Abstract
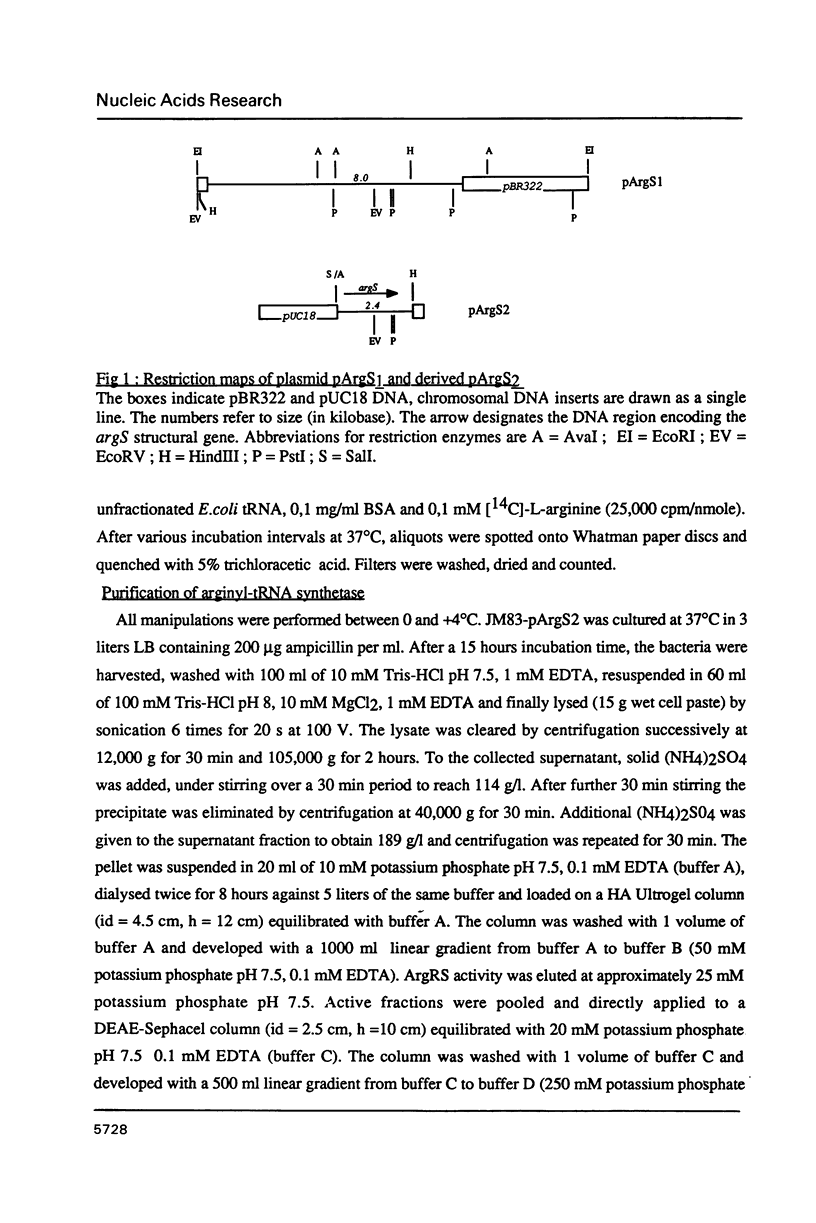
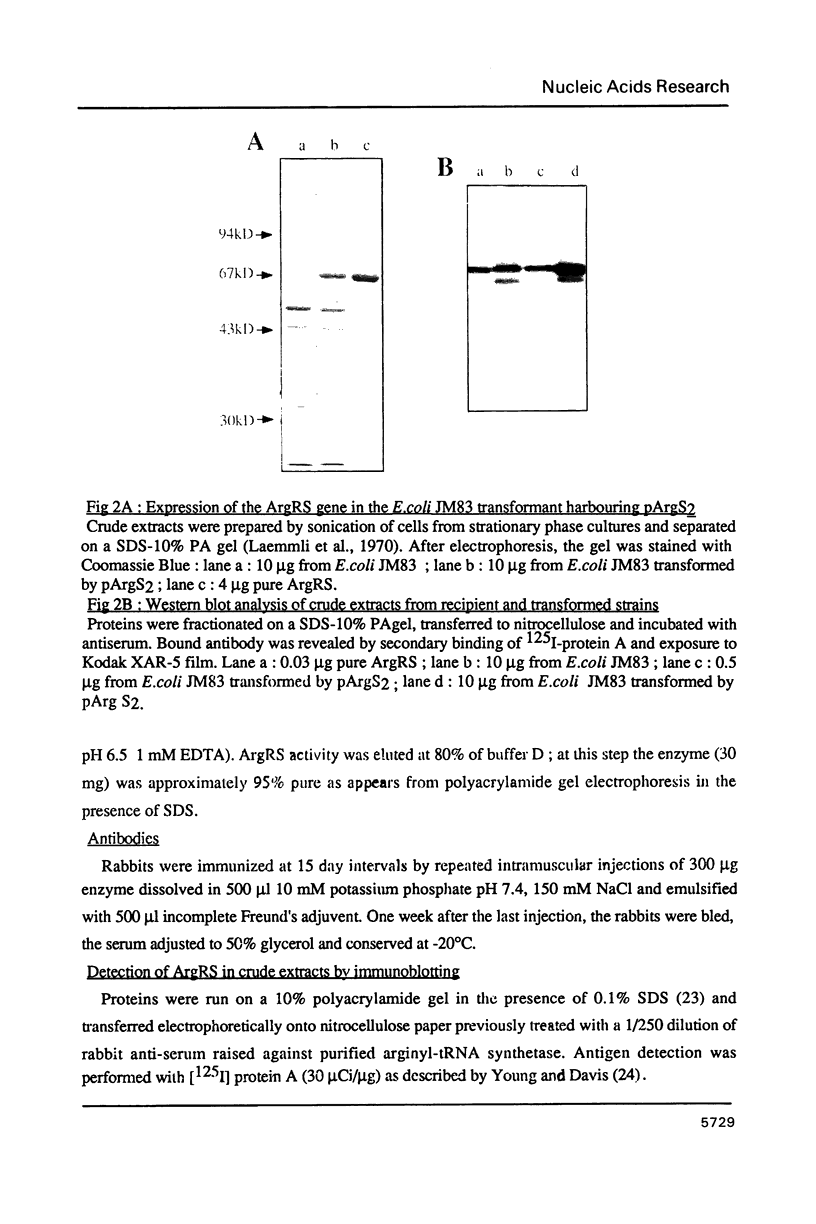
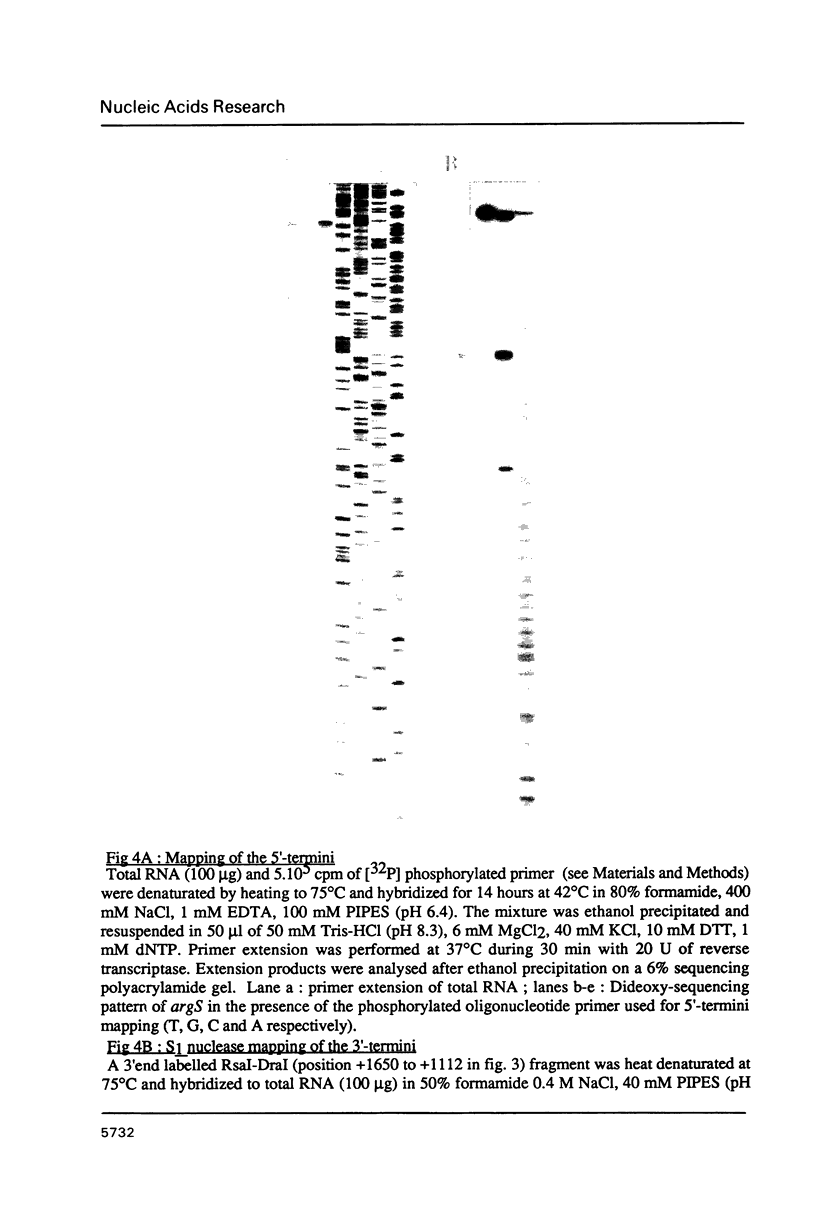
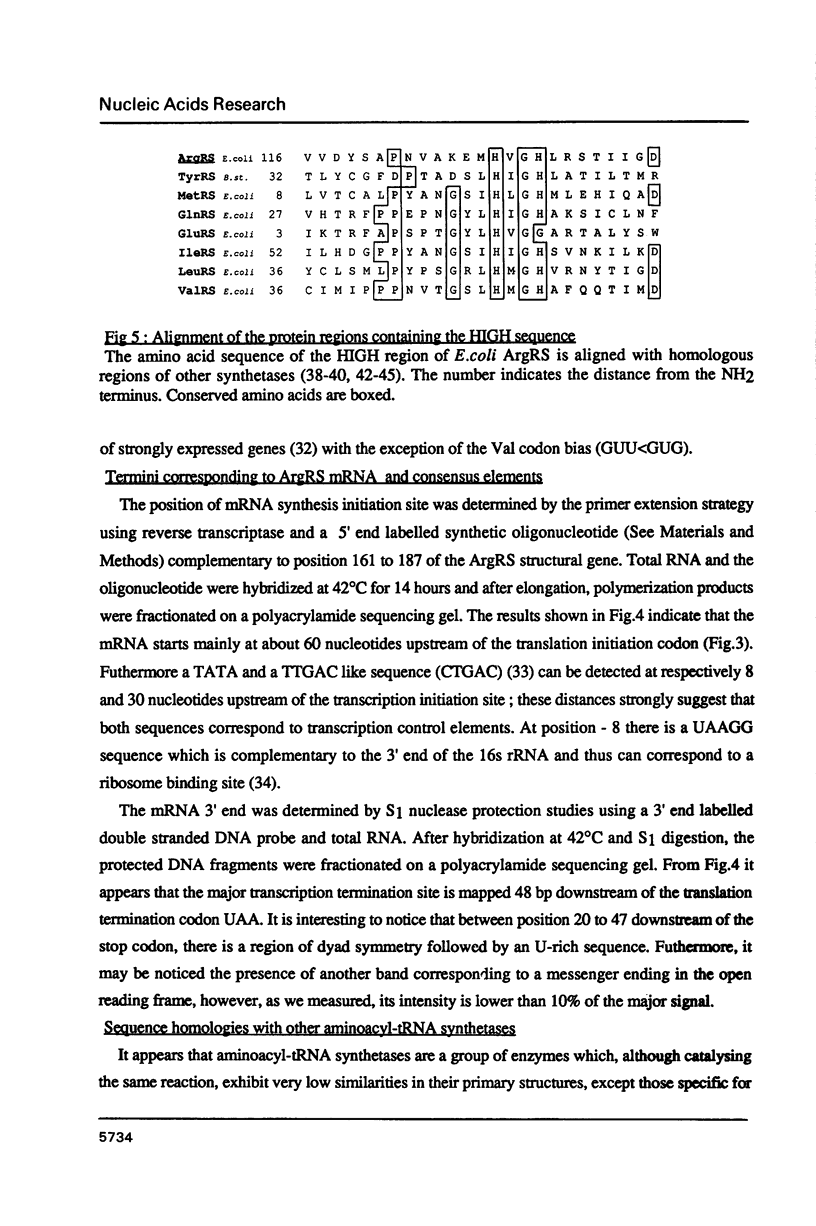
The gene coding for Escherichia coli arginyl-tRNA synthetase (argS) was isolated as a fragment of 2.4 kb after analysis and subcloning of recombinant plasmids from the Clarke and Carbon library. The clone bearing the gene overproduces arginyl-tRNA synthetase by a factor 100. This means that the enzyme represents more than 20% of the cellular total protein content. Sequencing revealed that the fragment contains a unique open reading frame of 1734 bp flanked at its 5' and 3' ends respectively by 247 bp and 397 bp. The length of the corresponding protein (577 aa) is well consistent with earlier Mr determination (about 70 kd). Primer extension analysis of the ArgRS mRNA by reverse transcriptase, located its 5' end respectively at 8 and 30 nucleotides downstream of a TATA and a TTGAC like element (CTGAC) and 60 nucleotides upstream of the unusual translation initiation codon GUG; nuclease S1 analysis located the 3'-end at 48 bp downstream of the translation termination codon. argS has a codon usage pattern typical for highly expressed E. coli genes. With the exception of the presence of a HVGH sequence similar to the HIGH consensus element, ArgRS has no relevant sequence homologies with other aminoacyl-tRNA synthetases.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. G., Ebel J. P., Jakes R., Bruton C. J. Methionyl-tRNA synthetase from Escherichia coli. Primary structure of the active crystallised tryptic fragment. Eur J Biochem. 1982 Oct;127(3):449–457. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Berger S. L., Wallace D. M., Puskas R. S., Eschenfeldt W. H. Reverse transcriptase and its associated ribonuclease H: interplay of two enzyme activities controls the yield of single-stranded complementary deoxyribonucleic acid. Biochemistry. 1983 May 10;22(10):2365–2372. doi: 10.1021/bi00279a010. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bhat T. N., Blow D. M., Brick P., Nyborg J. Tyrosyl-tRNA synthetase forms a mononucleotide-binding fold. J Mol Biol. 1982 Jul 15;158(4):699–709. doi: 10.1016/0022-2836(82)90255-8. [DOI] [PubMed] [Google Scholar]
- Breton R., Sanfaçon H., Papayannopoulos I., Biemann K., Lapointe J. Glutamyl-tRNA synthetase of Escherichia coli. Isolation and primary structure of the gltX gene and homology with other aminoacyl-tRNA synthetases. J Biol Chem. 1986 Aug 15;261(23):10610–10617. [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Cooper P. H., Hirshfield I. N., Maas W. K. Map location of arginyl-tRNA synthetase mutations in Escherichia coli K-12. Mol Gen Genet. 1969 Aug 15;104(4):383–390. doi: 10.1007/BF00334238. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Dietrich A., Giege R., Comarmond M. B., Thierry J. C., Moras D. Crystallographic studies on the aspartyl-tRNA synthetase-tRNAAsp system from yeast. The crystalline aminoacyl-tRNA synthetase. J Mol Biol. 1980 Mar 25;138(1):129–135. doi: 10.1016/s0022-2836(80)80008-8. [DOI] [PubMed] [Google Scholar]
- Ehresmann B., Imbault P., Weil J. H. Spectrophotometric determination of protein concentration in cell extracts containing tRNA's and rRNA's. Anal Biochem. 1973 Aug;54(2):454–463. doi: 10.1016/0003-2697(73)90374-6. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Gangloff J., Dirheimer G. Reaction pathway and rate-determining step in the aminoacylation of tRNAArg catalyzed by the arginyl-tRNA synthetase from yeast. Biochemistry. 1978 Sep 5;17(18):3740–3746. doi: 10.1021/bi00611a011. [DOI] [PubMed] [Google Scholar]
- Gren E. J. Recognition of messenger RNA during translational initiation in Escherichia coli. Biochimie. 1984 Jan;66(1):1–29. doi: 10.1016/0300-9084(84)90188-3. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck J. D., Hatfield G. W. Valyl-tRNA synthetase gene of Escherichia coli K12. Primary structure and homology within a family of aminoacyl-TRNA synthetases. J Biol Chem. 1988 Jan 15;263(2):868–877. [PubMed] [Google Scholar]
- Hirshfield I. N., Bloemers H. P. The biochemical characterization of two mutant arginyl transfer ribonucleic acid synthetases from Escherichia coli K-12. J Biol Chem. 1969 Jun 10;244(11):2911–2916. [PubMed] [Google Scholar]
- Hoben P., Royal N., Cheung A., Yamao F., Biemann K., Söll D. Escherichia coli glutaminyl-tRNA synthetase. II. Characterization of the glnS gene product. J Biol Chem. 1982 Oct 10;257(19):11644–11650. [PubMed] [Google Scholar]
- Hountondji C., Blanquet S., Lederer F. Methionyl-tRNA synthetase from Escherichia coli: primary structure at the binding site for the 3'-end of tRNAfMet. Biochemistry. 1985 Feb 26;24(5):1175–1180. doi: 10.1021/bi00326a018. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Dessen P., Blanquet S. Sequence similarities among the family of aminoacyl-tRNA synthetases. Biochimie. 1986 Sep;68(9):1071–1078. doi: 10.1016/s0300-9084(86)80181-x. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Lederer F., Dessen P., Blanquet S. Escherichia coli tyrosyl- and methionyl-tRNA synthetases display sequence similarity at the binding site for the 3'-end of tRNA. Biochemistry. 1986 Jan 14;25(1):16–21. doi: 10.1021/bi00349a003. [DOI] [PubMed] [Google Scholar]
- Härtlein M., Frank R., Madern D. Nucleotide sequence of Escherichia coli valyl-tRNA synthetase gene valS. Nucleic Acids Res. 1987 Nov 11;15(21):9081–9082. doi: 10.1093/nar/15.21.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlein M., Madern D. Molecular cloning and nucleotide sequence of the gene for Escherichia coli leucyl-tRNA synthetase. Nucleic Acids Res. 1987 Dec 23;15(24):10199–10210. doi: 10.1093/nar/15.24.10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin S. X., Shi J. P., Cheng X. D., Wang Y. L. Arginyl-tRNA synthetase from Escherichia coli, purification by affinity chromatography, properties, and steady-state kinetics. Biochemistry. 1988 Aug 23;27(17):6343–6348. doi: 10.1021/bi00417a022. [DOI] [PubMed] [Google Scholar]
- Looman A. C., van Knippenberg P. H. Effects of GUG and AUG initiation codons on the expression of lacZ in Escherichia coli. FEBS Lett. 1986 Mar 3;197(1-2):315–320. doi: 10.1016/0014-5793(86)80349-0. [DOI] [PubMed] [Google Scholar]
- Nazario M., Evans J. A. Physical and kinetic studies of arginyl transfer ribonucleic acid ligase of Neurospora. A sequential ordered mechanism. J Biol Chem. 1974 Aug 10;249(15):4934–4936. [PubMed] [Google Scholar]
- Neidhardt F. C., Vaughn V., Phillips T. A., Bloch P. L. Gene-protein index of Escherichia coli K-12. Microbiol Rev. 1983 Jun;47(2):231–284. doi: 10.1128/mr.47.2.231-284.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfait R., Grosjean H. Arginyl-transfer ribonucleic-acid synthetase from Bacillus stearothermophilus. Purification, properties and mechanism of action. Eur J Biochem. 1972 Oct;30(2):242–249. doi: 10.1111/j.1432-1033.1972.tb02092.x. [DOI] [PubMed] [Google Scholar]
- Reddy P., Peterkofsky A., McKenney K. Translational efficiency of the Escherichia coli adenylate cyclase gene: mutating the UUG initiation codon to GUG or AUG results in increased gene expression. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5656–5660. doi: 10.1073/pnas.82.17.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M. J., Söll D. Discrimination between glutaminyl-tRNA synthetase and seryl-tRNA synthetase involves nucleotides in the acceptor helix of tRNA. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6627–6631. doi: 10.1073/pnas.85.18.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. R., Söll D. Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu Rev Biochem. 1979;48:601–648. doi: 10.1146/annurev.bi.48.070179.003125. [DOI] [PubMed] [Google Scholar]
- Schwartz S. A., Helinski D. R. Purification and characterization of colicin E1. J Biol Chem. 1971 Oct 25;246(20):6318–6327. [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Summers W. C. A simple method for extraction of RNA from E. coli utilizing diethyl pyrocarbonate. Anal Biochem. 1970 Feb;33(2):459–463. doi: 10.1016/0003-2697(70)90316-7. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebe R. Arginyl-tRNA synthetase from brewer's yeast. Purification, properties, and steady-state mechanism. Eur J Biochem. 1983 Feb 15;130(3):517–524. doi: 10.1111/j.1432-1033.1983.tb07180.x. [DOI] [PubMed] [Google Scholar]
- Webster T., Tsai H., Kula M., Mackie G. A., Schimmel P. Specific sequence homology and three-dimensional structure of an aminoacyl transfer RNA synthetase. Science. 1984 Dec 14;226(4680):1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]
- Winter G., Koch G. L., Hartley B. S., Barker D. G. The amino acid sequence of the tyrosyl-tRNA synthetase from Bacillus stearothermophilus. Eur J Biochem. 1983 May 2;132(2):383–387. doi: 10.1111/j.1432-1033.1983.tb07374.x. [DOI] [PubMed] [Google Scholar]
- Zelwer C., Risler J. L., Brunie S. Crystal structure of Escherichia coli methionyl-tRNA synthetase at 2.5 A resolution. J Mol Biol. 1982 Feb 15;155(1):63–81. doi: 10.1016/0022-2836(82)90492-2. [DOI] [PubMed] [Google Scholar]