Abstract
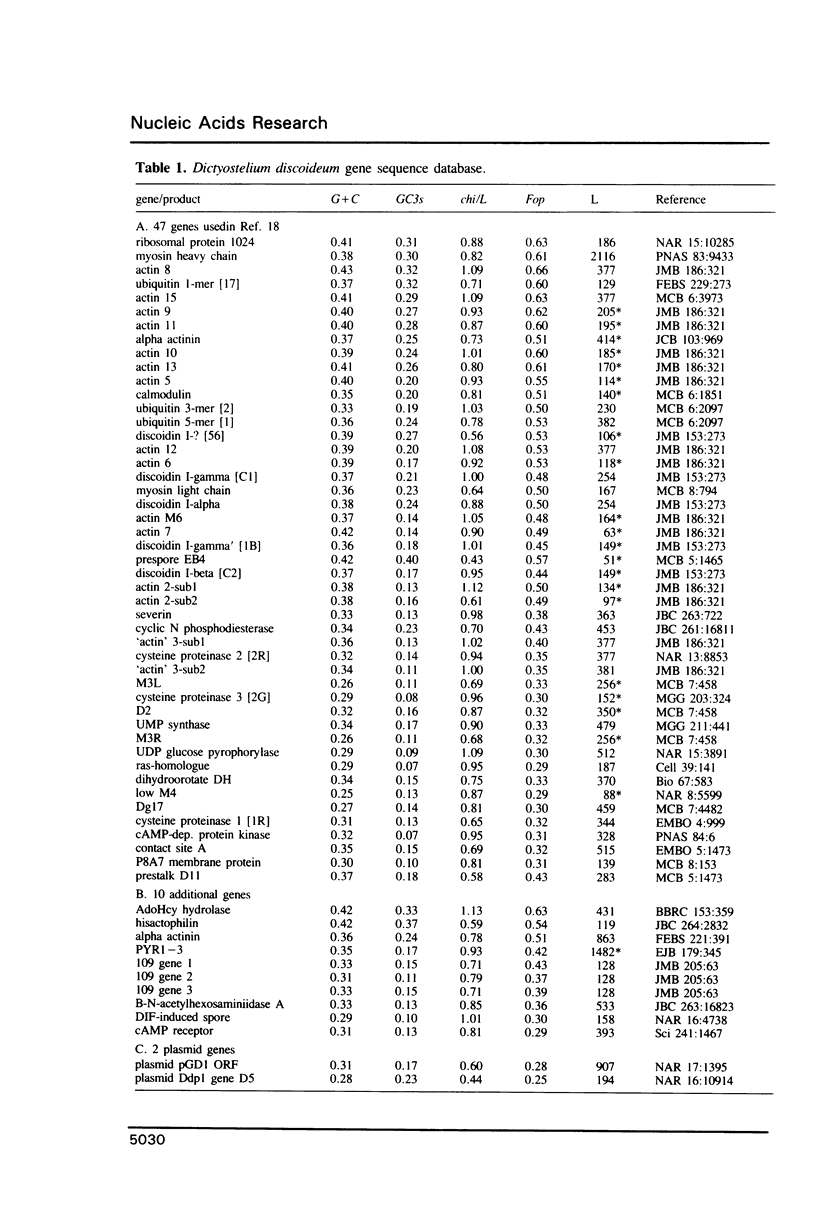
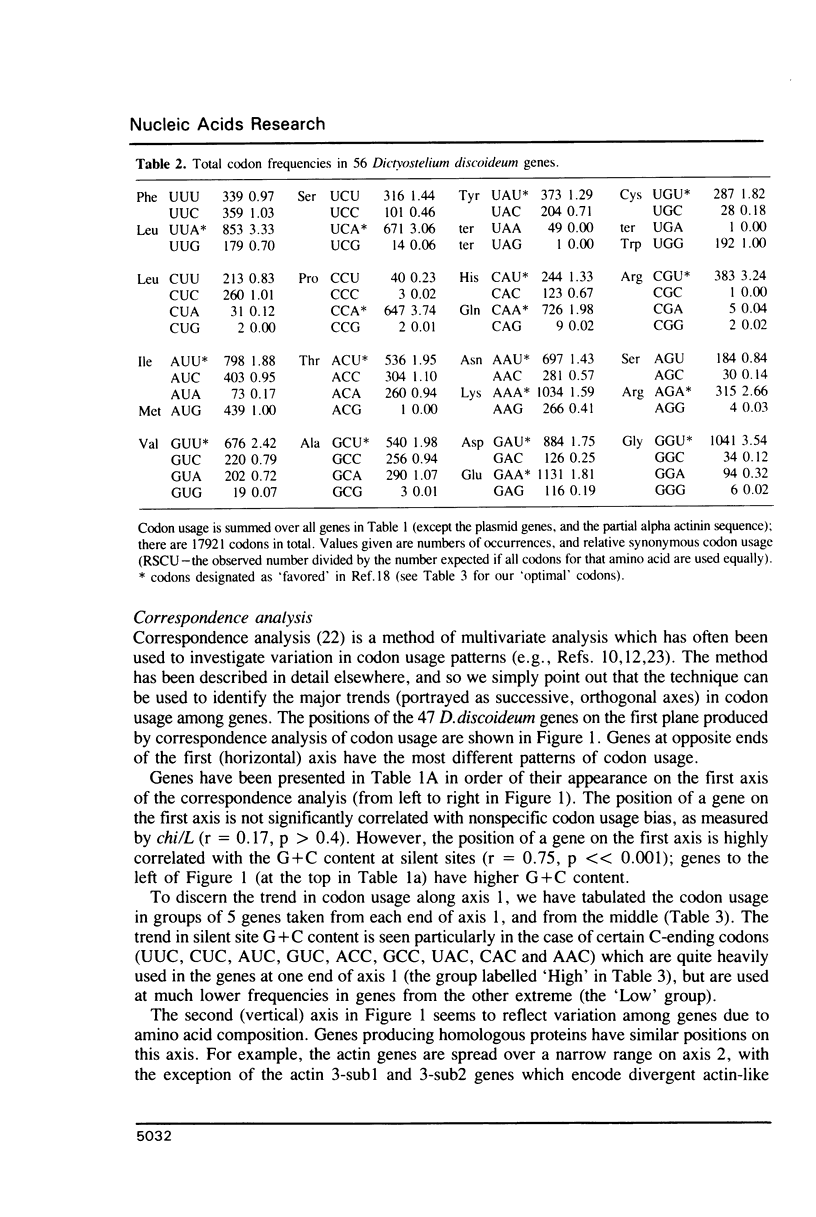
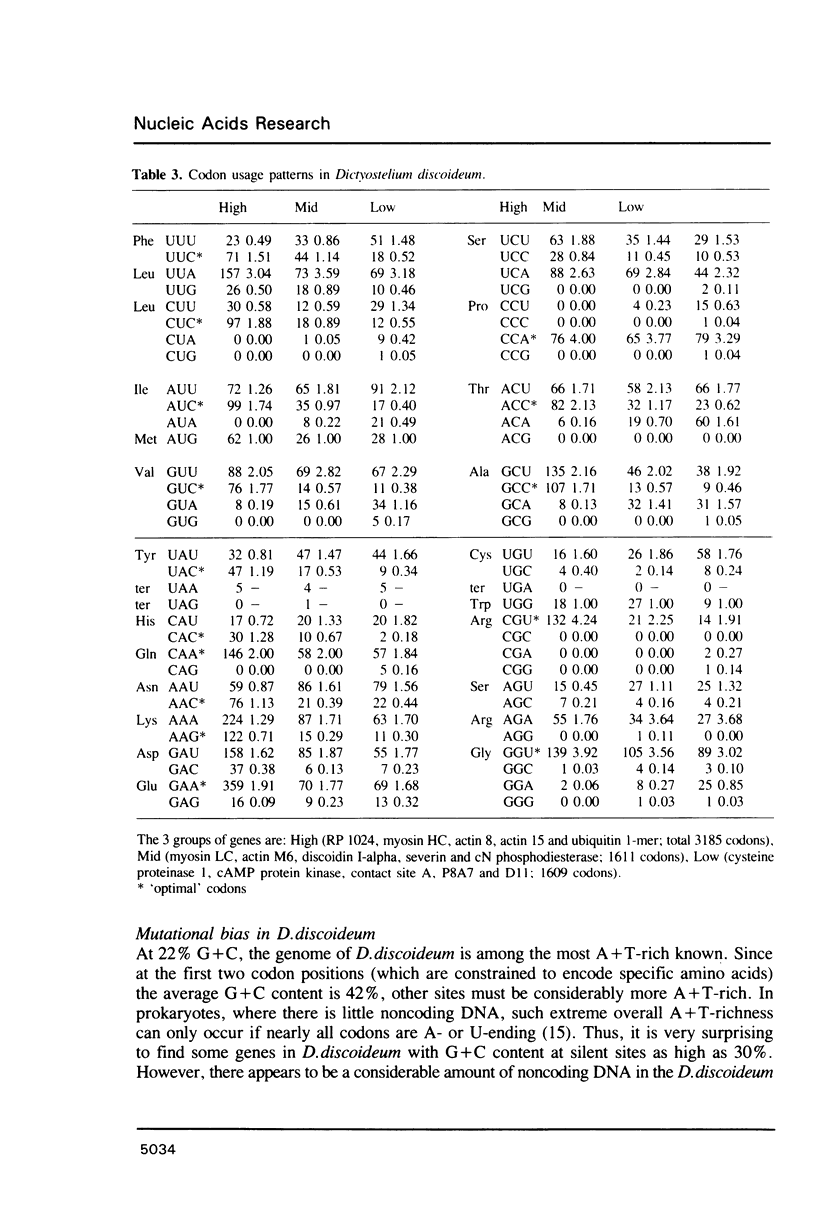
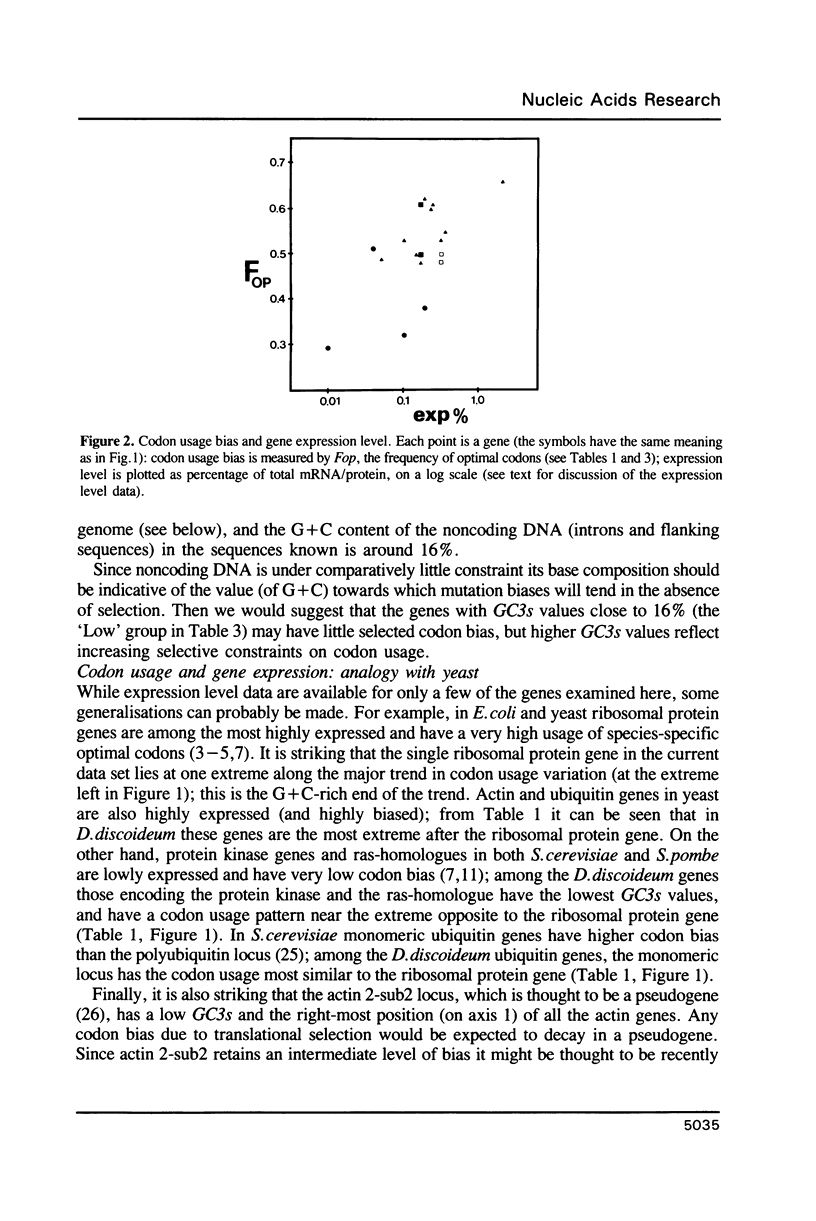
Codon usage patterns in the slime mould Dictyostelium discoideum have been re-examined (a total of 58 genes have been analysed). Considering the extreme A + T-richness of this genome (G + C = 22%), there is a surprising degree of codon usage variation among genes. For example, G + C content at silent sites varies from less than 10% to greater than 30%. It was previously suggested [Warrick, H.M. and Spudich, J.A. (1988) Nucleic Acids Res. 16: 6617-6635] that highly expressed genes contain fewer 'optimal' codons than genes expressed at lower levels. However, it appears that the optimal codons were misidentified. Multivariate statistical analysis shows that the greatest variation among genes is in relative usage of a particular subset of codons (about one per amino acid), many of which are C-ending. We have identified these as optimal codons, since (i) their frequency is positively correlated with gene expression level, and (ii) there is a strong mutation bias in this genome towards A and T nucleotides. Thus, codon usage in D. discoideum can be explained by a balance between the forces of mutational bias and translational selection.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aota S., Ikemura T. Diversity in G + C content at the third position of codons in vertebrate genes and its cause. Nucleic Acids Res. 1986 Aug 26;14(16):6345–6355. doi: 10.1093/nar/14.16.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bilofsky H. S., Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861–1863. doi: 10.1093/nar/16.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M., Bazari W. L., Kayman S. C. Isolation and properties of calmodulin from Dictyostelium discoideum. J Bacteriol. 1980 Jan;141(1):397–400. doi: 10.1128/jb.141.1.397-400.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M., Spudich J. A. Biochemical and structural studies of actomyosin-like proteins from non-muscle cells. Isolation and characterization of myosin from amoebae of Dictyostelium discoideum. J Mol Biol. 1974 Jun 25;86(2):209–222. doi: 10.1016/0022-2836(74)90013-8. [DOI] [PubMed] [Google Scholar]
- Giorda R., Ohmachi T., Ennis H. L. Organization of a gene family developmentally regulated during Dictyostelium discoideum spore germination. J Mol Biol. 1989 Jan 5;205(1):63–69. doi: 10.1016/0022-2836(89)90364-1. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C., Attimonelli M., Lanave C., di Paola G. ACNUC--a portable retrieval system for nucleic acid sequence databases: logical and physical designs and usage. Comput Appl Biosci. 1985 Sep;1(3):167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J. E., Sims P. F. Anomalous dinucleotide frequencies in both coding and non-coding regions from the genome of the human malaria parasite Plasmodium falciparum. Gene. 1987;61(2):177–187. doi: 10.1016/0378-1119(87)90112-0. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Kasir J., Aksamit R. R., Backlund P. S., Jr, Cantoni G. L. Amino acid sequence of S-adenosyl-L-homocysteine hydrolase from Dictyostelium discoideum as deduced from the cDNA sequence. Biochem Biophys Res Commun. 1988 May 31;153(1):359–364. doi: 10.1016/s0006-291x(88)81231-2. [DOI] [PubMed] [Google Scholar]
- Kimmel A. R., Firtel R. A. Intervening sequences in a Dictyostelium gene that encodes a low abundance class mRNA. Nucleic Acids Res. 1980 Dec 11;8(23):5599–5610. doi: 10.1093/nar/8.23.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel A. R., Firtel R. A. Sequence organization in Dictyostelium: unique structure at the 5'-ends of protein coding genes. Nucleic Acids Res. 1983 Jan 25;11(2):541–552. doi: 10.1093/nar/11.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K., Gerisch G., Fromme I., Mayer H., Tsugita A. A membrane glycoprotein of aggregating Dictyostelium cells with the properties of contact sites. Eur J Biochem. 1979 Sep;99(2):419–426. doi: 10.1111/j.1432-1033.1979.tb13271.x. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H., Muto A., Yamao F., Ohama T., Andachi Y. Role of directional mutation pressure in the evolution of the eubacterial genetic code. Cold Spring Harb Symp Quant Biol. 1987;52:777–789. doi: 10.1101/sqb.1987.052.01.087. [DOI] [PubMed] [Google Scholar]
- Post L. E., Nomura M. DNA sequences from the str operon of Escherichia coli. J Biol Chem. 1980 May 25;255(10):4660–4666. [PubMed] [Google Scholar]
- Romans P., Firtel R. A., Saxe C. L., 3rd Gene-specific expression of the actin multigene family of Dictyostelium discoideum. J Mol Biol. 1985 Nov 20;186(2):337–355. doi: 10.1016/0022-2836(85)90109-3. [DOI] [PubMed] [Google Scholar]
- Saul A., Battistutta D. Codon usage in Plasmodium falciparum. Mol Biochem Parasitol. 1988 Jan 1;27(1):35–42. doi: 10.1016/0166-6851(88)90022-9. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E., Higgins D. G., Shields D. C., Wolfe K. H., Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988 Sep 12;16(17):8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24(1-2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. Codon usage in regulatory genes in Escherichia coli does not reflect selection for 'rare' codons. Nucleic Acids Res. 1986 Oct 10;14(19):7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. C., Sharp P. M., Higgins D. G., Wright F. "Silent" sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol. 1988 Nov;5(6):704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- Shields D. C., Sharp P. M. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res. 1987 Oct 12;15(19):8023–8040. doi: 10.1093/nar/15.19.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu C. H., Lerner R. A., Ma G., Firtel R. A., Loomis W. F. Developmentally regulated proteins of the plasma membrane of Dictyostelium discoideum. The carbohydrate-binding protein. J Mol Biol. 1976 Jan 15;100(2):157–178. doi: 10.1016/s0022-2836(76)80146-5. [DOI] [PubMed] [Google Scholar]
- Uyemura D. G., Brown S. S., Spudich J. A. Biochemical and structural characterization of actin from Dictyostelium discoideum. J Biol Chem. 1978 Dec 25;253(24):9088–9096. [PubMed] [Google Scholar]
- Warrick H. M., Spudich J. A. Codon preference in Dictyostelium discoideum. Nucleic Acids Res. 1988 Jul 25;16(14A):6617–6635. doi: 10.1093/nar/16.14.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Sharp P. M., Li W. H. Mutation rates differ among regions of the mammalian genome. Nature. 1989 Jan 19;337(6204):283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Pardee J. D., Reidler J., Stryer L., Spudich J. A. Mechanism of interaction of Dictyostelium severin with actin filaments. J Cell Biol. 1982 Dec;95(3):711–719. doi: 10.1083/jcb.95.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]


