Abstract
Background & objectives:
Diverse mechanisms have been identified in enteric bacteria for their adaptation and survival against multiple classes of antimicrobial agents. Resistance of bacteria to the most effective fluoroquinolones have increasingly been reported in many countries. We have identified that most of the enterotoxigenic Escherichia coli (ETEC) were resistant to several antimicrobials in a diarrhoea outbreak at Ahmedabad during 2000. The present study was done to identify several genes responsible for antimicrobial resistance and mobile genetic elements in the ETEC strains.
Methods:
Seventeen ETEC strains isolated from diarrhoeal patients were included in this study. The antimicrobial resistance was confirmed by conventional disc diffusion method. PCR and DNA sequencing were performed for the identification of mutation in the quinolone resistance-determining regions (QRDRs). Efflux pump was tested by inhibiting the proton-motive force. DNA hybridization assay was made for the detection of integrase genes and the resistance gene cassettes were identified by direct sequencing of the PCR amplicons.
Results:
Majority of the ETEC had GyrA mutations at codons 83 and 87 and in ParC at codon 80. Six strains had an additional mutation in ParC at codon 108 and two had at position 84. Plasmid-borne qnr gene alleles that encode quinolone resistance were not detected but the newly described aac(6’)-Ib-cr gene encoding a fluoroquinolne-modifying enzyme was detected in 64.7 per cent of the ETEC. Class 1 (intI1) and class 2 (intI2) integrons were detected in six (35.3%) and three (17.6%) strains, respectively. Four strains (23.5%) had both the classes of integrons. Sequence analysis revealed presence of dfrA17, aadA1, aadA5 in class 1, and dfrA1, sat1, aadA1 in class 2 integrons. In addition, the other resistance genes such as tet gene alleles (94.1%), catAI (70.6%), strA (58.8%), blaTEM-1(35.2%), and aphA1-Ia (29.4%) were detected in most of the strains.
Interpretation & conclusions:
Innate gene mutations and acquisition of multidrug resistance genes through mobile genetic elements might have contributed to the emergence of multidrug resistance (MDR) in ETEC. This study reinforces the necessity of utilizing molecular techniques in the epidemiological studies to understand the nature of resistance responsible for antimicrobial resistance in different species of pathogenic bacteria.
Keywords: Efflux system, ETEC, integrons, mutations, QRDR, resistance genes
Enterotoxigenic Escherichia coli (ETEC) is an important cause of diarrhoea due to their expression of either heat-labile (LT) or heat-stable (ST) or both the enterotoxins. In developing countries, ETEC is generally associated with sporadic infection especially among children and travelers1. The first outbreak of ETEC mediated diarrhoea in India was reported from Ahmedabad, Gujarat, during 2000 and majority of the strains expressed multidrug resistance2. In clinical practice, there is a great deal of speculation regarding therapeutic and subtherapeutic use of antimicrobials as it helps in accelerating the development and dissemination of antimicrobial-resistant bacterial pathogens1.
Quinolones and fluoroquinolones are the first-line drugs for the treatment of diarrhoea for many years. However, several studies have documented the emergence and spread of fluoroquinolone resistant enteric pathogens in India3–6. Quinolone resistance mechanisms have been described and characterized in a variety of bacteria7. These have mostly been related to specific mutations that lead to amino acid alterations in the quinolone-resistance determining regions (QRDRs) within the subunits constituting topoisomerases II (GyrA and GyrB) and IV (ParC and ParE), which are involved in DNA replication, recombination, transcription and in the partitioning the replicated chromosome7. Most of the quinolone resistant clinical isolates of E. coli have mutations in the QRDRs of both the gyrA and the parC. In addition, a novel mechanism of plasmid-mediated quinolone resistance (PMQR) was reported that involves DNA gyrase protection by a protein from the pentapeptide repeat family called Qnr8. Qnr determinant confers resistance to nalidixic acid and increases the MICs of fluoroquinolones by 4-8 folds and supplements the resistance with other mechanisms8. Recent studies stress the importance of the PMQR among different pathogenic bacteria of clinical origin9,10. The qnr gene alleles were also detected in bacterial strains that express extended spectrum β-lactamase (ESBL). In addition, involvement of many efflux pump mechanisms and presence of a novel gene, aac(6’)-Ib-cr encoding a fluoroquinolne-modifying enzyme (aminoglycoside acetyltransferase) have been detected in many enteric pathogens11.
Mobile genetic elements such as integrons have been identified in many multidrug resistant bacteria that play an important role in the acquisition and dissemination of several antibiotic resistance genes12. To date, at least eight classes of integrons have been described and each class is distinguished by differences in the sequences of the integrase gene. Majority of integrons identified among clinical isolates belongs to class 1 type13. Class 2 integrons exists in the transposon Tn7 or its derivatives, and the 3’-CS contains five tns genes involved in the movement of the transposon14. More than 60 different antibiotic-resistance genes, covering most antimicrobials presently in use, have been characterized in cassette structures. In all the reported cases, the cassettes have been inserted in the same orientation and are usually transcribed from a promoter in the 5’-CS15.
In this study, 17 ETEC strains isolated from a diarrhoeal outbreak in Ahmedabad during 2000, were analysed for mutation in the QRDR of gyrA and parC genes, resistance gene cassettes in the integrons, fluoroquinolone efflux and other genes mediating resistance to different antimicrobials.
Material & Methods
Bacterial strains: Seventeen E. coli isolates obtained from patients from a diarrhoeal outbreak during 2000 in Ahmedabad, Gujarat, were included in this study. The detailed description of the outbreak along with the phenotypic and genetic features of the ETEC strains were published elsewhere2. E. coli (J53), Klebsiella pneumonia (NK835), Morgonella morgonii (500914) and Shigella flexneri 3a (IDH6663), harbouring qnr gene alleles A, B, D and S, respectively were used as positive control in the PCR assay. The amplicons of these strains were confirmed by DNA sequencing. Vibrio cholerae O1 (SK-10) and E. coli strain carrying R483::Tn7 were used in the preparation of DNA probes that were the positive controls for the detection of intI1 and intI2, respectively. E. coli strain DH5α or C600 was used as negative control in all the PCR assays. The ATCC strains E. coli 25922 and Staphylococcus aureus 25923 were used as quality control in the antimicrobial susceptibility assay.
Antimicrobial susceptibility test: The susceptibility of ETEC strains to different antimicrobials was performed by disk diffusion method using commercially available discs (Becton, Dickinson & Co, Sparks, MD, USA). Disc containing ampicillin (A, 10 μg), chloramphenicol (Ch, 30 μg), co-trimoxazole (Co, 25 μg), gentamicin (G, 10 μg), neomycin (Ne, 30 μg), tetracycline (T, 30 μg), streptomycin (S, 10 μg), nalidixic acid (Na, 30 μg), cephalothin (Ce, 30 μg), amikacin (Am, 30 μg), ceftazidime (Cf 10 μg), kanamycin (K, 30 μg), ceftriaxone (Ci, 30 μg), ciprofloxacin (Cx, 5 μg), norfloxacin (Nx, 10 μg), cefotaxime (Ct, 30 μg) and ceftazidime (Tz, 30 μg) were used. Susceptibility patterns were based on the recommendations of the Clinical Laboratory Standards Institute16.
The minimal inhibitory concentrations (MICs) of nalidixic acid, ciprofloxacin, norfloxacin, cefotaxime and ceftazidime were determined by E-test (AB Biodisk, Solna, Sweden) or agar dilution technique17 using Mueller-Hinton agar (Difco, USA) supplemented with appropriate concentration of antimicrobials (Nacalai Tesque, Kyoto, Japan). All the ETEC strains were also tested for ESBL-production by modified double-disk synergy test, phenotypic confirmatory disk diffusion test and E-test with Ct and Tz strips.
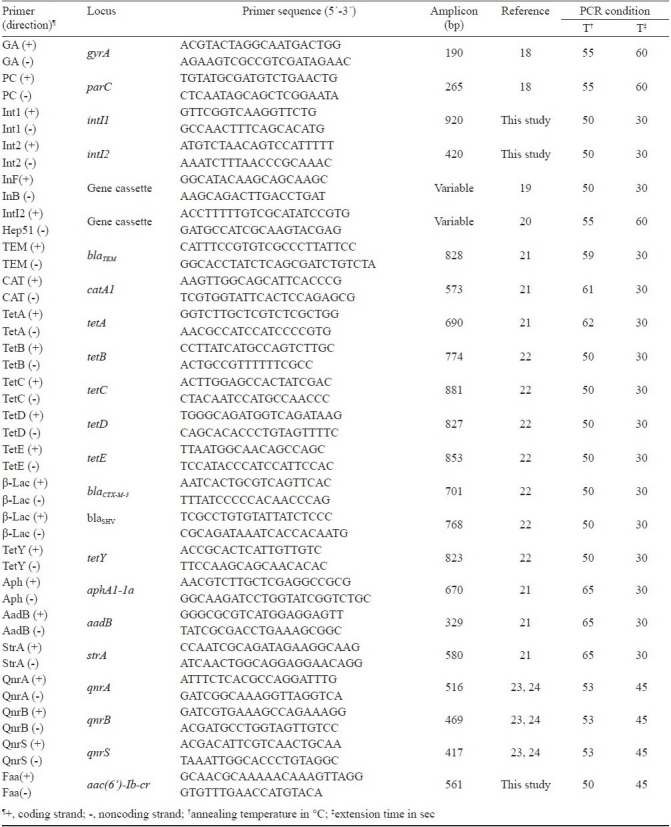
Polymerase chain reaction (PCR) assay: DNA templates were prepared from 200 μl of overnight culture of the test strains in Luria Bretani broth (LB, Difco, USA) by centrifugation using mircofuge tubes and resuspending the bacterial pellet to the initial volume with distilled water. The crude template DNA was prepared by boiling for 10 min and directly used in the PCR assay. The reactions were performed in a GeneAmp PCR system 9700 (Applied Biosystems, USA). The target genes, primer sequences, PCR conditions and amplified product sizes are given in Table I. The PCR products were analyzed after electrophoresis and staining with ethidium bromide.
Table 1.
List of PCR primer pairs used in this study
Nucleotide sequencing: The PCR products were purified either directly or using gel extraction kits (Qiagen, Hilden, Germany). Sequencing of both the DNA strands was performed with the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems) according to the manufacturers’ instructions. The DNA fragments were sequenced and analyzed using an automated sequencer (ABI PRISM 310 DNA, Applied Biosystems). The nucleotide and deduced protein sequences were analyzed with DNASIS (Hitachi, Yokohama, Japan) and DNASTAR (DNA Star Inc. Madison, USA) software. Nucleotide sequences were analyzed by searching the GenBank database of the National Center for Biotechnology Information via the BLAST network service (www.ncbi.nlm.nih.gov/blast).
Fluoroquinolone accumulation assay: E. coli cells were grown to mid-log phase in LB (OD600 = 0.4), harvested, and suspended in 0.2 M morpholinepropanesulphonic acid-Tris buffer (pH 7.0) to an optical density at 600 nm of 20 per ml. Cells were energized with 0.2 per cent glucose for 20 min. Fluoroquinolones were added at a concentration of 10 μg/ml. Aliquots of this mixture was harvested and suspended in 1 ml of 100 mM glycine-HCl (pH 3.0) and shaken for 1 h at room temperature. The amount of released fluoroquinolone was determined spectroflurometrically with excitation at 277 nm and emission at 448 nm. Experiments were done in triplicate and included repeated measurements after carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to the assay mixture as an inhibitor of the proton-motive force at a final concentration of 100 μM.
DNA hybridization analysis: Locations of intI1 and intI2 were identified by colony hybridization assay using Hybond+ nylon membranes (Amersham Biosciences Corp, Bucks, UK). DNA probes for intI1, intI2 and aac(6’)-1b-cr genes were prepared by PCR using primers described in Table I with V. cholerae (SK10), R483::Tn7 plasmid and ETEC strain AV195 as templates, respectively. The probe DNA (integron) was labelled by digoxigenin (DIG) labelling and detection kit (Roche Diagnostic GmbH, Manheim, Germany) and aac(6’)-1b-cr probe was labelled with alkaline phosphatase (GE Healthcare, Buckinghamshire. UK). The identities of the amplified products were confirmed by PCR direct sequencing. Plasmid and genomic DNA of the integron positive isolates were extracted following standard procedures. Genomic DNA was digested with EcoRI and EcoRV enzymes (Takara Suzo Co Ltd, Otsu, Japan). The plasmid and the digested genomic DNA were subjected to 0.7 per cent agarose gel electrophoresis and then blotted on a nylon membrane. Southern hybridization was performed to determine the location of intI1, intI2 and aac(6’)-1b-cr using the respective probes, under high-stringency conditions.
Nucleotide sequence accession numbers: The nucleotide sequence data reported here have been assigned the following GenBank accession numbers: DQ116793 to DQ116794 for gyrA, DQ116795 to DQ116798 for parC AB188258 to AB188268; AB188271 to AB188272 for resistance gene cassettes, and EF501991 to EF501992 for acc(6’)-Ib-cr.
Results
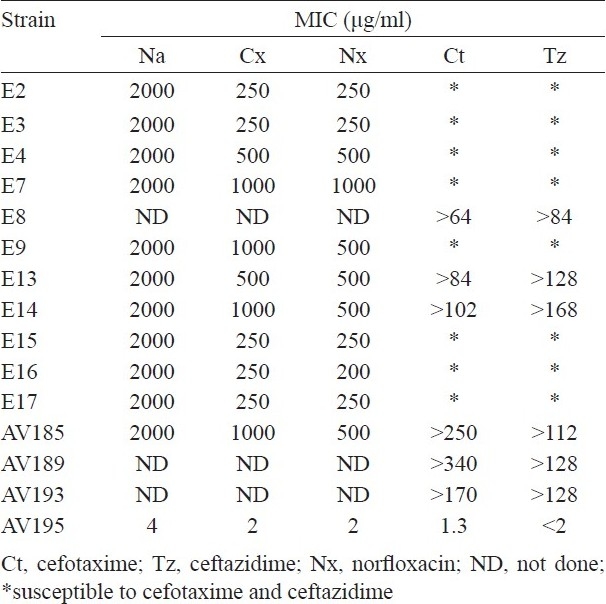
The antimicrobial susceptibility results showed that the ETEC strains were resistant to ampicillin (100%), cephalothin, tetracycline, co-trimoxazole, nalidixic acid, norfloxacin, ciprofloxacin (94% each), chloramphenicol (59%), streptomycin, kanamycin (52% each). In addition, most of the ETEC were also resistance to other antibiotics (Table II). The multidrug resistant ETEC were not associated with a particular serogroup but overall the serogroup O1 was comparatively high (53%). The MICs for nalidixic acid resistance was high (2000 μg/ml) and for fluoroquinolones, the MIC ranged between 250 and 1000 μg/ml. In six ETEC strains, the MIC for Ct and Tz were ranged from >64-340 and >84-168 μg/ml, respectively (Table III).
Table II.
Resistance profile, integrons and drug resistance gene cassettes in ETEC strains
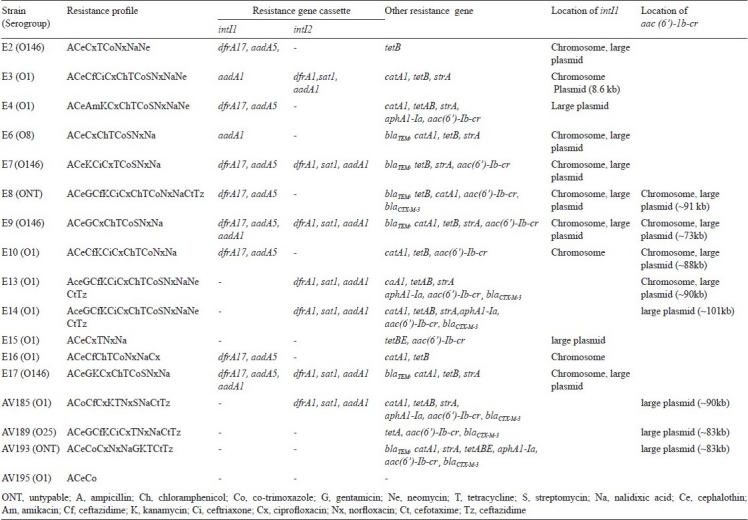
Table III.
MICs of fluroquinolone and ESBLs of ETEC strains
DNA sequencing of the 190-bp PCR product covering the QRDR of gyrA demonstrated presence of mutations at codons 83 in all the quinolone and fluoroquinolone resistant strains compared to the sensitive strain AV195 (Table IV). Mutation at codon 83 was a C→T transversion in the codon TCG that resulted substitution of leucine for serine. Except for one strain (E17), the second mutation was noted in all the strains at position 87 (G→A transversion of codon GAC), which resulted in an asparagine substitution for an aspartate. A 265-bp PCR amplicon of parC was also analyzed in this study. Majority of the quinolone resistant strains had a mutation at codon 80 (G→T translation of codon AGC), resulting substitution of isoleucine for serine. A second mutation at position 84 (E→G) in the parC was detected in E13 and AV185, in which glutamic acid was replaced by glycine. In six ETEC strains, mutation was detected at position 108 (A→V)), where alanine was substituted by valine. In three strains (E2, E14, and AV185) the mutation at position 108 was absent. Strain AV185 had a third mutation at position 90 (A→V), where alanine was replaced by valine. Interestingly, the strain AV195 that is susceptible for nalidixic acid (MIC 4μg/ml) exhibited low-levels of resistance to fluoroquinolones. Since none of the fluoroquinolone resistance mechanisms were identified in this strain, we assume that it may harbour a novel PMQR and/or other hitherto unknown functional gene(s).
Table IV.
Amino acid substitutions in the QRDRs and fluroquinolone efflux of ETEC strains
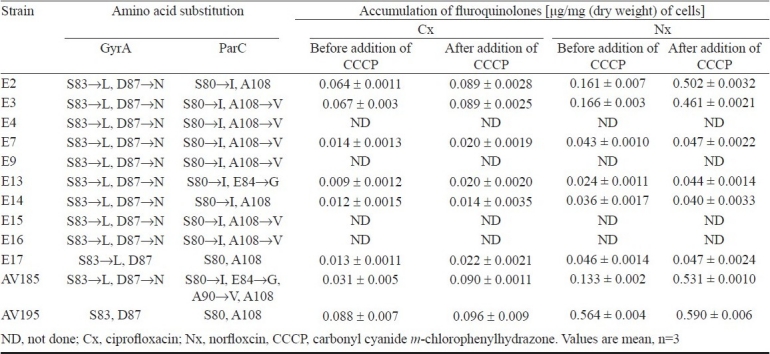
None of the ETEC strains were positive in the Qnr-PCR assay, indicating the absence of all the tested qnr alleles. In addition to the mutations in QRDR, efflux pumps play an important role in intrinsic resistance of E. coli to fluoroquinolones. The fluoroquinolone accumulation kinetics before and after the addition of CCCP was almost similar in most of the resistant strains (Table IV) indicating the involvement of other resistance mechanism(s). Most of the ETEC strains (64.7%) harboured the newly described aac(6’)-Ib-cr gene, which encodes the fluoroquinolone-modifying enzyme aminoglycoside acetyltransferase (Table II). In majority of ETEC strains the acc(6’)-Ib-cr gene was located in large plasmids of varying sizes and seven ETEC strains had this gene in the chromosome (Table II). All the six ETEC strains, which were resistant to Ct and Tz harboured blaCTX-M-3 (Table III).
In the colony hybridization assay, class 1 and class 2 integrons were detected in 35.3 and 17.6 per cent of the ETEC strains, respectively. Four strains (23.5%) had both the classes of integrons (Table II). ETEC strains carrying the integrons were further screened for the presence of contiguous resistance gene cassettes, using specific primers inF/inB and intl2/Hep51, respectively. Of the 10- intI1 probe positive strains, six carried two cassettes and the resistance genes detected in these strains were dfrA17 and aadA5, which confer resistance to trimethoprim and spectinomycin/streptomycin using dihydrofolatereductase and adenylytransferase, respectively.
In addition to these resistance gene cassettes, two strains (E9 and E17) gave an additional amplicon with a size of 1009 bp, which was identified as aadA1, conferring resistance for aminoglycosides. E3 and E6 strains carried 1009 and about 800 bp with complete and incomplete aadA1 gene cassette, respectively. In one intI1 probe positive strain (E15), the CS specific primers did not gave amplicon. In seven ETEC strains, intl2 was detected, of which four were also positive for intI1. All the intI2 positive strains carried three cassettes (2449 bp) as those found in Tn7, namely dfrA1, sat1, and aadA1 (Table II). The sat gene encodes for streptothricin acetyltransferase.
The plasmid patterns and the location of the intI1 gene in the plasmids were not uniform (Table II). The restriction enzymes, EcoRI and EcoRV were used for digestion of chromosomal DNA, since the intI1 did not have the corresponding restriction sites for these enzymes. Southern hybridization results showed that class 1 integron was detected both in the chromosome and large plasmid in most of the strains (Table II), however, class 2 integron was detected only in the chromosome. In E15, intI1 was detected in the large plasmid, however, this strain did not carry any resistance gene cassette.
The alleles of genes such as tet, catA1, strA, blaTEM, and aphA1-Ia encoding for resistance for tetracycline, chloramphenicol, streptomycin, ampicillin, and kanamycin, were detected in 94.1, 70.6, 58.8, 35.2, 29.4 per cent of the ETEC strains, respectively. An analysis on the presence of different resistance gene combinations showed that ETEC harbouring blaTEM gene was detected with catA1 gene in 5 out of 6 strains and the strains harbouring aphA1-Ia were also positive for tetAB or tetABE. In one streptomycin resistant strain (E2) strA gene was negative but carried aadA5, which was associated with the class 1 integron structure. The four strains, E2, E8, E10 and E16 were susceptible to streptomycin and did not harbour aadA1 or strA. In 6 gentamicin resistant strains, aadB, which confer gentamicin resistance, was not identified.
Among tet gene alleles, tetB was detected in 52.9 per cent of the ETEC strains. Combination of tetAB was detected among four strains, and tetBE, tetABE, tetA was detected in one strain each (Table II). The other tested tet gene types such as tetC, tetD and tetY were not detected in any of the ETEC strains (data not shown). When tested for the MIC of tetracycline by agar dilution technique with strains harbouring different tet gene classes, tetA or tetB alone had the MIC of 100 μg/ml. ETEC strains with combination of tetBE and tetABE had the MIC of 150 μg/ml (data not shown).
Discussion
All the ETEC strains isolated from the patients with diarrhoea during the outbreak were resistant to several antibiotics. The differences found in their XbaI PFGE patterns indicate that most of the ETEC strains in this study are distinct clones2. The unusual multidrug resistance profiles of the ETEC strains prompted us to undertake an in depth study on mechanisms of antimicrobial resistance using the molecular tools.
Fluoroquinolones have been used for the treatment of variety of infectious diseases. In the present study, most of the ETEC were highly resistant to fluoroquinolones. Prior to 1990, resistance of E. coli to fluoroquinolones was rare25. As reported with other pathogens, use of fluoroquinolones in India may have progressively favoured emergence of MDR-ETEC strains4–6. Majority of the ETEC investigated in the present study were resistant to several other antibiotics as well. It has been demonstrated that fluoroquinolone resistance was related with resistance to at least one non-fluoroquinolone antibiotic26. In this study, the fluoroquinolone resistant ETEC strains had mutations in gyrA at positions 83 and 87. The gyrA mutations that frequently affect residue serine at 83 and aspartate at 87 are common among fluoroquinolone resistant E. coli27. In addition to the mutations in gyrA, mutations were detected in the parC. For the expression of high-level resistance, acquisition of a second gyrA mutation and a parC mutation seems important. A similar resistance mechanism has been observed in E. coli strains28. With the sequenced regions of QRDR in this study, association could not be made with the increased MIC of quinolone/fluoroquinolones and amino acid substitutions in topoisomerases. Although the quinolone resistance is due to mutations in gyrA and parC, studies indicate that resistance can also be transferred on plasmids carrying the qnr gene alleles7,8.
Although E. coli has shown to have intrinsic proton-dependent multidrug-resistant efflux pump systems, the specific activity of any one pump in its natural state and level has not been established29. CCCP is a well-known inhibitor for fluoroquinolone efflux pump in many bacteria. After the addition of this proton motive force uncoupler, the expected accumulation did not occur in most of the fluoroquinolone resistant ETEC. In view of the large number of proven or putative efflux pumps30, involvement of more than one mechanism seems possible. We assume that additional non-efflux resistance mechanism(s) may also contribute to the high level fluoroquinolone resistance.
Class 1 and class 2 integrons are widely prevalent in most of the clinical strains of Gram-negative bacteria. Despite the differences in the integrase proteins, the two classes of integrons can include identical gene cassettes31. Our finding has shown that the dfr and aad gene alleles encoding dihydrofolate reductases and adenyltransferase, respectively were present in both the classes of integrons among ETEC. In this study, dfr cassettes (dfrA1 and dfrA17) that confer resistance to trimethoprim were detected in majority of the ETEC strains. In 50 per cent of the intl1 positive ETEC, dfr17 was detected along with aadA5 cassette. The dfr and aadA cassettes were shown to be common among members of the family Enterobacteriaceae32. The dfr was detected behind the 5’-conserved segment, which is closest to the promoter, thereby providing high-level and conditional resistance. It is likely that selection for cassettes carrying dfr genes has occurred among ETEC as trimethoprim in combination with sulphamethoxazole (co-trimoxazole) is used frequently in the treatment of diarrhoea and other infections. Presence of aadA5 did not have any influence towards resistance of streptomycin, as the aad cassette confers resistance only to low level of streptomycin33. One ETEC strain (E15), which was intI1 probe positive, did not amplify any resistance gene cassette regions, possibly due to the variable nature of a 3’-conserved segment.
In intI2 positive ETEC strains, we have detected dfrA1, sat1 and aadA1 gene cassettes conferring resistance to trimethoprim, streptothricin, and streptomycin/spectinomycin, respectively. Existence of sat1 has not been reported in ETEC. Class 2 integron carrying dfrA1, sat1 and aadA1 was also reported among members of the family Enterobacteriaceae due to the defective integrase gene in class 2 integron20.
Within the integrons, the genes responsible for resistance to beta-lactams, tetracycline, and chloramphenicol were not mapped in this study. The tetB gene that encodes an efflux protein, which confers resistance to tetracycline, was predominant in majority of our ETEC strains. As reported in other findings, combination of tet genes was detected in six ETEC strains22. Except for one strain (AV189), tetA was always detected with tetB in this study. Interestingly, ETEC harbouring combination of tetA, tetB or tetA, tetB, and tetE expressed high MIC for tetracycline than tetB alone or tetB and tetE. None of the six gentamicin resistant strains carried the aadB gene. Possibly, the other variant genes encoding aminoglycoside adenyltransferase may play a role in resistance towards gentamicin. We have tested the ETEC strains for ESBL genes such as blaSHV and blaCTX-M-3genes for blaTEM, which were widely distributed in ETEC22. Due to co-selection, blaTEM was found in association with the catA1 in most of the ETEC strains22. Similarly, aphA1-Ia was detected along with different tet alleles other than tetB. We have detected the aminoglycosides (neomycin and kanamycin) resistance gene (aphA1-Ia) in five strains (29.4%), even though 59 per cent of the ETEC were resistant to kanamycin. This may be due to cross-resistance caused by aminoglycoside resistance genes. In this study, 64.7 per cent of the ETEC strains harboured aac(6’)-Ib-cr, of which, 73 per cent of the strains concomitantly had ESBL genes. The synergistic effect of these genes on increased resistance toward fluoroquinolones has been established in many members of Enterobacteriaceae34.
Even though the antimicrobial therapy is generally supportive for the treatment of ETEC mediated diarrhoea, its rational use is difficult to implement in diarrhoea endemic countries especially during outbreaks. As evidenced from this study, innate gene mutations and acquisition of multidrug resistance genes through mobile genetic elements might have contributed to the emergence of MDR-ETEC. This study reinforces the necessity of utilizing molecular techniques in the epidemiological studies to understand the nature of resistance responsible for antimicrobial resistance in different species of pathogenic bacteria.
Acknowledgments
Authors thank Dr G.A. Jacoby for providing us the E. coli strain J53, A. Forslund and V. Falbo for the Vibrio cholerae and E. coli strains harbouring intI1 and intI2 genes, respectively. This work was supported in part by the Indian Council of Medical Research (Project No. 5/3/3/5/99-ECD-I), Ministry of Education, Culture, Sports, Science and Technology of Japan, Grand-in-aid (17406012) and the National Academy Science, India, Allahabad.
References
- 1.World Health Organization. New frontiers in the development of vaccines against enterotoxigenic (ETEC) and enterohaemorrhagic (EHEC) E. coli infections. Part I. Wkly Epidemiol Res. 1999;74:98–101. [PubMed] [Google Scholar]
- 2.Chakraborty S, Deokule JS, Garg P, Bhattacharya SK, Nandy RK, Nair GB, et al. Concomitant infection of enterotoxigenic Escherichia coli in an outbreak of cholera caused by Vibrio cholerae O1 and O139 in Ahmedabad, India. J Clin Microbiol. 2001;39:3241–6. doi: 10.1128/JCM.39.9.3241-3246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranwal S, Dey K, Ramamurthy T, Nair GB, Kundu M. Role of active efflux with in association with target gene mutations in fluoroquinolone resistance in clinical isolates of Vibrio cholerae. Antimicrob Agents Chemother. 2002;46:2676–8. doi: 10.1128/AAC.46.8.2676-2678.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg P, Sinha S, Chakraborty R, Bhattacharya SK, Nair GB, Ramamurthy T, et al. Emergence of fluoroquinolone-resistance strains of Vibrio cholerae O1 biotype El Tor among hospitalized patients with cholera in Calcutta, India. Antimicrob Agents Chemother. 2001;45:1605–6. doi: 10.1128/AAC.45.5.1605-1606.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazhani GP, Sarkar B, Ramamurthy T, Bhattacharya SK, Takeda Y, Niyogi SK. Clonal multidrug-resistance Shigella dysenteriae type 1 strains associated with epidemic and sporadic dysenteries in Eastern India. Antimicrob Agents Chemother. 2004;48:681–4. doi: 10.1128/AAC.48.2.681-684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha S, Chattopadhyay S, Bhattacharya SK, Nair GB, Ramamurthy T. An unusually high level of quinolone resistance associated with type II topoisomerase mutations in quinolone resistance-determining regions of Aeromonas caviae isolated from diarrhoeal patients. Res Microbiol. 2004;55:827–9. doi: 10.1016/j.resmic.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Hooper DC. Mechanisms of fluroquinolone resistance. Drug Resist Update. 1999;2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Martinez L, Pascual A, Garcia I, Tran J, Jacoby GA. Interaction of plasmid and host quinolone resistance. J Antimicrob Chemother. 2003;51:1037–9. doi: 10.1093/jac/dkg157. [DOI] [PubMed] [Google Scholar]
- 9.Mendez Arancibia E, Pitart C, Ruiz J, Marco F, Gascón J, Vila J. Evolution of antimicrobial resistance in enteroaggregative Escherichia coli and enterotoxigenic Escherichia coli causing traveller's diarrhoea. J Antimicrob Chemother. 2009;64:343–7. doi: 10.1093/jac/dkp178. [DOI] [PubMed] [Google Scholar]
- 10.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22:664–89. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machado E, Coque TM, Canton R, Baquero F, Sousa JC, Peixe L. Dissemination in Portugal of CTX-M-15, OXA-1, and TEM-1-producing Enterobacteriaceae strains containing the aac(6’)-Ib-cr gene, which encodes aminoglycoside and fluoroquinolone-modifying enzyme. Antimicrob Agents Chemother. 2006;50:3220–1. doi: 10.1128/AAC.00473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall RM. Mobile gene cassettes and integrons moving antibiotic resistance genes in gram-negative bacteria. Ciba Found Symp. 1997;207:192–202. doi: 10.1002/9780470515358.ch12. [DOI] [PubMed] [Google Scholar]
- 13.Sallen B, Rajoharison A, Desvarenne S, Mabilat C. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of Enterobacteriaceae. Microb Drug Resist. 1995;1:195–202. doi: 10.1089/mdr.1995.1.195. [DOI] [PubMed] [Google Scholar]
- 14.Hall RM, Brookes DE, Stokes HW. Site-specific insertion of genes into integrons: role of the 59-bp element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–59. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 15.Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–83. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 16.Performance standards for antimicrobial disk susceptibility tests. 9th ed. Wayne, PA: CLSI; 2006. Clinical Laboratory Standards Institute. Approved standard CLSI document M2-A9. [Google Scholar]
- 17.Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 7th ed. Wayne, PA: CLSI; 2006. Clinical Laboratory Standards Institute. Approved standard CLSI document M7-A7. [Google Scholar]
- 18.Everett MJ, Jin YF, Ricci V, Piddock LJ. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from human and animals. Antimicrob Agents Chemother. 1996;40:2380–6. doi: 10.1128/aac.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Shi L, Li L, Guo S, Zhang X, Yamasaki S, et al. Identification and characterization of class 1 integron resistance gene cassettes among Salmonella strains isolated from healthy humans in China. Microbiol Immunol. 2004;48:639–45. doi: 10.1111/j.1348-0421.2004.tb03473.x. [DOI] [PubMed] [Google Scholar]
- 20.White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the Enterobacteriaceae. Antimicro Agents Chemother. 2001;45:2658–61. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidhof H, Guerra B, Abbas S, Elsheikha HM, Whittam TS, Beutin L. A multiresistance clone of Shiga toxin-producing Escherichia coli O118:[H16] is spread in cattle and humans over different European countries. Appl Environ Microbiol. 2002;68:5834–42. doi: 10.1128/AEM.68.12.5834-5842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maynard C, Fairbrother JM, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, et al. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob Agents Chemother. 2003;47:3214–21. doi: 10.1128/AAC.47.10.3214-3221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother. 2003;47:2242–8. doi: 10.1128/AAC.47.7.2242-2248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicro Agents Chemother. 2006;50:2872–4. doi: 10.1128/AAC.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kresken M, Jansen A, Wiedemann B. Prevalence of resistance of aerobic gram-negative bacilli to broad-spectrum antibacterial agents: results of a multicentre study. J Antimicrob Chemother. 1990;25:1022–4. doi: 10.1093/jac/25.6.1022. [DOI] [PubMed] [Google Scholar]
- 26.Sahm DF, Critchley IA, Kelly LJ, Karlowsky JA, Mayfield DC, Thornsberry C, et al. Evaluation of current activities of fluoroquinolones against gram-negative bacilli using centralized in vitro testing and electronic surveillance. Antimicrob Agents Chemother. 2001;45:267–74. doi: 10.1128/AAC.45.1.267-274.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vila J, Vargas M, Ruiz J, Corachan M, De Anta MTJ, Gascon J. Quinolone resistance in enterotoxigenic Escherichia coli causing diarrhea in travelers to India in comparison with other geographical areas. Antimicrob Agents Chemother. 2000;44:1731–3. doi: 10.1128/aac.44.6.1731-1733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–85. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viveiros M, Jesus A, Brito M, Leandro C, Martins M, Ordway D, et al. Inducement and reversal of tetracycline resistance in Escherichia coli K-12 and expression of proton gradient-dependent multidrug efflux pump genes. Antimicrob Agents Chemother. 2005;49:3578–82. doi: 10.1128/AAC.49.8.3578-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saier MH, Jr, Paulsen IT, Sliwinski MK, Pao SS, Skurray RA, Nikaido H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–74. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 31.Hall RM, Brown HJ, Brookes DE, Stokes HW. Integrons found in different locations have identical 5’ ends but variable 3’ ends. J Bacteriol. 1994;176:6286–94. doi: 10.1128/jb.176.20.6286-6294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazel D, Dychinco B, Webb VA, Davies J. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob Agents Chemother. 2000;44:1568–74. doi: 10.1128/aac.44.6.1568-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collis CM, Hall RM. Expression of resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–62. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin SY, Kwon KC, Park JW, Song JH, Ko YH, Sung JY, et al. Characteristics of aac(6’)-Ib-cr gene in extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from Chungnam area. Korean J Lab Med. 2009;29:541–50. doi: 10.3343/kjlm.2009.29.6.541. [DOI] [PubMed] [Google Scholar]


