Abstract
Mammalian Sun1 belongs to an evolutionarily conserved family of inner nuclear membrane proteins, which are known as SUN domain proteins. SUN domain proteins interact with KASH domain partners to form bridging complexes, so-called LINC complexes, that physically connect the nuclear interior to the cytoskeleton. LINC complexes are critical for nuclear integrity and play fundamental roles in nuclear positioning, shaping and movement. The mammalian genome codes for at least five different SUN domain proteins used for the formation of a number of different LINC complexes. Recently, we reported on the identification of several Sun1 isoforms, which tremendously enlarges the alternatives to form functional LINC complexes. We now confirmed that Sun1 actually exists in at least seven distinct splice variants. Besides that, we observed that expression of individual Sun1 isoforms remarkably depends on the cell type, suggesting a cell type-specific adaption of Sun1 dependent LINC complexes to specific cellular and physiological requirements.
Key words: Sun1, SUN domain protein, LINC complex, mouse, nuclear envelope, isoform
LINC (linker of nucleoskeleton and cytoskeleton) complexes are highly conserved nuclear envelope spanning protein assemblies formed by SUN (Sad1p/Unc84 homology) and KASH (Klarsicht/Anc1/Syne1 homology) domain proteins that interact with each other within the perinuclear space.1–5 SUN domain proteins as inner nuclear membrane (INM) components of LINC complexes provide a link to nucleoplasmic structures (i.e., the lamina), while KASH proteins, the outer nuclear membrane (ONM) partners, connect to the cytoskeleton. That way, LINC complexes form a solid scaffold for integrating nuclei into the cellular environment.5,6 Besides just functioning passively in nuclear positioning and anchorage, LINC complexes were shown to play major roles in active processes like movement of chromosomes and the entire nucleus. Furthermore, recent studies indicated that they have a central function in directed nuclear shaping and deformation as well.7–10
In a recent study, we reported on the identification of two novel LINC complexes and suggested their involvement in a quite exceptional dynamic cellular process, the shaping of the mammalian sperm head.10 In that particular study we have analyzed different LINC components and their behavior during sperm differentiation. An important outcome of our assays was that within the given cellular context two SUN domain proteins, Sun3 and Sun1, distinguish between KASH partners to form discrete LINC complexes which could be assigned for distinct tasks. Like germ cells, mammalian somatic cells contain different SUN and KASH domain proteins as well. They express at least two SUN domain proteins, i.e., Sun1 and Sun2, and up to 4 different KASH domain partners (known as nesprins 1, 2, 3 and 4).11–14 This variability allows for the assembly of diverse LINC complexes that connect nuclear structures to different cytoskeletal elements (for recent overview see Starr and Fridolfson 2010).6 Interestingly, in our previous study we could demonstrate that the Sun1 gene itself encodes not only one single transcript, but at least seven distinguishable isoforms.10 This matter is quite remarkable, as it tremendously enlarges the alternatives to form functional LINC complexes, which in turn could be considered for LINC adaption to cell type-specific physiological requirements.
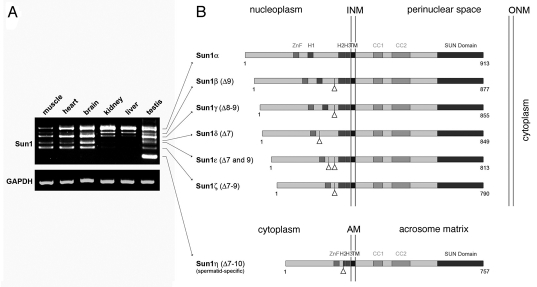
To verify these initial experiments, we now repeated the RT-PCR experiments for Sun1 using selected tissues obtained form adult male mice. Consistent with our previous findings,10 we could amplify seven distinguishable Sun1 isoforms (Fig. 1A) that showed striking differences concerning their expression pattern in different tissues. While the shortest isoform, Sun1η, could be detected exclusively in the testis but not in any of the somatic tissues tested, the expression profile of the other six isoforms is not that restricted. Even though all of them are expressed in a variety of different cell types, they actually show distinct peculiarities as well. Besides germ cell specific Sun1η, in testis we could detect all six additional isoforms. Similarly, in the muscle and heart—with exception of Sun1η—expression of six isoforms could be verified. By contrast, kidney apparently expresses not more than four variants and, most striking, under the very same stringent PCR conditions in the liver we could identify only three different isoforms (Fig. 1A). Hence, the few selected examples shown here yet clearly evidence that Sun1 isoforms expression is not equably, rather it appears quite variable between different tissue types. This finding is supported by our previous results that disclosed comparable variations in a number of additional tissues.10 Interestingly, a more detailed comparison of the isoform expression profile between the different tissue types revealed a general tendency. The smaller isoforms appear to be restricted to selected tissues and are expressed in the testis, the brain and, albeit to a lesser extent, in muscle and heart tissues, but not in the other tissues tested. By contrast, larger splice forms turned out to be ubiquitously expressed and are the dominant products in the kidney and the liver. Taken together, our data presented here validated that the murine Sun1 gene codes for at least seven distinct isoforms (see also below). Furthermore, our detailed analysis disclosed quite overt differences regarding their individual expression patterns.
Figure 1.
Differential expression of murine Sun1 isoforms. (A) Presence of different Sun1 isoforms in selected mouse tissues was analyzed by RT-PCR. To amplify the recently described N-terminal Sun1 variants with deletions between exon 7 and 10, primers were selected which specifically annealed in exon 6 and exon 11 (forward: 5′-CAG CAA TGG ATA CAC TT G CCG TG-3′; reverse: 5′-CCA GAA GGT TCC CGA GGC TG-3′; annealing at 60°C; 30 cycles). Amplification of GAPDH served as control for RN A fidelity (forward: 5′-GGG CCC ACT TGA AGG GTG GAG C-3′; reverse: 5′-GGC ACC ATA AAG AAT GTT CTA TTT CCT TGG ATC C-3′; annealing at 58°C; 25 cycles). (B) Schematic illustration of the mouse Sun1 isoforms as yet identified. Triangles mark positions of deleted exons. INM , inner nuclear membrane; ONM outer nuclear membrane; AM acrosomal membrane; ZnF, zinc finger; H1, H2, H3, hydrophobic domains; TM , transmembranen domain; CC, coiled coil domain.
To uncover molecular differences of the single isoforms we sequenced each of them. We found that the identified isoforms are distinguished by variable deletions between exon seven and ten, thus affecting the N-terminal part of the protein. Notably, RT-PCR experiments aimed for detecting putative splice forms concerning the C-terminal protein parts revealed no overt splice variations within this region. Consistent with this, amplification of entire Sun1 cDNAs resulted in seven transcripts with sizes that are fully in line with the expected seven isoforms (not shown). As the longest isoform we could identify the initially described Sun1 (reviewed in ref. 4), which to our yet introduced nomenclature will be referred to as Sun1α. Sun1α is a typical type II transmembrane protein with the N-terminal region localizing to the nucleus and the C-terminus extending into the NE lumen (Fig. 1B).4,15 Within its nucleoplasmic domain Sun1α contains characteristic features that are a zinc finger motif as well as three hydrophobic domains. Remarkably, due to alternative splicing the other isoforms—termed according to their size Sun1β to Sun1η—lack nucleoplasmic regions between the zinc finger motif and the TM domain (Fig. 1B). While in two of the N-terminally shortened variants, i.e., Sun1β and γ, all three hydrophobic domains are retained, Sun1δ, ε and ζ are characterized by the lack of hydrophobic region H1. Most prominent, however, is Sun1η (GenBank accession number: HQ402597) as in this splice variant exons seven to ten are removed, leading to complete loss of H1 and part of H2 (Fig. 1B).
What might be the significance of such a variability within the N-terminal region of Sun1? Interestingly, in a previous study it was demonstrated that the hydrophobic regions located within the nucleoplasmic part of Sun1 (i.e., Sun1α) are crucial for effective INM targeting and membrane retention and, moreover, deletions of nucleoplasmic hydrophobic motifs severely affect the dynamic properties of the molecule.15 Thus, it appears quite conceivable that the natural deletions as determined in Sun1δ, ε, ζ and η provoke slightly different properties for the respective isoforms. Consistent with this, in our recent study we found that testis-specific Sun1η, which represents the shortest isoform and lacks nucleoplasmic domains including H1 and part of H2 (Fig. 1B), is not part of the nuclear envelope of spermatids, but instead localizes to the acrosomal membrane system.10 Exon deletions as found in the shortened Sun1 isoforms, however, do not only affect hydrophobic regions but also significant parts between them (Fig. 1B). Since via the nucleoplasmic region SUN domain proteins interact with nuclear components such as lamins and/or chromatin,4,8,16 lack of large parts of the N-terminus as evident in the small isoforms could impede binding of selected nuclear partners, hence leading to modified nucleocytoskeletal linkage. Anyhow, coexpression of different Sun1 isoforms in effect allows for variable assembly of Sun1 dependent LINC complexes that can be assigned for distinct tasks. Accordingly, the observed differences in Sun1 isoform expression between the tissue types may reflect a cell type-specific adaption of nuclear envelope bridging complexes to meet the particular physiological requirements.
Acknowledgments
This study was supported by the German Research Foundation (DFG; grant Al 1090/1-1), the Graduate School GK 1048 of the University of Würzburg and by the funding programme Open Access Publishing of the DFG and the University of Würzburg.
Abbreviations
- INM
inner nuclear membrane
- KASH
Klarsicht/Anc1/Syne1 homology
- LINC
linker of nucleoskeleton and cytoskeleton
- NE
nuclear envelope
- SUN
Sad1p/Unc84 homology
References
- 1.Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 3.Starr DA, Fischer JA. KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- 4.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUNKASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olins AL, Hoang TV, Zwerger M, Herrmann H, Zentgraf H, Noegel AA, et al. The LINC-less granulocyte nucleus. Eur J Cell Biol. 2009;88:203–214. doi: 10.1016/j.ejcb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong H, Rivero F, Euteneuer U, Mondal S, Mana-Capelli S, Larochelle D, et al. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic. 2008;9:708–724. doi: 10.1111/j.1600-0854.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- 9.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, et al. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Göb E, Schmitt J, Benavente R, Alsheimer M. Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS ONE. 2010;5:12072. doi: 10.1371/journal.pone.0012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J Biol Chem. 2000;275:31986–31995. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- 12.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, et al. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 14.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, et al. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci USA. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, et al. Functional association of Sun1 with nuclear pore complexes. J Cell Biol. 2007;178:785–798. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, et al. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina und the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]