions (Supporting Information Figure S3). Among them, three were consistently found with much reduced peak intensity in the supernatant samples (Figure 2c), including m/z 1640. The other two are m/z 961 (K)FQNALLVR (403–410), and m/z 2045 (K)VFDEFKPLVEEPQNLIK (373–389).
Figure 2.
MALDI-MS spectra of released peptides when HSA was (a) crosslinked and (b) only adsorbed to the PAA-Fe3O4 NPs. Analysis of supernatant from the crosslinking and non-crosslinking samples were shown in (c) and (d), respectively.
Because the circular dichroism (CD) spectra (Supporting Information Figure S4) showed negligible change to the secondary structure of HSA after being adsorbed to the NPs, we mapped these three peptides on the crystal structure of HSA (PDB ID: 2VUF). Interestingly, they all belong to the subdomain IIIA of HSA, and locate quite close to each other.25 Therefore, they could be near the NP surface during protein adsorption and have all been crosslinked to the NPs. It is unknown at this point that why only m/z 1640 was released by hydrolysis. We hypothesized it may be related to its consecutive lysine residues, K413 and K414. K413 was the tryptic digestion site on this fragment, and K414 could be the crosslinking site which was also the N-terminal of this fragment. This location possibly makes the amide bond formed between the lysine side chain K413 and the PAA quite easily be attacked by the amine during hydrolysis. Obviously, more investigation is needed to test this possibility and detailed method development is required to improve the peptide release efficiency.
Nevertheless, the persistent identification of particular peptides in our crosslinking experiment strongly supports that those peptides, especially peptide KVPQVSTPTLVEVSR (414–428) with an m/z value of 1640, should be part of the binding site of the PAA-coated Fe3O4 NPs on HSA, and their approaching to the NPs surface was not a random event. We examined the solvent accessible surface areas of all lysine residues on HSA using the on-line tool of Getarea (http://curie.utmb.edu/getarea.html). This software obtains a percentage value to represent the solvent accessibility of individual residue. Residues are considered to be solvent exposed if the Getarea value exceeds 50% and to be buried if it is less than 20%. It turns out that the digestion sites for m/z 961 and m/z 2045, K402 and K372, both have the calculated percentages over 55%, and considered to be exposed to the solvent; but that of K413 and K414 was only 23.9% and 8.0%, respectively. Therefore, K413 and K414 in fact have very limited solvent accessible areas. They could be crosslinked to the NPs solely because they located near the NP surface during NP-protein interaction, but not because they were more exposed than other lysine residues.
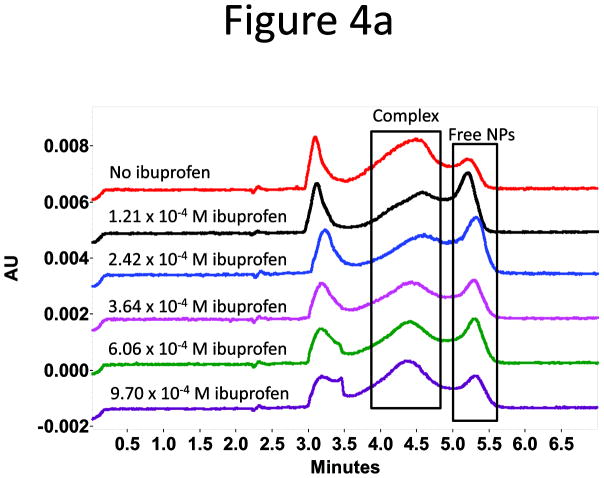
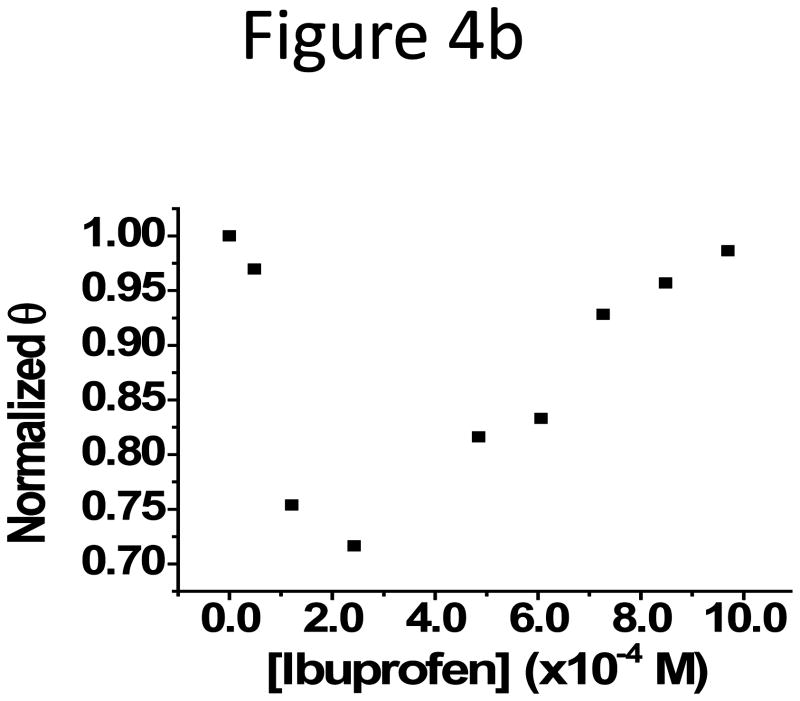
The subdomain IIIA is a putative binding site, the so-called drug-binding site 2, for drugs with acidic or electronegative features like ibuprofen and naproxen (Figure 3).28 Ibuprofen binds to the site 2 on HSA with an equilibrium constant around 106 M−1.32,33 The binding can improve the solubility of the drug in serum and help its transportation in the circulation system, affecting the pharmacokinetics of the drug.28,34 Because of the colocalization of the binding sites of ibuprofen and the PAA-coated Fe3O4 NPs on HSA, competitive binding between the NPs and ibuprofen was expected. To verify this possibility, we incubated 1.19 × 10−4 M HSA with ibuprofen at concentrations starting from 4.84 × 10−5 M for 30 min, and then added a constant amount of NPs to the HSA-ibuprofen mixture. The HSA concentration was selected so that a large portion of NPs was bound to HSA with the complex peak well observed under the experimental conditions; and the drug concentration range was chosen accordingly so that all HSA molecules should have been bound to ibuprofen. The mixture was analyzed with capillary electrophoresis (CE) after another round of incubation with the NPs in present at room temperature for overnight. Since the NP-HSA complexes could be well separated from the free HSA and NPs,30 the fraction of protein-bound NPs, θ, obtained from the peak area ratio of the protein- bound and free NPs, was used to judge the NP-HSA interaction strength: a smaller θ representing weaker binding. The selected electropherograms and the plot of the normalized θ (θ obtained with no drug present was seen as 1 in normalization) vs. drug concentration were shown in Figure 4. A decrease in θ was observed with increasing ibuprofen concentration, and the reduction reached a maximum of 28% at 3 × 10−4 M ibuprofen. Since binding of ibuprofen induces very little conformational change in HSA,28 inhibition of NP-HSA interaction by ibuprofen could not be due to the exposure of new epitopes on HSA upon drug binding, but rather from the occupation of ibuprofen in site 2. We also confirmed that there was no interaction between the drug and the NPs by the same CE method (Supporting information Figure S5a). However, when the drug concentration increased further, θ started to increase back but the complex peak shifted slightly towards the injection peak in CE. At even higher drug concentrations (Supporting Information Figure S5b), the free NP peak disappeared; and the complex peak profile changed completely with a much earlier migration time. It is possible that at higher drug concentrations, ibuprofen might bind to HSA at sites other than the drug binding site 2 and increase the hydrophobicity of the protein, which in turn enhance the binding between the protein and the NPs and form a three-party complex with both the drug and the NP binding simultaneously on HSA. Certainly, more exploration is needed to find out the reason for the observed results. On the contrary, a drug, fusidic acid, that binds further away from the subdomain IIIA35 (PDB ID: 2VUF), exhibited no effect on the NP-HSA interaction (Supporting Information Figure S5c & 5d).
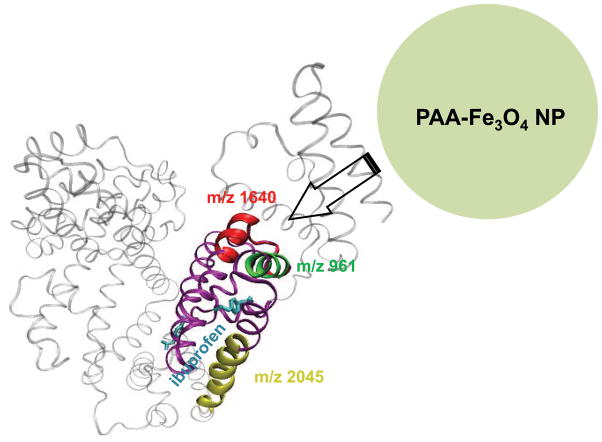
Figure 3.
Crystal structure of HSA with ibuprofen bound (PDB ID: 2BXG), with the peptides in interest highlighted in red, green, and yellow, respectively.
Figure 4.
(a) Representative electropherograms attained at 288 nm for the study of competitive binding to HSA between ibuprofen and PAA-Fe3O4 NPs. HSA peak appeared at 3.0 min, and the free ibuprofen showed up at around 3.4 min. (b) Normalized θ (the fraction of protein-bound NPs) was plotted against the drug concentration.
The particular binding of the PAA-coated Fe3O4 NPs towards the drug binding site 2 could be determined by the carboxyl groups of the PAA coating. It has been known that the drug binding site 2 has a pre-formed hydrophobic cavity with distinct polar features and a single main polar patch at the pocket entrance.28 Distribution of basic and polar residues on the largely hydrophobic interior walls attracts the electronegative drugs, attributed to this binding pocket.28 For example, ibuprofen has a carboxyl side chain (Supporting Information Figure S6). The carboxyl group could be attracted towards site 2, helping the aromatic rings be adapted into the hydrophobic pocket. Similarly, the PAA molecules on the NPs surface carried a large number of carboxyl groups that could be pulled close to the basic residues of R410, Y411, and K414. However, it is unclear at this point if the Fe3O4 surface was exposed and contributed to the interaction with HSA.
Conclusion
Identification of the binding sites of NPs on proteins can help us predict the possible biological consequences after the attachment. The specific binding of the PAA-coated NPs to the drug binding site 2 on HSA may be closely related to the biodistribution and cell internalization of the NPs, which should be explored further in future studies. The present work demonstrated that crosslinking chemistry coupled with MS was a convenient approach to probe the possible binding sites of NPs on proteins. Still, it is not clear if this was the only binding site, and only one peptide was discovered. Further development is needed to increase coverage of all binding sites and to mimic interaction under physiological conditions. For example, interaction occurred in 1 × PBS should be studied, but more optimization is desired to improve signal intensity because the weaker affinity in PBS leads to less protein bound. Additionally, different proteases and crosslinking chemistry can be employed to target functional groups other than lysine. Moreover, complimentary peptide identification methods like LC-MS/MS can be used to target smaller peptides; and different release methods can be explored to improve the release efficiency. All of these are currently under investigation in our lab.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Environmental Health Sciences Grant No. 1R21ES017870-01A1. The authors are also grateful for the support of Dr. Yadong Yin on nanoparticle synthesis, and the kind assistance from Mr. Yongsheng Xiao and Dr. Yinsheng Wang on MS analysis.
Footnotes
Note
The sequence numbers for peptides used here were adopted from PDB, because we used the crystal structure downloaded from PDB to map out the location of these peptides. These numbers were 24 residues different than those reported in UniProtKB/Swiss-Prot, protein ID P02768. For example, using UniProtKB sequence number, m/z 1640 should be 438–452.
Supporting Information Available
Supporting information includes characterization of the PAA-coated Fe3O4 NPs, MALDI-Q-TOF-MS/MS spectrum for peptide sequence identification, MALDI-TOF-MS spectrum for HSA digest, CD spectra for HSA and NP-bound HSA, CE study of drug inhibition on NP-HSA binding at high ibuprofen concentrations and interaction between ibuprofen and the NPs, crystal structure of HSA bound with fusidic acid and result of the HSA binding competition between fusidic acid and NPs, and the structures of the two drug molecules used in our study. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Mu Q, Li Z, Li X, Mishra SR, Zhang B, Si Z, Yang L, Jiang W, Yan B. J Phys Chem C. 2009;113:5390–5395. [Google Scholar]
- 2.Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Adv Drug Delivery Rev. 2009;61:428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch I, Cedervall T, Lundqvist M, Cabaleiro-Lago C, Linse S, Dawson KA. Adv Colloid Interface Sci. 2007;134–135:167–174. doi: 10.1016/j.cis.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Z-J, Carboni R, Quercio JMJ, Yan B, Miranda OR, Anderton DL, Arcaro KF, Rotello VM, Vachet RW. Small. 2010;6:2261–2265. doi: 10.1002/smll.201000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lundqvist M, Sethson I, Jonsson B-H. Langmuir. 2004;20:10639–10647. doi: 10.1021/la0484725. [DOI] [PubMed] [Google Scholar]
- 6.Shang W, Nuffer JH, Muniz-Papandrea VA, Colon W, Siegel RW, Dordick JS. Small. 2009;5:470–476. doi: 10.1002/smll.200800995. [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Jia HF, Wang P. Biotechnol Adv. 2006;24:296–308. doi: 10.1016/j.biotechadv.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 8.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Nat Chem. 2009;1:461–465. S461/461–S461/426. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long MS, Keating CD. Anal Chem. 2006;78:379–386. doi: 10.1021/ac051882t. [DOI] [PubMed] [Google Scholar]
- 10.Corot C, Robert P, Idee J-M, Port M. Adv Drug Delivery Rev. 2006;58:1471–1504. doi: 10.1016/j.addr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Neuberger T, Schoepf B, Hofmann H, Hofmann M, Von RB. J Magn Magn Mater. 2005;293:483–496. [Google Scholar]
- 12.Oh E, Lee D, Kim Y-P, Cha SY, Oh D-B, Kang HA, Kim J, Kim H-S. Angew Chem, Int Ed. 2006;45:7959–7963. doi: 10.1002/anie.200601948. [DOI] [PubMed] [Google Scholar]
- 13.Delfino I, Cannistraro S. Biophys Chem. 2009;139:1–7. doi: 10.1016/j.bpc.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 14.De M, You C-C, Srivastava S, Rotello VM. J Am Chem Soc. 2007;129:10747–10753. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 15.Cliffel DE, Turner BN, Huffman BJ. Wiley Interdisciplinary Rev : Nanomed Nanobiotechnol. 2009;1:47–59. doi: 10.1002/wnan.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayraktar H, You C-C, Rotello VM, Knapp MJ. J Am Chem Soc. 2007;129:2732–2733. doi: 10.1021/ja067497i. [DOI] [PubMed] [Google Scholar]
- 17.Calzolai L, Franchini F, Gilliland D, Rossi F. Nano Lett. 2010;10:3101–3105. doi: 10.1021/nl101746v. [DOI] [PubMed] [Google Scholar]
- 18.Atzrodt J, Derdau V, Fey T, Zimmermann J. Angew Chem Int Ed. 2007;46:7744–7765. doi: 10.1002/anie.200700039. [DOI] [PubMed] [Google Scholar]
- 19.Tsutsui Y, Wintrode PL. Curr Med Chem. 2007;14:2344–2358. doi: 10.2174/092986707781745596. [DOI] [PubMed] [Google Scholar]
- 20.Bennett KL, Matthiesen T, Roepstorff P. Methods Mol Biol. 2000;146:113–131. doi: 10.1385/1-59259-045-4:113. [DOI] [PubMed] [Google Scholar]
- 21.Chakravarti B, Lewis SJ, Chakravarti DN, Raval A. Curr Proteomics. 2006;3:1–21. [Google Scholar]
- 22.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miernyk JA, Thelen JJ. Plant J. 2008;53:597–609. doi: 10.1111/j.1365-313X.2007.03316.x. [DOI] [PubMed] [Google Scholar]
- 24.Rana S, Yeh Y-C, Rotello VM. Curr Opin Chem Biol. 2010;14:828–834. doi: 10.1016/j.cbpa.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 26.Singha N, Jenkinsa GJS, Asadib R, Doaka SH. Nano Rev. 2010;1 doi: 10.3402/nano.v3401i3400.5358. [DOI] [Google Scholar]
- 27.Liu SJ, Han YC, Qiao RR, Zeng JF, Jia QJ, Wang YL, Gao MY. J PhysChem C. 2010;114:21270–21276. [Google Scholar]
- 28.Ghuman J, Zunszain PA, Petitpas I, Bhattacharya AA, Otagiri M, Curry S. J Mol Biol. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 29.Ge J, Hu Y, Biasini M, Dong C, Guo J, Beyermann WP, Yin Y. Chem Eur J. 2007;13:7153–7161. doi: 10.1002/chem.200700375. [DOI] [PubMed] [Google Scholar]
- 30.Li N, Zeng S, He L, Zhong W. Anal Chem. 2010;82:7460–7466. doi: 10.1021/ac101627p. [DOI] [PubMed] [Google Scholar]
- 31.Hermanson GT. Bioconjugation Techniques. 2. Academic Press; New York: 2008. [Google Scholar]
- 32.Jin L, Choi DY, Liu H, Row KH. Bull Korean Chem Soc. 2005;26:136–138. [Google Scholar]
- 33.Yamasaki K, Rahman MH, Tsutsumi Y, Maruyama T, Ahmed S, Kragh-Hansen U, Otagiri M. AAPS PharmSciTech. 2000;1 doi: 10.1007/BF02830527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ascenzi P, Fasano M. Biophys Chem. 2010;148:16–22. doi: 10.1016/j.bpc.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Zunszain PA, Ghuman J, McDonagh AF, Curry S. J Mol Biol. 2008;381:394–406. doi: 10.1016/j.jmb.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.