Mice lacking all three Rb genes in the liver develop tumors resembling specific subgroups of human hepatocellular carcinomas, and Notch activity appears to suppress the growth and progression of these tumors.
Abstract
Hepatocellular carcinoma (HCC) is the third cancer killer worldwide with >600,000 deaths every year. Although the major risk factors are known, therapeutic options in patients remain limited in part because of our incomplete understanding of the cellular and molecular mechanisms influencing HCC development. Evidence indicates that the retinoblastoma (RB) pathway is functionally inactivated in most cases of HCC by genetic, epigenetic, and/or viral mechanisms. To investigate the functional relevance of this observation, we inactivated the RB pathway in the liver of adult mice by deleting the three members of the Rb (Rb1) gene family: Rb, p107, and p130. Rb family triple knockout mice develop liver tumors with histopathological features and gene expression profiles similar to human HCC. In this mouse model, cancer initiation is associated with the specific expansion of populations of liver stem/progenitor cells, indicating that the RB pathway may prevent HCC development by maintaining the quiescence of adult liver progenitor cells. In addition, we show that during tumor progression, activation of the Notch pathway via E2F transcription factors serves as a negative feedback mechanism to slow HCC growth. The level of Notch activity is also able to predict survival of HCC patients, suggesting novel means to diagnose and treat HCC.
Hepatocellular carcinoma (HCC) results in >600,000 deaths per year worldwide (Caldwell and Park, 2009). Although the major risk factors have been identified, including infection with hepatitis viruses B or C, mechanisms that are at work during the development of HCC remain poorly understood, hindering the development of novel therapeutic approaches (Farazi and DePinho, 2006).
The extended retinoblastoma (RB) pathway is comprised of p16INK4a and p21CIP1 family members, which inhibit the kinase activity of Cyclin–Cyclin-dependent kinase (CDK) complexes; these complexes in turn normally inactivate the RB protein and its two family members p107 and p130 by hyperphosphorylation during the G1/S transition of the cell cycle, thereby activating E2F transcription factors (Burkhart and Sage, 2008). Accumulating evidence suggests an almost universal inactivation of the RB pathway in HCC, including by promoter hypermethylation of the p16INK4a (∼50% of cases) or p15INK4b (∼15% of cases) genes, and amplification of the gene coding for Cyclin D1 (∼30% of cases). Mutations in the RB gene itself are rare, but the RB protein is often undetected in HCC cells (Edamoto et al., 2003; Laurent-Puig and Zucman-Rossi, 2006; Knudsen and Knudsen, 2008). Furthermore, Gankyrin, an E3 ligase that triggers degradation of RB family members, is overexpressed in the majority of HCC tumors (Higashitsuji et al., 2000). Finally, the RB pathway is also inactivated in liver cells by viral proteins: for example, hepatitis B virus infection results in the constitutive expression of viral proteins such as preS2 and HBx, which directly or indirectly activate Cyclin–CDK complexes (Park et al., 2006; Hsieh et al., 2007; Wang et al., 2008; Kim et al., 2010b). Similarly, recruitment of the E6AP ubiquitin ligase by the hepatitis C virus protein NS5B leads to the degradation of RB and potentially p107 and p130 (Munakata et al., 2007). These observations suggest a yet untested model in which overall inactivation of the RB pathway by simultaneous decreased function of the three RB family members may be necessary for HCC development. This idea is supported by the observation that deletion of the Rb (Rb1) gene in the mouse liver is not sufficient to drive HCC development (Williams et al., 1994; Mayhew et al., 2005).
Many of the events resulting in the functional inactivation of the RB pathway in human HCC occur early in the course of the disease, suggesting that the RB pathway may play an important role in the prevention of HCC initiation (Zhang et al., 2006; Nordenstedt et al., 2010). However, identifying the cell type or types from which HCC may originate is not currently possible in patients. Mature hepatocytes (parenchymal cells) in the liver have the capacity to proliferate and could therefore serve as the cell of origin for HCC (Oertel and Shafritz, 2008; Duncan et al., 2009; Benhamouche et al., 2010). In addition, the portal triad, a structure composed of a bile duct, a portal vein, and a hepatic artery, is thought to serve as a niche for several populations of stem and progenitor cells (nonparenchymal cells; Susick et al., 2001; Schmelzer et al., 2006; Zhang et al., 2008). These progenitor cells, often termed oval cells, also have the capacity to expand in response to stress and injury (Erker and Grompe, 2008; Duncan et al., 2009). All of these different liver cell populations may serve as a cell of origin for HCC. Unfortunately, the current lack of a comprehensive knowledge of the different stages of differentiation in the hepatocytic lineage has hampered a thorough investigation of the cellular origin of HCC (Lee and Thorgeirsson, 2006; Zhang et al., 2008; Duncan et al., 2009; Mishra et al., 2009; Dorrell et al., 2011; Shin et al., 2011).
In this study, we sought to address the molecular and cellular mechanisms of HCC development upon loss of RB pathway function. We show that Rb family triple KO (TKO) mice, in which the RB pathway is genetically inactivated, develop liver cancer with histological and molecular similarities to human HCC. In addition, we show that loss of RB family function allows the expansion of normally quiescent populations of stem/progenitor cells, and we identify activation of Notch signaling as a suppressor feedback mechanism during HCC progression.
RESULTS
Genetic ablation of the Rb gene family in the liver of adult mice results in the development of tumors similar to human HCC
To model the functional inactivation of the RB pathway found in human HCC, we specifically deleted the three Rb family genes in the liver of adult mice by performing intrasplenic injection of adenovirus expressing the Cre recombinase (Ad-Cre) in conditional TKO (cTKO; Rblox/lox;p130lox/lox;p107−/−; Viatour et al., 2008) adult mice. In this protocol, the adenovirus predominantly delivers the Cre recombinase to hepatocytes as well as nonparenchymal cells in the liver (Fig. S1 A). All of the Ad-Cre–infected cTKO mice analyzed and none of the control mice infected with Ad-GFP developed multiple liver lesions after a latency of 3–4 mo and had to be euthanized within 5–8 mo because of tumor burden (Fig. 1 A); these mice did not develop any other visible pathology outside the liver (not depicted). Quantitative RT-PCR (RT-qPCR) analysis with RNA extracted from macro-dissected tumors showed decreased expression of Rb and p130, confirming deletion of these two genes (Fig. 1 B). The presence of one p130 or p107 allele was sufficient to prevent the development of liver tumors at the same time points (Fig. S1 B and not depicted). Cells in these lesions were actively cycling, as visualized by immunostaining for BrdU incorporation (Fig. 1 C) and by immunoblot analysis for known markers of proliferation (Fig. S1 C). Finally, TKO liver tumors could be serially transplanted in immunocompromised mice, confirming their tumorigenic potential (Fig. S1, D–F).
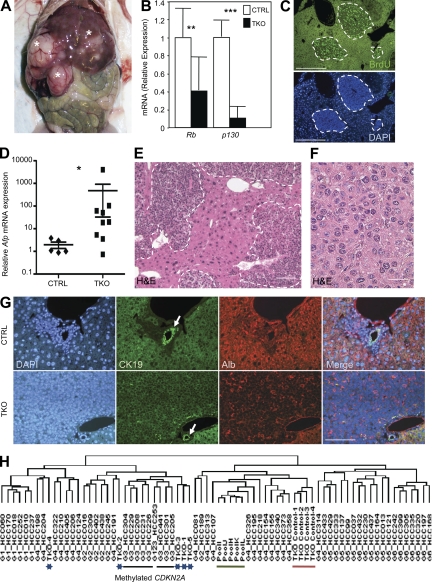
Figure 1.
Genetic inactivation of the Rb gene family in the mouse liver results in HCC development. (A) One representative (n > 20) TKO mouse with tumors (asterisks) in the liver is shown 4 mo after intrasplenic Ad-Cre injection. All experiments were performed on TKO mice 3–4 mo after Ad-Cre injection. (B) RT-qPCR analysis of Rb and p130 messenger RNA (mRNA) expression in TKO tumors (n = 9) and control livers (n = 5; CTRL, Rblox/lox;p130lox/lox;p107−/−). (C) Immunostaining on TKO liver sections for the DNA replication marker BrdU. Areas of proliferation are circled with dashed lines. (D) RT-qPCR analysis of Afp mRNA levels in TKO tumors (n = 9) and CTRL livers (n = 5). (E) H&E staining of TKO liver sections with multiple independent tumors (delineated by dashed lines). (F) At higher magnification, tumor cells resemble small hepatocytes. (G) Representative sections (n > 20) from CTRL and TKO livers were stained with DAPI, CK19, and Albumin (Alb). The white arrows point to a bile duct. Merged pictures are shown on the right. (H) Nonsupervised hierarchical clustering of gene expression profiles from human HCCs and mouse TKO tumors (blue asterisks). The black bar marks human tumors in the group G3, which are characterized in part by methylation of the CDKN2A locus. The green bar marks normal human liver samples, and the red bar marks CTRL mouse livers. Error bars indicate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars: (C, E, and G) 50 µm; (F) 5 µm.
TKO tumors expressed high levels of Afp, a molecular marker of HCC (Fig. 1 D), and were classified histopathologically as trabecular lesions with features of hepatocytes (Fig. 1, E and F). Immunostaining experiments further indicated that TKO tumors were composed of cells expressing heterogeneous levels of Albumin (a hepatocyte marker) but undetectable levels of CK19 or EpCAM (cholangiocyte markers; Fig. 1 G and Fig. S1 G), similar to most human HCCs. To further assess the relevance of the TKO model to human HCC, we performed a microarray analysis comparing TKO tumors to human HCCs. A previous analysis of gene expression profiles from a large set of human HCCs identified six HCC subgroups (G1–G6), each with different molecular features (Boyault et al., 2007). We found that TKO tumors clustered preferentially with the G1–G3 groups (Fig. 1 F). Interestingly, some TKO tumors clustered with human G3 tumors, which are characterized in part by the expression of several genes implicated in cell cycle control, including E2F target genes, and by the methylation of the CDKN2A locus, which codes for p16INK4a, an upstream regulator of the entire RB family. Together, these observations indicate that the tumors growing in the liver of TKO mice provide a novel model for human HCC.
Expansion of the stem/progenitor compartment upon deletion of Rb family genes in the liver of adult mice
The early stages of cancer initiation are challenging to study in HCC patients, who are often diagnosed with advanced disease, but are readily accessible to in vivo analysis in mouse models. We examined tumor development 1–5 wk after Cre-mediated inactivation of Rb family genes in the liver of adult cTKO mice. At these time points, the mutant liver appeared macroscopically normal (not depicted), but liver sections showed the progressive appearance of foci of small cells growing next to the portal triad; these cells had a high nuclear/cytoplasm ratio and often displayed an oval shape as well as some histopathological features of hepatocytes (Fig. 2 A and Fig. S2 A). Immunostaining for glutamine synthase (GS), a marker of centrilobular hepatocytes (Gebhardt and Hovhannisyan, 2010), confirmed that the small lesions were growing next to portal triads, away from central veins (Fig. 2 B). The adult liver is a quiescent organ, and very few cells stained positive for the cell cycle marker Ki67 on liver sections from control mice 2–4 wk after Ad-Cre infection (not depicted). In contrast, immunostaining for Ki67 on liver sections from TKO mice showed that cells in the small lesions were proliferative; strikingly, very little proliferation was observed in populations of mature hepatocytes surrounding the early lesions (Fig. 2 C; see a second example in Fig. S2, C and E). Little, if any apoptotic cell death was observed by immunostaining for cleaved caspase 3 on sections from control and TKO mice at these early time points (not depicted). This analysis suggested that loss of RB family members in the liver of adult mice leads rapidly and specifically to the proliferation of progenitor-like cells (Roskams et al., 2003; Dorrell et al., 2008).
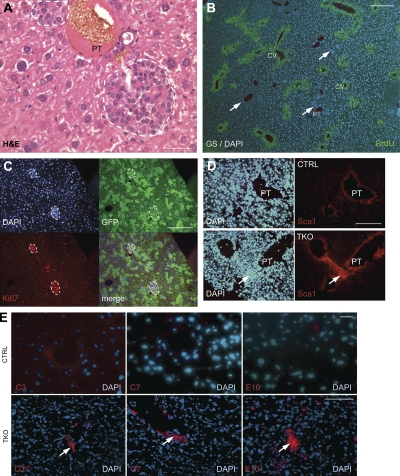
Figure 2.
Inactivation of the Rb gene family in the adult liver results in the expansion of cells with features of stem/progenitor cells. (A) Representative H&E staining (n > 20) of the liver of a cTKO mouse infected with Ad-Cre (4 wk after the injection) shows that TKO early liver lesions are composed of small cells. White dashed lines indicate lesions. All mice used were between 2 and 4 mo of age at the time of injection. PT, portal triad. (B) Sections from TKO liver were stained antibodies against GS (green), a marker of hepatocytes around the central vein (CV). Immunofluorescence images were merged with DAPI (blue) images. The white arrows indicate early TKO lesions. (C) Early lesions in cTKO;Rosa26LSL-YFP mice infected with Ad-Cre were immunostained for Ki67 and GFP. White dashed lines indicate lesions. (D) Control (CTRL) and TKO liver sections were stained with Sca1 antibodies. The white arrows point to Sca1-positive cells. (E) CTRL and TKO livers were stained with C3, C7, and E10 antibodies. Immunofluorescence images were merged with DAPI images. White arrows indicate early lesions. Bars: (A–D and E, top) 5 µm; (E, bottom) 50 µm.
Evidence suggests that CK19 and EpCAM, which are markers of normal cholangiocytes, may also be expressed in certain populations of liver progenitor cells (Libbrecht, 2006; Schmelzer et al., 2006; Yovchev et al., 2008; Zhang et al., 2008; Duncan et al., 2009). However, we did not detect expression of these markers by immunostaining in the small lesions growing in the liver of TKO mice. The A6 antigen is also a marker of both liver progenitor cells and bile duct cells (Engelhardt et al., 1993; Haybaeck et al., 2009), and we found an expansion of A6+ cells around the portal triad of cTKO mice 2–3 wk after Cre induction (Fig. S2, F–K). Several cells in the TKO lesions were also positive for Sox9, a recently identified marker of liver progenitors (Fig. S2, L–N; Dorrell et al., 2011). Similarly, we used antibodies against Sca1, a marker expressed at the surface of mouse stem/progenitor cells in different tissues (Holmes and Stanford, 2007), including in the liver (Clayton and Forbes, 2009), and found an expansion of Sca1+ cells within the small lesions by immunostaining (Fig. 2 D). A panel of antibodies that recognize specific surface epitopes on cells activated during an oval cell response initiated by DDC (3,5-diethoxycarbonyl-1,4-dihydrocollidine) treatment was recently developed (Dorrell et al., 2008). Among the nine monoclonal antibodies that we tested, only MIC1-1C3 (C3), OC2-3C7 (C7), and OC2-6E10 (E10) consistently stained TKO early lesions, albeit with variable frequency and intensity (Fig. 2 E and Fig. S2, O–Q). The difference in signal intensity suggests that progenitor cells expanding in the DDC and TKO models are only partially overlapping, which is supported by the different histological structures developed in the two models (Fig. S2, R and S). Thus, cells expanding in the liver of TKO mice express markers that are characteristic of subpopulations of adult liver stem/progenitor cells, providing additional support to a model in which loss of RB family function leads to the expansion of populations of liver progenitor cells.
To control for reactive effects to the adenoviral infection in the liver, we used intraperitoneal injection of tamoxifen (Tam) in Rosa26CreER cTKO mice to delete Rb and p130. In this model, the CreER fusion protein is broadly expressed and can be activated upon Tam injection to inactivate the RB family in multiple tissues and organs (Viatour et al., 2008; Burkhart et al., 2010). We found that induction of Cre activity by Tam in this system was often more efficient than with Ad-Cre, as judged by the analysis of a YFP reporter (Fig. S2, T–W); we also found an expansion of cells with progenitor cell markers in Rosa26CreER TKO mice (Fig. S2 B and not depicted). In this Rosa26CreER cTKO system as with Ad-Cre cTKO mice, we did not observe any increase in liver size at preneoplastic stages (not depicted).
Rosa26CreER TKO and Ad-Cre TKO mice were used interchangeably to measure the expression of the Sca1, C3, C7, and E10 surface markers by flow cytometry. These experiments confirmed that both Sca1+ cells and C3/C7/E10+ cells (where the three antibodies were pooled in the FACS analysis) increased in frequency in TKO livers shortly after Cre activation (Fig. 3, A and B). Interestingly, only a minor fraction of cells were double positive for Sca1 and C3/C7/E10 (Fig. 3 A), indicating that these antigens marked distinct populations of cells. Because the expansion of progenitor cells in TKO mice could be caused by a non–cell autonomous, reactive activation of these cells, we measured p130 levels in Sca1+ and/or C3/C7/E10+ cells isolated from TKO mice compared with control mice. We found decreased levels of p130, suggesting that the expanding progenitor cells were indeed mutant for the Rb family (Fig. 3 C). These data confirmed the observation made with the inducible GFP reporter that Cre had been activated in cells within early lesions (Fig. 2 C). We next sought to gain more insights into the biology of the TKO progenitor cells by examining some characteristics of stem/progenitor cells in these populations. Compared with C3/C7/E10+ cells, we found that Sca1+ cells were cycling more slowly (Fig. 3 D and Fig. S3, A and B), had higher expression levels of the stem cell marker Bmi1 (Fig. 3 E; Valk-Lingbeek et al., 2004), and expressed lower levels of the Ck19 and Albumin differentiation markers (Fig. 3 F).
Figure 3.
HCC development is associated with an expansion of the progenitor compartment in TKO mice. (A) Representative FACS analysis (n = 4) of nonparenchymal cells 2 wk after Cre-mediated recombination in TKO mice compared with controls (CTRL). The nonhematopoietic fraction (CD45low) was analyzed for Sca1 and C3/C7/E10 expression. (B) Numbers of Sca1+ and C3/C7/E10+ cells in the nonparenchymal fractions of TKO livers (n = 4 for each genotype). (C) The expression of p130 was assessed by RT-qPCR in Sca1+ and C3/C7/E10+ subsets of control and TKO nonparenchymal cells (n = 3). (D) Cell cycle activity in TKO C3/C7/E10+ cells compared with TKO Sca1+ cells was measured by propidium iodide staining of fixed cells isolated by FACS (n = 4). (E) Expression of Bmi1 in Sca1+ and C3/C7/E10+ populations from control and TKO mice in early lesions, as assessed by RT-qPCR (n = 3). (F) Expression of Albumin and CK19 in C3/C7/E10+ or Sca1+ TKO cells (n = 3). (G) Colony-forming activity of unfractioned nonparenchymal cells from either control or TKO mice. Colonies were either grown in the presence or absence of EGF and HGF. Plates were stained with crystal violet after 8 d before counting (mean of four independent experiments). (H and I) Colony-forming activity of control and TKO Sca1+ and C3/C7/E10+ populations. Colony assay was performed in 24-well plates with sorted CTRL and TKO cells 2 wk after Cre-mediated recombination. (H) Representative pictures of colonies formed by C3/C7/E10+ (top) and Sca1+ (bottom) TKO cells. (I) Quantification of H. Colonies were fixed and stained after 8 d before counting (mean of three independent experiments). Error bars indicate SEM. *, P < 0.05; **, P < 0.01; ns, not significant.
One common characteristic of stem/progenitor cells is their ability to form colonies in defined culture conditions. When placed in culture, TKO nonparenchymal cells produced more colonies than nonparenchymal cells from control mice (Fig. 3 G). Under these conditions, although control cells absolutely required epidermal growth factor (EGF) and hepatocyte growth factor (HGF) to form colonies, cells from TKO livers formed colonies in the absence of these growth factors, although not to the extent of cells with EGF and HGF. Nonparenchymal TKO cells were able to form lesions resembling the lesions growing in the liver of TKO mice when transplanted into the liver of immunodeficient mice (Fig. S3 C), but they failed to form tumors when transplanted under the skin (not depicted). This observation and the partial growth factor independence suggest that, at this early stage, TKO liver cells in the small lesions still require an appropriate microenvironment to expand. When we used flow cytometry to further identify the cells in the liver of TKO mice that had the ability to form colonies in culture, we found that Sca1/C3/C7/E10− cells from TKO mice reproducibly failed to produce colonies under these conditions (unpublished data). In contrast, both Sca1+ and/or C3/C7/E10+ TKO cells formed colonies, although Sca1+ cells formed fewer and smaller colonies (Fig. 3, H and I), which could be related to their slower cell cycle (Fig. 3 D). Together, these data show that inactivation of the RB pathway in the liver of adult mice specifically leads to the expansion of populations of stem/progenitor cells, suggesting that these cells initiate HCC development.
Up-regulation of E2F and Myc activities in TKO HCC cells
To understand the molecular changes that occur during the development of HCC in TKO mice, we first ran the SAM (significance analysis of microarrays) statistical method to compare the TKO dataset with a control liver dataset. The three most induced genes in TKO tumors, H19, Tff3, and Rrm2, are molecular markers of human HCC (Sohda et al., 1998; Okada et al., 2005; Satow et al., 2010). This analysis also showed increased expression of genes involved in the regulation of cell cycle, DNA synthesis, and DNA repair in the TKO dataset, including known E2F target genes (see Table S1 for selected genes and Table S2 for the entire list). DAVID (Database for Annotation, Visualization, and Integrated Discovery) analysis indicated that many of the genes up-regulated in the TKO dataset have a known role in cell cycle progression, DNA repair, and chromosome maintenance (Table S1). Nonsupervised hierarchical clustering showed that TKO HCC tumors clustered closely with mouse liver tumors initiated by the overexpression of E2F1 and c-Myc (Fig. 4 A; Conner et al., 2000; Lee et al., 2004). Accordingly, c-Myc levels were higher in TKO liver tumors than in control livers (Fig. 4 B), and a gene set enrichment analysis (GSEA) showed significant enrichment in the TKO dataset for genes in a Myc-centered regulatory network independent of a core embryonic stem cell program (Fig. 4 C and not depicted; false discovery rate [FDR] q-value 0.045; Kim et al., 2010a). GSEA with the TKO dataset and a library of curated gene sets (see Materials and methods and Table S3) also showed significant enrichments for c-Myc, E2F1, and E2F3 (Fig. 4 D and supplemental text). Together, the observations that E2F and Myc activities are elevated in TKO tumors, previous findings that ectopic expression of either E2F1 or c-Myc is sufficient to initiate HCC (Conner et al., 2000; Lee et al., 2004), and the rapid appearance of multiple lesions in the liver of TKO mice suggest that loss of RB family function is sufficient to initiate HCC development in the liver of mice by promoting the proliferation of the mutant cells.
Figure 4.
Activation of cellular signaling pathways in TKO HCC cells. (A) Nonsupervised hierarchical clustering of datasets from various mouse HCC models (TKO, c-Myc overexpression, and E2F1 overexpression). Datasets from different platforms were first normalized before analysis (see Materials and methods). A cluster of genes specifically up-regulated in the TKO dataset was identified (cluster M; supplemental text). The color scale is drawn relative to each gene (each row) with blue representing the lowest expression and red the highest. (B) RT-qPCR of c-Myc expression in five control livers (CTRL) and nine TKO HCC samples. (C) GSEA shows a significant enrichment in the TKO dataset of a predefined gene list that is specific for a core Myc signature (Myc module). (D) GSEA analysis was performed by comparing the TKO dataset (Table S2) with the entire list of curated gene sets available. The entire list of gene sets enriched in the TKO dataset is displayed in Table S3. The gene sets (pathways) are ranked from the highest to the lowest score (NES: most significant, left). Significant p-value is <0.05, and significant FDR is <0.25. (E) The genes in cluster M were processed through DAVID analysis for GO annotations, and the top enrichment scores and p-values are shown. (F) RT-qPCR analysis of Notch1 and the Notch pathway target Nrarp in TKO HCCs compared with c-Myc–induced HCCs and control liver samples (n = 3). Error bars indicate SEM. *, P < 0.05; **, P < 0.01.
Transcriptional up-regulation of the Notch signaling pathway in TKO HCC cells
GSEA also identified enrichment for signaling pathways other than E2F and Myc in TKO HCCs compared with control livers (Fig. 4 D), including pathways such as the Wnt, p38 MAPK (mitogen-activated protein kinase), and Ras pathways, which are known to be involved in human HCC development (Laurent-Puig and Zucman-Rossi, 2006; Villanueva et al., 2007; Whittaker et al., 2010; Min et al., 2011) and could cooperate with E2F and c-Myc activities in the development of TKO tumors. Potential changes in the Notch signaling pathway were also detected by GSEA. Although Notch signaling has been shown to play a critical role in cell fate decisions during liver development, little is known regarding its possible role in liver cancer development (Li et al., 1997; Qi et al., 2003; Geisler et al., 2008; Zong et al., 2009; Hofmann et al., 2010). Changes in the expression of genes in the Notch pathway in TKO HCCs were supported by the presence of Notch pathway members in a cluster of genes that are specifically up-regulated in the TKO HCC model compared with other HCC mouse models; this cluster includes Notch receptors (Notch1–4), as well as a downstream transcriptional target of the Notch pathway (Nrarp; Fig. 4, A and E; and supplemental text; Pirot et al., 2004). RT-qPCR analysis confirmed that Notch1 and Nrarp were specifically up-regulated in TKO tumors compared with c-Myc–induced HCC (Fig. 4 F), suggesting that this up-regulation was specific to tumors with inactivation of the RB pathway.
RT-qPCR analysis further showed that the expression of several components of the Notch pathway, including Notch1, Notch3, and Notch4, as well as two other canonical Notch target genes, Hes1 and Hey1, was significantly increased in TKO HCC cells compared with control liver cells (Fig. 5 A). This increase in the transcription of several genes in the Notch pathway in Rb family mutant cells raised the possibility that it was a direct consequence of the activation of E2F transcription factors. Indeed, reporter gene assays showed that promoters of the mouse Hes1 gene and the human NOTCH1 gene could be activated by E2F1 and E2F3, two transactivating members of the E2F family (Fig. 5 B). Chromatin immunoprecipitation (ChIP) assays showed direct binding of E2F1 and E2F3 to the proximal promoter regions of Dtx3, Dtx4, Notch1, Notch3, Hes1, and Hey1 in a cell line derived from a TKO HCC (Fig. 5 C and not depicted). E2F binding to the promoter region of Notch pathway genes was also observed in WT mouse adult livers; in particular, we found increased binding of E2F4, a repressor E2F, to several of the promoters tested in these quiescent liver cells (Fig. 5 D), correlating with the lower transcription of Notch pathway genes in normal liver (Fig. 5 A). To determine whether this finding was specific to mouse cells, we performed similar experiments with the human HCC cell lines Hep3B and SNU-449 and found binding of E2F1, E2F3, and E2F4 to the same Notch pathway gene promoters (Fig. 5 E and not depicted). Reintroduction of RB in mouse HCC cells led to the repression of the E2F target gene Ccna2 as well as the Notch pathway member Hey1, although not all Notch pathway members responded to RB reintroduction under these specific conditions (Fig. 5 F). Together, these experiments indicate a direct control of the Notch signaling pathway in HCC cells by the RB/E2F axis, providing one explanation why loss of Rb family genes results in the enhanced transcription of Notch pathway genes in these tumor cells.
Figure 5.
Notch signaling is regulated in HCC cells by E2F transcription factors. (A) RT-qPCR analysis was performed on five control livers (CTRL) and nine TKO HCC samples to detect the expression of members of the Notch pathway. (B) Reporter gene assays to measure the transactivation of the murine Hes1 promoter (left; light gray) and the human NOTCH1 promoter (right; gray) by 0.2 and 1 µg E2F1 or 1 µg E2F3. Data are representative of three independent experiments performed in Saos cells. (C–E) ChIP analysis for E2F1, E2F3 (activating E2Fs), and E2F4 (repressor E2F) binding to the promoter of members of the Notch pathway in a cell line derived from a TKO HCC (C), WT primary mouse liver cells (D), and a human HCC cell line, Hep3B (E). A p16 antibody was used as a negative control. Binding to the Cyclin A (Ccna2) promoter, a well-known E2F target, was used as a positive control. Displayed images are representative of three independent experiments. (F) Reintroduction of RB in mouse HCC cells. Two cell lines derived from TKO HCCs were transfected with a plasmid expressing GFP (empty vector) or a fusion protein composed of GFP and the large pocket domain of RB (RB reintroduction). GFP-positive cells were isolated by FACS 40 h after transfection, and the expression of Ccna2, Hey1, and Hes1 was assessed by RT-qPCR (n = 3). Error bars indicate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Notch signaling inhibits the expansion of HCC cells
Potentially as a result of this transcriptional activation by E2F, Notch signaling was activated in TKO HCC, as illustrated by the increased level of the cleaved active form of the Notch1 (intracellular domain [ICD] of Notch [NICD]) protein compared with control liver cells (Fig. 6 A) and the induction of the Notch target Nrarp in TKO tumor cells (Fig. 4 F). Furthermore, immunostaining on tumor sections revealed that the Notch target HES1 and Notch1 itself (but not Notch2) were expressed in the nucleus of TKO HCC cells in vivo, which is a sign of Notch pathway activation (Fig. S4).
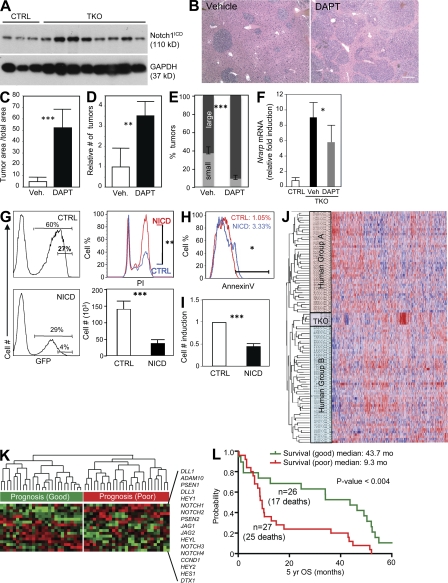
Figure 6.
The Notch signaling pathway inhibits the expansion of HCC cells. (A) Immunoblotting analysis of activated Notch1 (Notch1ICD) in three control livers (CTRL) and eight independent TKO tumors. GAPDH serves as a loading control. (B) H&E staining of liver sections from TKO mice treated with DAPT (n = 4) or vehicle control (n = 4). Bar, 50 µm. (C) Quantification of tumor area compared with the total area in DAPT- and vehicle-treated (Veh) TKO mice (n = 4). (D) Quantification of tumor numbers by surface area (n = 4). (E) Quantification of small and large tumors. The cutoff was arbitrarily set at 10,000 µm2, which corresponds to the size of early TKO lesions (n = 4). (F) Analysis of Nrarp mRNA levels in tumors upon DAPT treatment by RT-qPCR (n = 4). (G) Four HCC cell lines (two TKO cell lines, human cells SNU-449 and HepG2) were infected with MigR1-IRES-GFP retroviruses either empty (CTRL) or expressing NICD, and GFPhigh cells were isolated by FACS. (left) GFP expression in control and NICD-expressing cells. (right) Sorted cells were analyzed for propidium iodide incorporation and growth in culture. The number of cells was quantified 90 h after plating 20,000 cells. Data are representative of two independent experiments. (H) Five HCC cell lines (TKO1, TKO2, C3a, HepG2, and SNU-449) were infected with MigR1-IRES-GFP either empty (CTRL) or expressing NICD. GFPhigh cells were isolated by FACS and allowed to rest in culture for 72 h, after which survival was measured by Annexin V staining. Data are representative of two independent experiments. (I) C3/C7/E10+ nonparenchymal cells were FACS sorted, and 5,000 cells were plated in culture. Cells were infected with MigR1-IRES-GFP either empty (CTRL) or expressing NICD and counted 7 d after infection (n = 4). (J) The TKO dataset clusters with the subgroup B of human HCC after nonsupervised hierarchical clustering analysis of 3,497 mouse and human orthologues. (K) Nonsupervised hierarchical clustering of a curated gene set representing the Notch pathway (17 genes, including ligands, receptors, and target genes). Expression of Notch pathway genes was retrieved from a dataset of 53 primary human HCCs with good (group B) and bad (group A) prognosis. (L) A curated set of 17 genes in the Notch pathway (same as in K) predicts survival in HCC patients (OS, overall survival in months over a 5-yr period). Error bars indicate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine the functional role of the Notch pathway in TKO mice, we treated TKO mice 75 d after Ad-Cre injection, at a time when small tumors were present, with DAPT, a γ-secretase inhibitor which blocks the Notch pathway by preventing the cleavage of the internal domain of the Notch receptor (Hellström et al., 2007). 12 d after the beginning of treatment, we found that the liver from mice treated with DAPT exhibited macroscopically visible tumors that were not present in vehicle-treated mice (not depicted). Sections from vehicle-treated and DAPT-treated mice showed a significant increase in tumor area and tumor numbers (Fig. 6, B–E). Nrarp expression was decreased in DAPT-treated tumors compared with vehicle-treated tumors, indicating that DAPT treatment did inhibit Notch signaling, as expected (Fig. 6 F). To directly test whether activation of Notch signaling was sufficient to block the proliferation of HCC cells in a cell intrinsic manner, we overexpressed the constitutively active ICD of Notch1 in liver tumor cell lines derived either from TKO mice (TKO1 and TKO2) or from human HCC (C3a, SNU-449, and HepG2). We found that expression of Notch1 ICD was sufficient to inhibit the proliferation of three out of the four cell lines tested (TKO1, TKO2, and SNU-449 but not HepG2; Fig. 6 G and not depicted) and also induced an apoptotic response in four out of the five cell lines tested (TKO1, C3a, HepG2, and SNU-449 but not TKO2; Fig. 6 H). These findings indicate that activation of the Notch signaling pathway after activation of E2F in RB family mutant HCC cells serves as a negative feedback mechanism to slow the expansion of the tumor cells.
Activation of Notch signaling is critical for biliary differentiation of liver progenitors (Li et al., 1997; Geisler et al., 2008; Hofmann et al., 2010; Roskams et al., 2010; Onoyama et al., 2011). This observation raised the question of the status of Notch signaling in the liver progenitor populations from which TKO HCCs may arise. RT-qPCR analysis from FACS-sorted Sca1+ and C3/C7/E10+ liver progenitor cells and hepatocytes showed that Notch1 signaling was significantly higher in progenitor cell populations compared with mature liver cells (Fig. S5 A). Immunostaining for HES1 and nuclear Notch1 confirmed the activation of the Notch pathway in early TKO lesions (Fig. S4). However, although the Notch pathway is transcriptionally activated in TKO hepatocytes compared with their controls, loss of the three RB family proteins did not further induce the expression of Notch1, Hes1, Hey1, or Nrarp in TKO progenitors (Fig. S5 A). These findings suggest that transcriptional control of Notch pathway genes by RB family members is cell type dependent and evolves during tumor progression, being more apparent in cells with low Notch pathway activity. We found no differences in TKO progenitor cell numbers or in their cell cycle activity upon treatment with DAPT in vivo (Fig. S5, B–D), possibly because high Notch activity in these cells renders them less sensitive to partial inhibition of the Notch pathway by DAPT. However, the colony-forming activity of TKO liver progenitor cells in culture was significantly impaired when these cells were sorted from mice and infected with a Notch1 ICD retrovirus (Fig. 6 I), similar to what we had observed with HCC cells (Fig. 6 G). This observation suggests that hyperactivation of Notch signaling also inhibits the proliferation of liver progenitor cells.
To address the relevance of the findings that activation of Notch signaling may serve as a mechanism to dampen the proliferation of HCC cells to HCC patients, we took advantage of a dataset in which gene expression profiles are linked to patient survival (Lee et al., 2004). In this study, clustering analysis had led to the identification of two major HCC molecular groups, with group B patients showing a better survival than group A patients. We found that the TKO dataset preferentially clustered with human tumors of the B subgroup (Fig. 6 J), similar to what had been previously observed for tumors expressing Myc and E2F1 (Lee et al., 2004). Interestingly, the expression level of the human NOTCH1 and HES1 genes was significantly higher in group B versus group A (P < 0.05). However, removal of these Notch pathway genes from the analysis did not change the clustering of TKO tumors to group B (unpublished data), indicating that other genes or pathways may be involved in the enhanced survival of patients in group B. The analysis of recent genome-wide ChIP experiments indicates that RB may directly bind to the promoters of other genes in the Notch pathway such as JAG1 and PSEN2 in senescent fibroblasts (Chicas et al., 2010; unpublished data). These observations and our data in HCC cells led us to test the role of a broader Notch pathway signature in HCC. We found that a curated list of 17 genes used as a signature of the Notch pathway independently predicted increased survival in HCC patients (Fig. 6, J and K; and Fig. S5 E). Therefore, activation of Notch signaling in HCC may act as a mechanism to limit the expansion of HCC cells in patients.
DISCUSSION
In this study, we describe the consequences of genetically inactivating the Rb gene family in the liver of adult mice. We found that Rb family TKO mice develop HCC, providing a novel tractable mouse model for this deadly human cancer. Using this genetically defined model, we identified adult progenitor cells as initiating populations in HCC development upon inactivation of the RB pathway and we uncovered a potential tumor suppressor role for Notch signaling in human HCC.
RB family members display extensive functional overlap in many cell types, including in liver cells (Reed et al., 2009). In fact, we found that the presence of one WT allele of p107 or p130 was sufficient to prevent HCC development for at least 6–8 mo after Cre activation, similar to our observation in the hematopoietic compartment, indicating that reintroduction of one Rb family allele significantly impairs tumor development (Viatour et al., 2008). We cannot exclude the possibility that these mice will eventually develop HCC at later time points; indeed two of the cell lines that we have isolated thus far from TKO mice have retained a WT p130 allele (unpublished data). Although it is difficult to directly compare the genetic deletion of Rb family genes in the mouse system to the protein inactivation by degradation or phosphorylation observed in human HCC, these results support the observation that most events targeting the RB pathway in HCC simultaneously inactivate the three RB family proteins (see introduction).
An important question in the liver cancer field is whether HCC initiates from hepatocytes and/or earlier progenitors in the adult liver, a question which is challenging to address in patients. Cre activation in cTKO mice results in the proliferation and rapid expansion of cells with characteristics of liver stem/progenitor cells, including their morphology, their localization close to the portal triad, and their expression of specific markers. At the same time point, we observed only limited DNA replication in mature hepatocytes and no evidence of clonal expansion. These observations, together with the fact that a fraction enriched for nonparenchymal progenitor cells can recapitulate tumor development upon transplantation, strongly suggest that TKO HCC tumors initiate from the stem/progenitor compartment in the liver of mice. The stem/progenitor cell hypothesis is supported by evidence that Cullin 3, a regulator of Cyclin E (an inhibitor of the RB family), prevents liver cancer development from liver progenitors (Kossatz et al., 2010).
Although EpCAM and CK19 have been proposed to mark some liver progenitors (Schmelzer et al., 2006; Zhang et al., 2008), immunostaining and FACS analysis (unpublished data) for these markers did not identify EpCAM+ or CK19+ cells in TKO early lesions. One interpretation of this observation is that TKO lesions initiate from progenitors that are distinct from EpCAM+ or CK19+ progenitors, which would underscore the heterogeneity of liver stem/progenitor cell populations. This idea is supported by the observation that progenitor cells expanding upon inactivation of the NF2 tumor suppressor display different characteristics than progenitor cells expanding in TKO mice (Benhamouche et al., 2010). In addition, we found that some TKO liver progenitors are positive for markers that were originally developed in a model of DDC-induced liver progenitor expansion. However, the paucity of C3+, C7+, or E10+ cells in TKO mice compared with DDC treatment (compare Fig. 2 E with Fig. S2, L–N), as well as the lack of organized structure in TKO livers versus the formation of atypical bile ducts in the DDC model (Fig. S2, O and P) further suggest that the populations of progenitor cells involved in the expansion in the two models are not equivalent. Nevertheless, the observation that some cells in early TKO lesions express Sox9 indicates that there are some similarities between the two systems. Future experiments will continue to compare markers expressed by adult liver progenitor cells identified in different systems to better define distinct subpopulations of these cells.
In the TKO model, we found that the colony-forming potential was restricted to two distinct mutant stem/progenitor cell populations in TKO mice, C3/C7/E10+ cells and Sca1+ cells. The decreased expression of Bmi1 and the increased expression of lineage markers, as well as the increased cell cycle and colony-forming activities observed in TKO C3/C7/E10+ cells suggest that these cells may represent a more mature population of progenitors when compared with Sca1+ cells. C3/C7/E10+ or Sca1+ cells were not able to recapitulate tumors upon transplantation, suggesting that either they do not represent the cell of origin or that transplantation protocols need to be improved to allow a small number of purified stem/progenitor cells to colonize recipient liver.
Our data strongly suggest that TKO liver tumors arise from subpopulations of stem/progenitor cells. After exiting quiescence and an initial phase of expansion, these TKO cells undergo some differentiation, and these fairly differentiated proliferating cells represent the bulk of the HCC tumors. This model fits with increasing evidence suggesting that loss of RB function initiates cancer from specific populations of stem/progenitor cells (Macpherson, 2008; Viatour et al., 2008; Calo et al., 2010; Choi et al., 2010; Jiang et al., 2010), providing a general model for tumor suppression by the RB pathway. However, we cannot formally exclude at this point that an intermediate cell population (i.e., very late progenitor or early hepatocyte) could also be the cell of origin in TKO HCC. The recent identification of Cre-expressing mouse strains in liver progenitor populations (Dorrell et al., 2011; Shin et al., 2011) may provide novel means to perform lineage-tracing experiments and to formerly identify the cell of origin of the TKO tumors, including its differentiation potential and the role of the RB pathway in its cell fate.
Previous studies in neural progenitors have reported repression of Notch signaling by p107 (Vanderluit et al., 2004, 2007). In this study, we found that RB family members directly control the transcription of multiple components of the Notch pathway in human and mouse cells, identifying a strong link between these two central cellular pathways in liver cells. However, the lack of induction of Notch pathway genes in TKO liver progenitor cells suggests that this link is context dependent. More work is needed to understand the regulation of Notch pathway genes by E2F family members in different cell types. Functionally, our data showing that increased Notch activity leads to cell cycle arrest in G2 and/or cell death might be related to observations that inactivation of Rb family genes induces a G2 arrest in TKO fibroblasts (van Harn et al., 2010).
In vivo modulation of Notch pathway activity with DAPT, a γ-secretase inhibitor and potent inhibitor of Notch signaling, resulted in accelerated cancer development in TKO mice. Interestingly, liver-specific inactivation of Notch1 expression, although not sufficient to promote HCC, leads to the proliferation of hepatocytes in mice (Croquelois et al., 2005). In contrast, in vivo DAPT treatment failed to increase proliferation in TKO progenitor cells, either because these cells have already reached a very high level of cell cycle activity or because DAPT treatment is not sufficient to block Notch signaling in progenitor cells. In vitro, we found that activation of Notch signaling in mouse and human HCC cells was sufficient to block their expansion, as was reported in one human cell line before (Qi et al., 2003). We further show that HCC cells with enforced expression of NICD undergo cell cycle arrest in G2 and display increased apoptotic activity. The Notch pathway interacts with multiple molecular effectors, and there are probably multiple mechanisms involved in inhibiting tumor cell progression. In the future, thorough investigation of Notch effectors should bring more mechanistic insights into HCC development.
Our findings suggest a model in which Notch signaling acts as a tumor suppressor feedback mechanism in response to activation of E2F transcription factors in TKO liver cells. Although this negative feedback is clearly not sufficient to prevent cancer initiation, activation of Notch may significantly slow the expansion of tumor cells in vivo, and we found that expression of a Notch pathway signature of 17 genes strongly predicts survival in HCC patients. Because signaling downstream of Notch1 may play a tumor-suppressor role in different cancer types (Nicolas et al., 2003; Hanlon et al., 2010; Ranganathan et al., 2011), our observations indicate that local activation of the Notch pathway may provide novel treatment approaches in a large group of human cancers of epithelial origin.
MATERIALS AND METHODS
Mice and tumor experiments.
Rb family cTKO mice were described previously (Viatour et al., 2008) and were maintained in a mixed 129Sv/J;C57/BL6 background. For intrasplenic injections, mice were anesthetized and surgically opened on the upper left quadrant of the abdomen, followed by injection of adenovirus into the spleen. In Rosa26CreER TKO mice, Cre was induced by four consecutive injections of 1 mg Tam (T-5648; Sigma-Aldrich) in corn oil. Tam-injected Rosa26CreER TKO mice were euthanized and analyzed before their health deteriorated; although the exact cause of death in TKO mice is not known, it may be caused by metabolic and food absorption problems. Injections were performed in mice that were between 2 and 6 mo of age, most of the mice being 10–12 wk old. Rosa26LSL-YFP mice were obtained from the Jackson Laboratory (Gt(ROSA)26Sortm1(EYFP)Cos/J). Subcutaneous serial transplantation of cells was performed in SCID mice, obtained from the Stanford mouse colony. Limiting dilution transplantation experiments were performed by injecting a 50:50 mix of cells/Matrigel (Invitrogen) into the flank of SCID/Il2r−/− mice (The Jackson Laboratory). DAPT was solubilized in corn oil and injected at a concentration of 10 mg/kg/d in the peritoneal cavity. All the experiments with mice were approved by Stanford Institutional Animal Care and Use Committee.
Histology, immunostaining, and immunoblot analysis.
For paraffin sections, organs were fixed in 4% paraformaldehyde overnight at room temperature and then transferred to 70% ethanol before processing and embedding. For cryosections, organs were embedded and frozen in OCT using liquid nitrogen–cooled isopentane. For pathological analysis, paraffin sections were stained with hematoxylin and eosin (H&E). Tumor quantification was performed with the BIOQUANT Life Science software (BIOQUANT Image Analysis Corp.). Antigen retrieval on paraffin sections was performed using the Trilogy solution (Cell Marque) for 15 min in a pressure cooker. Cryosections were thawed at room temperature for at least 20 min and fixed with 4% formaldehyde for 15 min. Sections labeled with C3, C7, and E10 antibodies were fixed in 100% acetone at −20°C for 10 min. All sections were then in brief washed in PBS + 0.1% Tween 20 (PBST) and then blocked for 1 h at room temperature in PBST with 10% normal horse serum and 1% bovine serum Albumin. Sections were incubated with primary antibodies in block solution overnight at room temperature, washed in PBST, and then incubated with secondary antibodies diluted 1:500 in block solution for 1 h at room temperature. Primary antibodies used were anti-Ki67 (BD), anti-CK19 (Dako), anti-Albumin (Bethyl Laboratories, Inc.), anti-A6 (a gift from V. Factor, National Cancer Institute, National Institutes of Health, Bethesda, MD), anti-Sca1 (eBioscience), anti-EpCAM (BD), anti-GS (BD), and rat antibodies marking oval cells (Dorrell et al., 2008). Immunohistochemistry for Notch pathway components was performed as described previously (Mazur et al., 2010a,b) using antibodies recognizing active Notch1 (Abcam), Notch2 (Development Studies Hybridoma Bank), and HES1 (gift from T. Sudo, Toray Industries, Inc., Kamakura, Japan). Protein expression was analyzed by immunoblotting, as described previously (Ho et al., 2009). Primary antibodies used were anti-GAPDH (Santa Cruz Biotechnology, Inc.) and anti–cleaved Notch1 (Cell Signaling Technology).
Cell preparation and culture.
To obtain single cell suspensions, livers were first perfused with a mixture of 1 mg/ml Collagenase A/D and 1 mg/ml DNaseI (Roche), mechanically minced in small pieces, and allowed to incubate for 1 h with the Collagenase/DNaseI mixture. Cells were then dispersed by manual pipetting and filtered, and red blood cells were lysed with the ACK buffer (NH4Cl/KHCO3). Liver cells were cultured as described previously (Dorrell et al., 2011) in 6- or 24-well plates coated with Collagen IV (BD). EGF was added at 20 µg/ml and HGF at 5 µg/ml. Colonies were stained with 1% crystal violet 8 d after plating. To obtain nonparenchymal cells, single cell suspensions were centrifuged at low speed (400 rpm); the pellet, enriched for mature hepatocytes (parenchymal fraction), was discarded. The supernatant was then collected and processed for centrifugation at normal speed (1,200 rpm) to pellet nonparenchymal cells. Tumor cell lines were derived from subcutaneously transplanted primary tumors from TKO mice and grown in RPMI supplemented with bovine growth serum; one cell line was confirmed to be TKO by RT-PCR, whereas one WT p130 allele was retained in the second line. HepG2, SNU-449, and Hep3B human cell lines were grown in RPMI supplemented with bovine growth serum. Retroviral infection of liver cells was performed by transfecting 293T cells with MigR1-IRES-GFP or MigR1-ICN-IRES-GFP (a gift of W.S. Pear, University of Pennsylvania, Philadelphia, PA), together with packaging plasmids (Zweidler-McKay et al., 2005). Infected HCC cells were sorted on a FACSVantage cell sorter (BD) for GFP expression 1 wk after infection. Reporter assays were performed by transfecting 1 µg of reporter plasmid (mHes1-Luc [a gift from R.S. Slack, University of Ottawa, Ottawa, Ontario, Canada] and hNotch1-Luc [a gift from T. Kiyono, National Cancer Center Research Institute, Tokyo, Japan]; Yugawa et al., 2007) in SNU-449 cells, together with 0.2 or 1 µg of expression vectors for E2F1 and E2F3 in 12-wells plates. Transfection efficiency was normalized by cotransfection of the Renilla vector, and luminescence was detected with the Dual Reporter Assay System kit (Promega). Each experiment was performed in triplicate, and three independent experiments were conducted. The RB reintroduction experiment was performed using a plasmid expressing a fusion protein between GFP and a large pocket fragment of RB with mutations in seven CDK phosphorylation sites (GFP-RB7LP [a gift of E.S. Knudsen, Thomas Jefferson University, Philadelphia, PA]; Jiao et al., 2006).
ChIP analysis.
ChIP was performed as described previously (O’Geen et al., 2006) with cultured hepatic TKO tumor cells. Antibodies used for immunoprecipitations were as follows: E2F1 (sc-193X), E2F3 (sc-878X), E2F4 (sc-1082X), and p16 (sc-467). Cyclin A, Notch1, Notch3, Notch4, Dtx3, Dtx4, and Jag2 promoter binding was assessed by quantitative PCR using SYBR green ER Master mix (Invitrogen).
RT-PCR and microarray analysis.
RNA was extracted either from frozen tumors or from cells sorted directly in TRIZOL. RNAs were processed, and qPCR reactions were prepared as described previously (Viatour et al., 2008). Microarray data were normalized with Expression Console software (Affymetrix), using robust means analysis algorithms. Low signals (<50) were filtered out using the PreprocessDataset module in GenePattern (http://www.broadinstitute.org/cancer/software/genepattern/). Differentially expressed genes in TKO mice were identified using SAM software (Tusher et al., 2001) with the cutoff FDR of 5%. Array files are available on the GEO DataSets database under accession no. GSE19004.
To compare TKO array (Affymetrix oligo) with mouse arrays from Lee et al. (2004), TKO data were collapsed to gene symbols and then converted to the format equivalent to two-color array by taking the log2 of expression ratio (TKO signal vs. mean of control signal). Log ratio data of genes present in both platforms were used in subsequent analysis. To further compare TKO arrays with human HCC microarrays (Lee et al., 2004; Boyault et al., 2007), we selected orthologous genes that were present in both arrays by using the mammalian orthology from the Mouse Genome Informatics database. In each tissue set, gene expression ratios were median centered for each gene. Hierarchical clustering (Eisen et al., 1998) was used to compare median-centered array data, using Pearson correlation in sample distance measure. DAVID analysis was performed as described previously (Huang et al., 2008). GSEA (Subramanian et al., 2005) was performed using GSEA version 2.0 software. Curated gene sets for GSEA analysis were obtained from http://www.broadinstitute.org/gsea/msigdb/collections.jsp#C2. Enrichment is considered significant when the p-value is <0.05 and the FDR is <0.25.
For the survival analysis, RNA from 53 human HCCs was obtained from Caucasian and Chinese patients, representing survival group A and B HCC patients. The HCC samples were hybridized to Illumina bead chips HumanRef8v2 and analyzed according to the manufacturer’s recommendation. All human tissue samples were obtained with approval by the Institutional Review Board of the National Institutes of Health and collaborating institutions on the condition that all samples were anonymized. The ability of the Notch family genes to predict overall survival of HCC patients were analyzed by retrieving the 17 genes in the Notch signature from the HCC dataset. Hierarchical cluster analysis identified two clinically relevant distinct subgroups of human HCCs based on the 17-gene expression profile. Kaplan-Meier plots and Mantel-Cox statistical analysis were applied in the survival analysis (Prism 5.01; GraphPad Software).
Flow cytometry.
Single cell suspensions were stained with Annexin V (Roche) or antibodies against Ter119, CD45, and Sca1 (eBioscience), as well as unconjugated antibodies recognizing oval cells (Dorrell et al., 2008). Anti–rat PE (eBioscience) was used as a secondary antibody. All analysis and sorting were performed as described previously (Viatour et al., 2008).
Statistical analysis.
Statistical significance was assayed by the two-tailed Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.005). Data are represented as mean ± SEM.
Online supplemental material.
Fig. S1 is related to Fig. 1 and further characterizes the tumors that develop in the liver of TKO mice after the inactivation of the Rb gene family. Fig. S2 is related to Fig. 2 and further defines the lesions that appear around the portal triad shortly after the inactivation of the Rb gene family. Fig. S3 is related to Fig. 3 and focuses on the proliferation and transplantation potential of liver progenitor cells in TKO mice. Fig. S4 is related to Figs. 5 and 6 and describes the expression of Notch pathway members in TKO and WT liver cells. Fig. S5 is related to Fig. 6 and shows the expression of Notch pathway members in early lesion and established tumors, the consequence of Notch signaling inhibition in early lesions, and clinical features of HCC patients based on Notch pathway expression. Table S1 displays a list of genes of interest that are dysregulated in TKO tumors versus control livers. Table S2, included as a separate Excel file, shows the entire list of genes that are dysregulated in TKO tumors versus control livers. Table S3, included as a separate Excel file, indicates the gene sets that are enriched in the TKO dataset. The supplemental text lists the genes included in the gene sets mentioned in Fig. 4 D and shows the genes that are included in group M in Fig. 4 A. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110198/DC1.
Acknowledgments
We thank Drs. S.E. Artandi, D.W. Felsher, T.A. Rando, D.I. Bellovin, and M. Cleary for critical reading and helpful discussions, Drs. V. Factor, T. Kiyono, W.S. Pear, R.S. Slack, and E.S. Knudsen for the generous gift of the A6 antibody, the hNotch1-Luc, the MigR1-ICN-GFP, the mHes1-Luc, and the RB-GFP constructs, respectively, and Dr. T. Sudo for the kind gift of the HES1 antibody. The Notch2 antibody was developed by Dr. S. Artavanis-Tsakonas.
This work was supported by the Lucile Packard Foundation for Children’s Health, the Damon Runyon Cancer Research Foundation, and National Institutes of Health–National Cancer Institute grant RO1 CA114102 (to J. Sage), as well as fellowships from Human Frontier Science Program, European Molecular Biology Organization, the Fonds de la Recherche Scientifique, and the Leon Fredericq Foundation (to P. Viatour) and a Dr. Mildred Scheel fellowship from the Deutsche Krebshilfe (to U. Ehmer).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CDK
- Cyclin-dependent kinase
- ChIP
- chromatin immunoprecipitation
- cTKO
- conditional TKO
- DDC
- 3,5-diethoxycarbonyl-1,4-dihydrocollidine
- EGF
- epidermal growth factor
- FDR
- false discovery rate
- GS
- glutamine synthase
- GSEA
- gene set enrichment analysis
- HCC
- hepatocellular carcinoma
- HGF
- hepatocyte growth factor
- ICD
- intracellular domain
- mRNA
- messenger RNA
- RB
- retinoblastoma
- RT-qPCR
- quantitative RT-PCR
- Tam
- tamoxifen
- TKO
- triple KO
References
- Benhamouche S., Curto M., Saotome I., Gladden A.B., Liu C.H., Giovannini M., McClatchey A.I. 2010. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 24:1718–1730 10.1101/gad.1938710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault S., Rickman D.S., de Reyniès A., Balabaud C., Rebouissou S., Jeannot E., Hérault A., Saric J., Belghiti J., Franco D., et al. 2007. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 45:42–52 10.1002/hep.21467 [DOI] [PubMed] [Google Scholar]
- Burkhart D.L., Sage J. 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer. 8:671–682 10.1038/nrc2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart D.L., Ngai L.K., Roake C.M., Viatour P., Thangavel C., Ho V.M., Knudsen E.S., Sage J. 2010. Regulation of RB transcription in vivo by RB family members. Mol. Cell. Biol. 30:1729–1745 10.1128/MCB.00952-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell S., Park S.H. 2009. The epidemiology of hepatocellular cancer: from the perspectives of public health problem to tumor biology. J. Gastroenterol. 44:96–101 10.1007/s00535-008-2258-6 [DOI] [PubMed] [Google Scholar]
- Calo E., Quintero-Estades J.A., Danielian P.S., Nedelcu S., Berman S.D., Lees J.A. 2010. Rb regulates fate choice and lineage commitment in vivo. Nature. 466:1110–1114 10.1038/nature09264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A., Wang X., Zhang C., McCurrach M., Zhao Z., Mert O., Dickins R.A., Narita M., Zhang M., Lowe S.W. 2010. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 17:376–387 10.1016/j.ccr.2010.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Curtis S.J., Roy D.M., Flesken-Nikitin A., Nikitin A.Y. 2010. Local mesenchymal stem/progenitor cells are a preferential target for initiation of adult soft tissue sarcomas associated with p53 and Rb deficiency. Am. J. Pathol. 177:2645–2658 10.2353/ajpath.2010.100306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E., Forbes S.J. 2009. The isolation and in vitro expansion of hepatic Sca-1 progenitor cells. Biochem. Biophys. Res. Commun. 381:549–553 10.1016/j.bbrc.2009.02.079 [DOI] [PubMed] [Google Scholar]
- Conner E.A., Lemmer E.R., Omori M., Wirth P.J., Factor V.M., Thorgeirsson S.S. 2000. Dual functions of E2F-1 in a transgenic mouse model of liver carcinogenesis. Oncogene. 19:5054–5062 10.1038/sj.onc.1203885 [DOI] [PubMed] [Google Scholar]
- Croquelois A., Blindenbacher A., Terracciano L., Wang X., Langer I., Radtke F., Heim M.H. 2005. Inducible inactivation of Notch1 causes nodular regenerative hyperplasia in mice. Hepatology. 41:487–496 10.1002/hep.20571 [DOI] [PubMed] [Google Scholar]
- Dorrell C., Erker L., Lanxon-Cookson K.M., Abraham S.L., Victoroff T., Ro S., Canaday P.S., Streeter P.R., Grompe M. 2008. Surface markers for the murine oval cell response. Hepatology. 48:1282–1291 10.1002/hep.22468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell C., Erker L., Schug J., Kopp J.L., Canaday P.S., Fox A.J., Smirnova O., Duncan A.W., Finegold M.J., Sander M., et al. 2011. Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 25:1193–1203 10.1101/gad.2029411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan A.W., Dorrell C., Grompe M. 2009. Stem cells and liver regeneration. Gastroenterology. 137:466–481 10.1053/j.gastro.2009.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edamoto Y., Hara A., Biernat W., Terracciano L., Cathomas G., Riehle H.M., Matsuda M., Fujii H., Scoazec J.Y., Ohgaki H. 2003. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int. J. Cancer. 106:334–341 10.1002/ijc.11254 [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 95:14863–14868 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt N.V., Factor V.M., Medvinsky A.L., Baranov V.N., Lazareva M.N., Poltoranina V.S. 1993. Common antigen of oval and biliary epithelial cells (A6) is a differentiation marker of epithelial and erythroid cell lineages in early development of the mouse. Differentiation. 55:19–26 10.1111/j.1432-0436.1993.tb00029.x [DOI] [PubMed] [Google Scholar]
- Erker L., Grompe M. 2008. Signaling networks in hepatic oval cell activation. Stem Cell Res. (Amst.). 1:90–102 10.1016/j.scr.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Farazi P.A., DePinho R.A. 2006. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer. 6:674–687 10.1038/nrc1934 [DOI] [PubMed] [Google Scholar]
- Gebhardt R., Hovhannisyan A. 2010. Organ patterning in the adult stage: the role of Wnt/beta-catenin signaling in liver zonation and beyond. Dev. Dyn. 239:45–55 [DOI] [PubMed] [Google Scholar]
- Geisler F., Nagl F., Mazur P.K., Lee M., Zimber-Strobl U., Strobl L.J., Radtke F., Schmid R.M., Siveke J.T. 2008. Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology. 48:607–616 10.1002/hep.22381 [DOI] [PubMed] [Google Scholar]
- Hanlon L., Avila J.L., Demarest R.M., Troutman S., Allen M., Ratti F., Rustgi A.K., Stanger B.Z., Radtke F., Adsay V., et al. 2010. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res. 70:4280–4286 10.1158/0008-5472.CAN-09-4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haybaeck J., Zeller N., Wolf M.J., Weber A., Wagner U., Kurrer M.O., Bremer J., Iezzi G., Graf R., Clavien P.A., et al. 2009. A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell. 16:295–308 10.1016/j.ccr.2009.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellström M., Phng L.K., Hofmann J.J., Wallgard E., Coultas L., Lindblom P., Alva J., Nilsson A.K., Karlsson L., Gaiano N., et al. 2007. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 445:776–780 10.1038/nature05571 [DOI] [PubMed] [Google Scholar]
- Higashitsuji H., Itoh K., Nagao T., Dawson S., Nonoguchi K., Kido T., Mayer R.J., Arii S., Fujita J. 2000. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat. Med. 6:96–99 10.1038/71600 [DOI] [PubMed] [Google Scholar]
- Ho V.M., Schaffer B.E., Karnezis A.N., Park K.S., Sage J. 2009. The retinoblastoma gene Rb and its family member p130 suppress lung adenocarcinoma induced by oncogenic K-Ras. Oncogene. 28:1393–1399 10.1038/onc.2008.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J.J., Zovein A.C., Koh H., Radtke F., Weinmaster G., Iruela-Arispe M.L. 2010. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development. 137:4061–4072 10.1242/dev.052118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes C., Stanford W.L. 2007. Concise review: stem cell antigen-1: expression, function, and enigma. Stem Cells. 25:1339–1347 10.1634/stemcells.2006-0644 [DOI] [PubMed] [Google Scholar]
- Hsieh Y.H., Su I.J., Wang H.C., Tsai J.H., Huang Y.J., Chang W.W., Lai M.D., Lei H.Y., Huang W. 2007. Hepatitis B virus pre-S2 mutant surface antigen induces degradation of cyclin-dependent kinase inhibitor p27Kip1 through c-Jun activation domain-binding protein 1. Mol. Cancer Res. 5:1063–1072 10.1158/1541-7786.MCR-07-0098 [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. 2008. DAVID gene ID conversion tool. Bioinformation. 2:428–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Deng T., Jones R., Li H., Herschkowitz J.I., Liu J.C., Weigman V.J., Tsao M.S., Lane T.F., Perou C.M., Zacksenhaus E. 2010. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J. Clin. Invest. 120:3296–3309 10.1172/JCI41490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W., Datta J., Lin H.M., Dundr M., Rane S.G. 2006. Nucleocytoplasmic shuttling of the retinoblastoma tumor suppressor protein via Cdk phosphorylation-dependent nuclear export. J. Biol. Chem. 281:38098–38108 10.1074/jbc.M605271200 [DOI] [PubMed] [Google Scholar]
- Kim J., Woo A.J., Chu J., Snow J.W., Fujiwara Y., Kim C.G., Cantor A.B., Orkin S.H. 2010a. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 143:313–324 10.1016/j.cell.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Jung J.K., Lee S.Y., Jang K.L. 2010b. Hepatitis B virus X protein overcomes stress-induced premature senescence by repressing p16(INK4a) expression via DNA methylation. Cancer Lett. 288:226–235 10.1016/j.canlet.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Knudsen E.S., Knudsen K.E. 2008. Tailoring to RB: tumour suppressor status and therapeutic response. Nat. Rev. Cancer. 8:714–724 10.1038/nrc2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossatz U., Breuhahn K., Wolf B., Hardtke-Wolenski M., Wilkens L., Steinemann D., Singer S., Brass F., Kubicka S., Schlegelberger B., et al. 2010. The cyclin E regulator cullin 3 prevents mouse hepatic progenitor cells from becoming tumor-initiating cells. J. Clin. Invest. 120:3820–3833 10.1172/JCI41959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent-Puig P., Zucman-Rossi J. 2006. Genetics of hepatocellular tumors. Oncogene. 25:3778–3786 10.1038/sj.onc.1209547 [DOI] [PubMed] [Google Scholar]
- Lee J.S., Thorgeirsson S.S. 2006. Comparative and integrative functional genomics of HCC. Oncogene. 25:3801–3809 10.1038/sj.onc.1209561 [DOI] [PubMed] [Google Scholar]
- Lee J.S., Chu I.S., Mikaelyan A., Calvisi D.F., Heo J., Reddy J.K., Thorgeirsson S.S. 2004. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat. Genet. 36:1306–1311 10.1038/ng1481 [DOI] [PubMed] [Google Scholar]
- Li L., Krantz I.D., Deng Y., Genin A., Banta A.B., Collins C.C., Qi M., Trask B.J., Kuo W.L., Cochran J., et al. 1997. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 16:243–251 10.1038/ng0797-243 [DOI] [PubMed] [Google Scholar]
- Libbrecht L. 2006. Hepatic progenitor cells in human liver tumor development. World J. Gastroenterol. 12:6261–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson D. 2008. Insights from mouse models into human retinoblastoma. Cell Div. 3:9 10.1186/1747-1028-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew C.N., Bosco E.E., Fox S.R., Okaya T., Tarapore P., Schwemberger S.J., Babcock G.F., Lentsch A.B., Fukasawa K., Knudsen E.S. 2005. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 65:4568–4577 10.1158/0008-5472.CAN-04-4221 [DOI] [PubMed] [Google Scholar]
- Mazur P.K., Einwächter H., Lee M., Sipos B., Nakhai H., Rad R., Zimber-Strobl U., Strobl L.J., Radtke F., Klöppel G., et al. 2010a. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc. Natl. Acad. Sci. USA. 107:13438–13443 10.1073/pnas.1002423107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur P.K., Grüner B.M., Nakhai H., Sipos B., Zimber-Strobl U., Strobl L.J., Radtke F., Schmid R.M., Siveke J.T. 2010b. Identification of epidermal Pdx1 expression discloses different roles of Notch1 and Notch2 in murine Kras(G12D)-induced skin carcinogenesis in vivo. PLoS ONE. 5:e13578 10.1371/journal.pone.0013578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L., He B., Hui L. 2011. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin. Cancer Biol. 21:10–20 10.1016/j.semcancer.2010.10.011 [DOI] [PubMed] [Google Scholar]
- Mishra L., Banker T., Murray J., Byers S., Thenappan A., He A.R., Shetty K., Johnson L., Reddy E.P. 2009. Liver stem cells and hepatocellular carcinoma. Hepatology. 49:318–329 10.1002/hep.22704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munakata T., Liang Y., Kim S., McGivern D.R., Huibregtse J., Nomoto A., Lemon S.M. 2007. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 3:e139 10.1371/journal.ppat.0030139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M., Wolfer A., Raj K., Kummer J.A., Mill P., van Noort M., Hui C.C., Clevers H., Dotto G.P., Radtke F. 2003. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 33:416–421 10.1038/ng1099 [DOI] [PubMed] [Google Scholar]
- Nordenstedt H., White D.L., El-Serag H.B. 2010. The changing pattern of epidemiology in hepatocellular carcinoma. Dig. Liver Dis. 42:S206–S214 10.1016/S1590-8658(10)60507-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H., Nicolet C.M., Blahnik K., Green R., Farnham P.J. 2006. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 41:577–580 10.2144/000112268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel M., Shafritz D.A. 2008. Stem cells, cell transplantation and liver repopulation. Biochim. Biophys. Acta. 1782:61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Kimura M.T., Tan D., Fujiwara K., Igarashi J., Makuuchi M., Hui A.M., Tsurumaru M., Nagase H. 2005. Frequent trefoil factor 3 (TFF3) overexpression and promoter hypomethylation in mouse and human hepatocellular carcinomas. Int. J. Oncol. 26:369–377 [PMC free article] [PubMed] [Google Scholar]
- Onoyama I., Suzuki A., Matsumoto A., Tomita K., Katagiri H., Oike Y., Nakayama K., Nakayama K.I. 2011. Fbxw7 regulates lipid metabolism and cell fate decisions in the mouse liver. J. Clin. Invest. 121:342–354 10.1172/JCI40725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.G., Chung C., Kang H., Kim J.Y., Jung G. 2006. Up-regulation of cyclin D1 by HBx is mediated by NF-kappaB2/BCL3 complex through kappaB site of cyclin D1 promoter. J. Biol. Chem. 281:31770–31777 10.1074/jbc.M603194200 [DOI] [PubMed] [Google Scholar]
- Pirot P., van Grunsven L.A., Marine J.C., Huylebroeck D., Bellefroid E.J. 2004. Direct regulation of the Nrarp gene promoter by the Notch signaling pathway. Biochem. Biophys. Res. Commun. 322:526–534 10.1016/j.bbrc.2004.07.157 [DOI] [PubMed] [Google Scholar]
- Qi R., An H., Yu Y., Zhang M., Liu S., Xu H., Guo Z., Cheng T., Cao X. 2003. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 63:8323–8329 [PubMed] [Google Scholar]
- Ranganathan P., Weaver K.L., Capobianco A.J. 2011. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer. 11:338–351 10.1038/nrc3035 [DOI] [PubMed] [Google Scholar]
- Reed C.A., Mayhew C.N., McClendon A.K., Yang X., Witkiewicz A., Knudsen E.S. 2009. RB has a critical role in mediating the in vivo checkpoint response, mitigating secondary DNA damage and suppressing liver tumorigenesis initiated by aflatoxin B1. Oncogene. 28:4434–4443 10.1038/onc.2009.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskams T.A., Libbrecht L., Desmet V.J. 2003. Progenitor cells in diseased human liver. Semin. Liver Dis. 23:385–396 10.1055/s-2004-815564 [DOI] [PubMed] [Google Scholar]
- Roskams T., Katoonizadeh A., Komuta M. 2010. Hepatic progenitor cells: an update. Clin. Liver Dis. 14:705–718 10.1016/j.cld.2010.08.003 [DOI] [PubMed] [Google Scholar]
- Satow R., Shitashige M., Kanai Y., Takeshita F., Ojima H., Jigami T., Honda K., Kosuge T., Ochiya T., Hirohashi S., Yamada T. 2010. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin. Cancer Res. 16:2518–2528 10.1158/1078-0432.CCR-09-2214 [DOI] [PubMed] [Google Scholar]
- Schmelzer E., Wauthier E., Reid L.M. 2006. The phenotypes of pluripotent human hepatic progenitors. Stem Cells. 24:1852–1858 10.1634/stemcells.2006-0036 [DOI] [PubMed] [Google Scholar]
- Shin S., Walton G., Aoki R., Brondell K., Schug J., Fox A., Smirnova O., Dorrell C., Erker L., Chu A.S., et al. 2011. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 25:1185–1192 10.1101/gad.2027811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohda T., Iwata K., Soejima H., Kamimura S., Shijo H., Yun K. 1998. In situ detection of insulin-like growth factor II (IGF2) and H19 gene expression in hepatocellular carcinoma. J. Hum. Genet. 43:49–53 10.1007/s100380050036 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susick R., Moss N., Kubota H., Lecluyse E., Hamilton G., Luntz T., Ludlow J., Fair J., Gerber D., Bergstrand K., et al. 2001. Hepatic progenitors and strategies for liver cell therapies. Ann. N. Y. Acad. Sci. 944:398–419 10.1111/j.1749-6632.2001.tb03851.x [DOI] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 98:5116–5121 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk-Lingbeek M.E., Bruggeman S.W., van Lohuizen M. 2004. Stem cells and cancer; the polycomb connection. Cell. 118:409–418 10.1016/j.cell.2004.08.005 [DOI] [PubMed] [Google Scholar]
- van Harn T., Foijer F., van Vugt M., Banerjee R., Yang F., Oostra A., Joenje H., te Riele H. 2010. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 24:1377–1388 10.1101/gad.580710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderluit J.L., Ferguson K.L., Nikoletopoulou V., Parker M., Ruzhynsky V., Alexson T., McNamara S.M., Park D.S., Rudnicki M., Slack R.S. 2004. p107 regulates neural precursor cells in the mammalian brain. J. Cell Biol. 166:853–863 10.1083/jcb.200403156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderluit J.L., Wylie C.A., McClellan K.A., Ghanem N., Fortin A., Callaghan S., MacLaurin J.G., Park D.S., Slack R.S. 2007. The Retinoblastoma family member p107 regulates the rate of progenitor commitment to a neuronal fate. J. Cell Biol. 178:129–139 10.1083/jcb.200703176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P., Somervaille T.C., Venkatasubrahmanyam S., Kogan S., McLaughlin M.E., Weissman I.L., Butte A.J., Passegué E., Sage J. 2008. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 3:416–428 10.1016/j.stem.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva A., Newell P., Chiang D.Y., Friedman S.L., Llovet J.M. 2007. Genomics and signaling pathways in hepatocellular carcinoma. Semin. Liver Dis. 27:55–76 10.1055/s-2006-960171 [DOI] [PubMed] [Google Scholar]
- Wang W.H., Hullinger R.L., Andrisani O.M. 2008. Hepatitis B virus X protein via the p38MAPK pathway induces E2F1 release and ATR kinase activation mediating p53 apoptosis. J. Biol. Chem. 283:25455–25467 10.1074/jbc.M801934200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker S., Marais R., Zhu A.X. 2010. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 29:4989–5005 10.1038/onc.2010.236 [DOI] [PubMed] [Google Scholar]
- Williams B.O., Schmitt E.M., Remington L., Bronson R.T., Albert D.M., Weinberg R.A., Jacks T. 1994. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 13:4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yovchev M.I., Grozdanov P.N., Zhou H., Racherla H., Guha C., Dabeva M.D. 2008. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology. 47:636–647 10.1002/hep.22047 [DOI] [PubMed] [Google Scholar]
- Yugawa T., Handa K., Narisawa-Saito M., Ohno S., Fujita M., Kiyono T. 2007. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol. Cell. Biol. 27:3732–3742 10.1128/MCB.02119-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Theise N., Chua M., Reid L.M. 2008. The stem cell niche of human livers: symmetry between development and regeneration. Hepatology. 48:1598–1607 10.1002/hep.22516 [DOI] [PubMed] [Google Scholar]
- Zhang Y.J., Rossner P., Jr, Chen Y., Agrawal M., Wang Q., Wang L., Ahsan H., Yu M.W., Lee P.H., Santella R.M. 2006. Aflatoxin B1 and polycyclic aromatic hydrocarbon adducts, p53 mutations and p16 methylation in liver tissue and plasma of hepatocellular carcinoma patients. Int. J. Cancer. 119:985–991 10.1002/ijc.21699 [DOI] [PubMed] [Google Scholar]
- Zong Y., Panikkar A., Xu J., Antoniou A., Raynaud P., Lemaigre F., Stanger B.Z. 2009. Notch signaling controls liver development by regulating biliary differentiation. Development. 136:1727–1739 10.1242/dev.029140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweidler-McKay P.A., He Y., Xu L., Rodriguez C.G., Karnell F.G., Carpenter A.C., Aster J.C., Allman D., Pear W.S. 2005. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 106:3898–3906 10.1182/blood-2005-01-0355 [DOI] [PMC free article] [PubMed] [Google Scholar]