Abstract
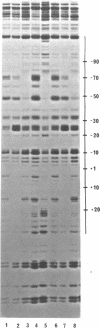
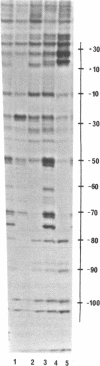
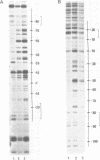
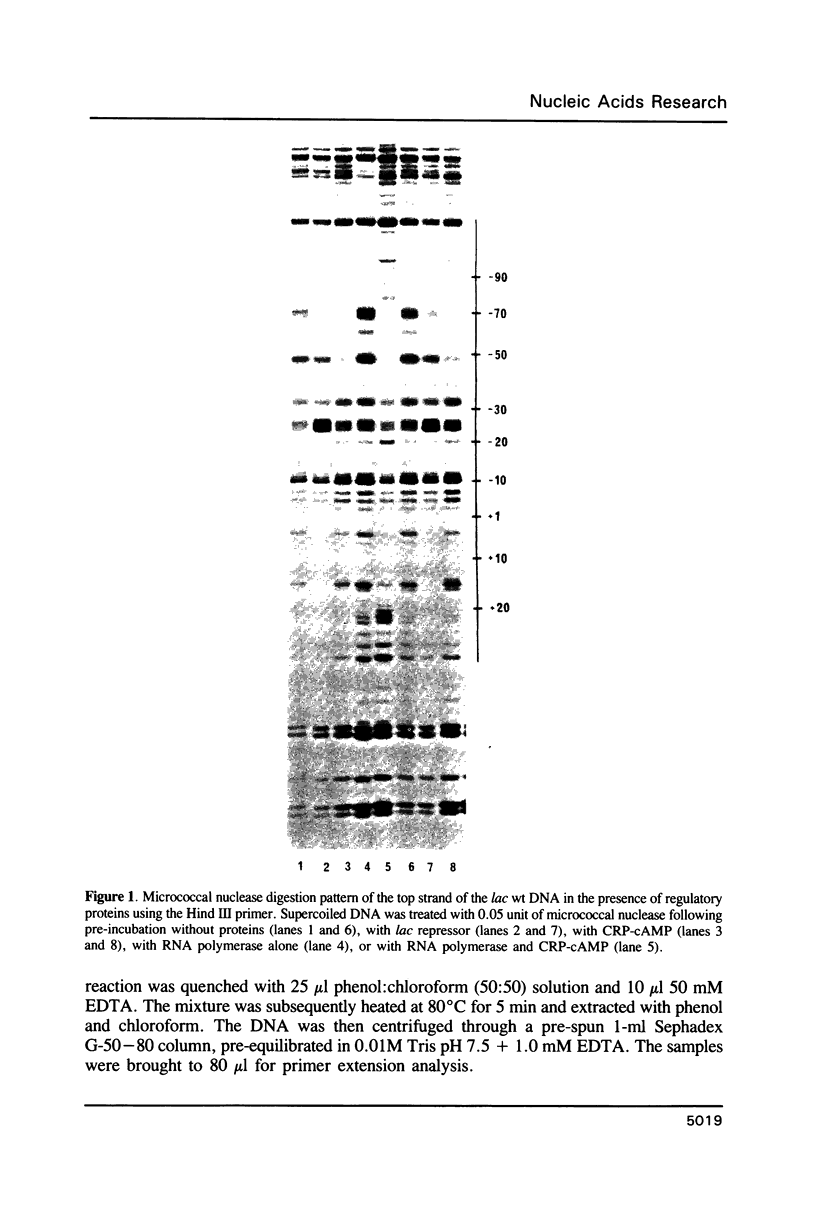
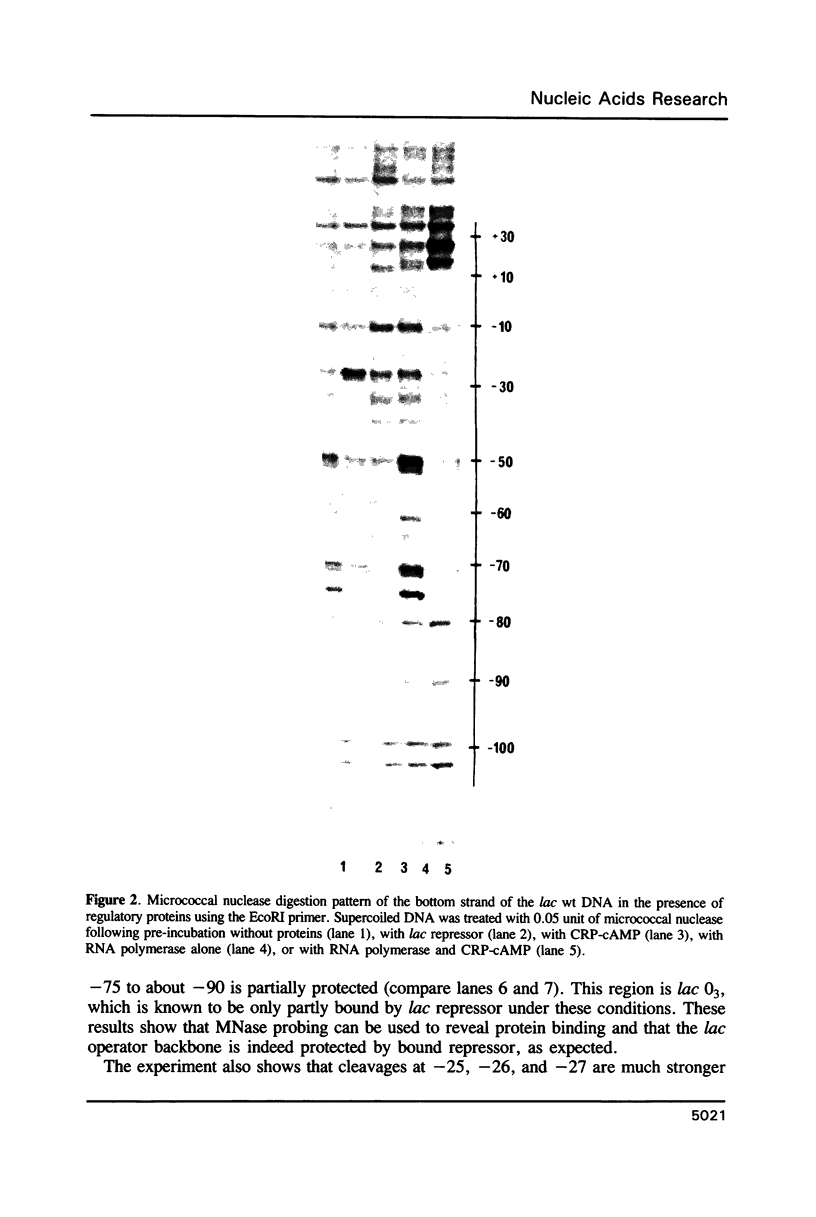
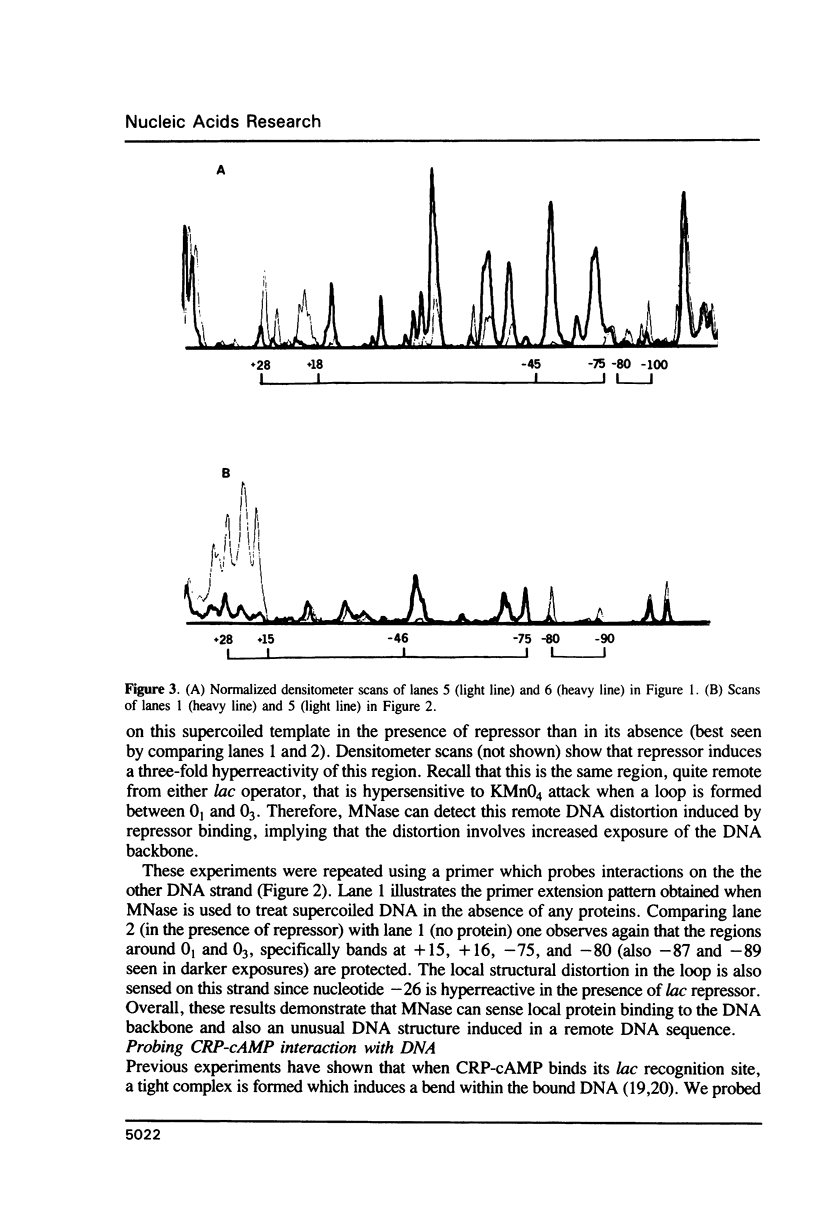
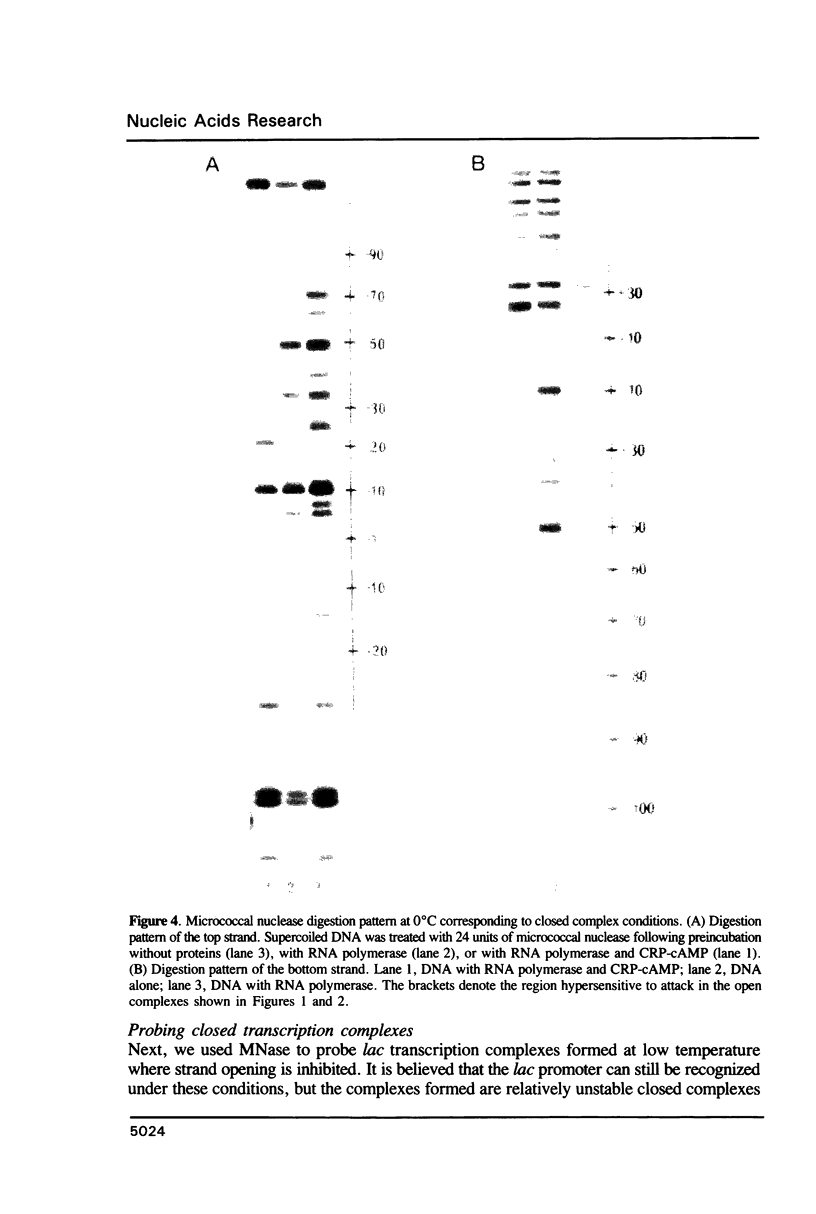
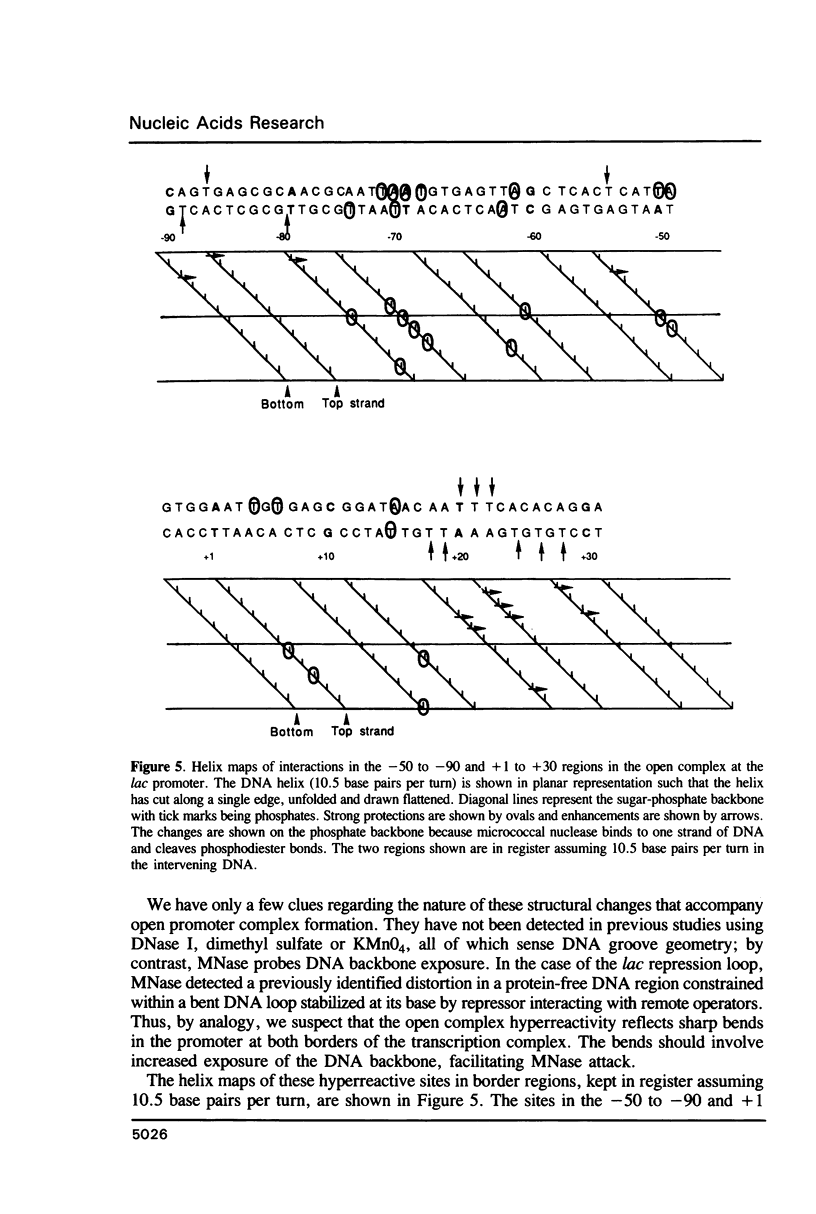
Micrococcal nuclease (MNase) is used to probe the structure of transcription and repression complexes at the lac regulatory region in vitro. Both the lac operator, 01, and the pseudo-operator, 03, are found to be protected from MNase digestion by the lac repressor on supercoiled DNA, and hypersensitive sites appear on both strands around nucleotide (nt) -26 between 01 and 03. This hyperreactive site is coincident with the site of the DNA kink shown previously to form within a loop caused by simultaneous repressor binding to 01 and 03. MNase hypersites are also observed both upstream from cAMP receptor protein (CRP) and downstream from bound RNA polymerase in open promoter complexes. In both open and closed complexes the binding of polymerase partially protects the backbone from MNase attack. Catabolite activator protein is shown to be required for both closed and open complex formation. Taken together with previous footprinting data, the results suggest that lac transcription complexes involve DNA bent towards a protein core consisting of RNA polymerase and catabolite activator protein.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker M. M., Wang J. C. Use of light for footprinting DNA in vivo. Nature. 1984 Jun 21;309(5970):682–687. doi: 10.1038/309682a0. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. High-resolution analysis of lac transcription complexes inside cells. Biochemistry. 1986 Sep 9;25(18):5051–5057. doi: 10.1021/bi00366a012. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Buc H., McClure W. R. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985 May 21;24(11):2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- Cockell M., Rhodes D., Klug A. Location of the primary sites of micrococcal nuclease cleavage on the nucleosome core. J Mol Biol. 1983 Oct 25;170(2):423–446. doi: 10.1016/s0022-2836(83)80156-9. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986 Sep 5;233(4768):1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Kovacic R. T. The 0 degree C closed complexes between Escherichia coli RNA polymerase and two promoters, T7-A3 and lacUV5. J Biol Chem. 1987 Oct 5;262(28):13654–13661. [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Malan T. P., Kolb A., Buc H., McClure W. R. Mechanism of CRP-cAMP activation of lac operon transcription initiation activation of the P1 promoter. J Mol Biol. 1984 Dec 25;180(4):881–909. doi: 10.1016/0022-2836(84)90262-6. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Meiklejohn A. L., Gralla J. D. Entry of RNA polymerase at the lac promoter. Cell. 1985 Dec;43(3 Pt 2):769–776. doi: 10.1016/0092-8674(85)90250-8. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A. Cyclic AMP receptor proteins interacts with lactose operator DNA. Nucleic Acids Res. 1981 Jan 24;9(2):277–292. doi: 10.1093/nar/9.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Spassky A., Sigman D. S. Nuclease activity of 1,10-phenanthroline-copper ion. Conformational analysis and footprinting of the lac operon. Biochemistry. 1985 Dec 31;24(27):8050–8056. doi: 10.1021/bi00348a032. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Dong Y. C., Ptashne M., Harrison S. C. Structure of a phage 434 Cro/DNA complex. Nature. 1988 Oct 27;335(6193):789–795. doi: 10.1038/335789a0. [DOI] [PubMed] [Google Scholar]