Polyketides isolated from fungi are typically derived from “iterative” Type I (multidomainal) polyketide synthases (PKSs), where individual catalytic domains are fused into single large proteins, and reused a fixed number of times in the “programmed” synthesis of a given product [1, 2]. The best known examples of iterative Type I enzymes are the yeast and animal fatty acid synthases (FASs) [3]. Animal fatty acid biosynthesis is initiated by acetyl-CoA, which is brought onto the enzyme by a bifunctional acyl transferase, malonyl-CoA:acyl-carrier protein (ACP) transacylase (MAT), which primarily shuttles units of malonyl-CoA for each successive two-carbon homologation leading to palmitate [4]. On the other hand, in fungi an α6β6 heterododecameric FAS complex harbors a specific acetyl transacylase for starter unit introduction [5]. In classical precursor incorporation experiments with fatty acids and fungal natural products it was commonly observed that an acetate starter unit would bear a higher specific incorporation of radiolabel from [14C]-acetate than the rest of the labeled sites in the molecule, in keeping with the intermediary conversion of acetyl-CoA to malonyl-CoA for each chain extension [6]. Conversely, if the complementary incorporation of [14C]-malonate were examined, the starter carbons would often be distinguishably less labeled than the remaining portions of the polyketide metabolite. This general pattern came to be known as the “starter unit effect” [7].
Application of the UMA algorithm to select interdomainal cut sites in PksA (GenBank accession no. AY371490), the iterative Type I PKS central to the biosynthesis of the mycotoxin aflatoxin, revealed two previously unrecognized, but clearly resolved domains [8]. The large N-terminal domain was recently established to be a starter unit:ACP transacylase (SAT) that selectively introduced hexanoyl starter units onto the PksA ACP to prime norsolorinic acid (1) biosynthesis [9]. Search of genome databases revealed that SAT domains were widespread among known and apparent nonreducing fungal PKSs. PksA is unusual among fungal PKSs in that the rare hexanoyl starter unit is supplied by a dedicated yeast-like pair of FAS subunits, HexA and HexB [10, 11]. The vast majority of fungal polyketides, however, are synthesized from acetyl-CoA and malonyl-CoA. These enzymes require no specialized apparatus to prepare a starter unit when these rudimentary building blocks are already available in the cell. In this paper we compare four related nonreducing iterative PKSs (none having an associated FAS pair) and demonstrate that the SAT domain in each selectively transfers acetyl-CoA in preference to higher CoA homologs or malonyl-CoA, while literature precedent and a representative example indicate the MAT domains present in these systems are restricted to malonyl transfer.
To test the proposed role of SAT domains, we examined established PKS pathways in Aspergillus nidulans, Colletotrichum orbiculare (also known as C. lagenarium), Cercospora nicotianae, and Gibberella fujikuroi. The presumed SAT monodomains from WA (Q03149), PKS1 (BAA18956), CTB1 (AAT69682), and PKS4 (CAB92399), required in the biosynthesis of the naphthopyrone YWA1 (2) [12], 1,3,6,8-tetrahydroxynaphthalene (THN, 4) [13], cercosporin (5) [14], and bikaverin (6) [15], respectively (Figure 1), were separately cloned, expressed in E. coli, and purified. The coding sequence for the SAT domains in these PKSs spans the first three exons. Exons 1 and 2 were codon-optimized to facilitate enhanced expression in E. coli [16, 17]. The third exon fragment was amplified from the gDNA, fused to the codon-optimized fragment by either ligation or overlap extension PCR, and inserted into pET24a for robust expression in BL21(DE3). Based on the available sequence data, the SAT domain in PKS4 (G. fujikuroi, CAB92399) lacks the coding sequence for the required Cys in the GXCXG motif expected for SAT domains. This Cys covalently tethers the starter unit during transfer [9]. Upon re-sequencing exons 1 and 2, a frame-shift mutation was discovered in the published sequence. Gene structure prediction using the revised sequence resulted in an alternative splicing pattern that translated through the GXCXG motif (FGENESH, www.softberry.com, Supporting Information).
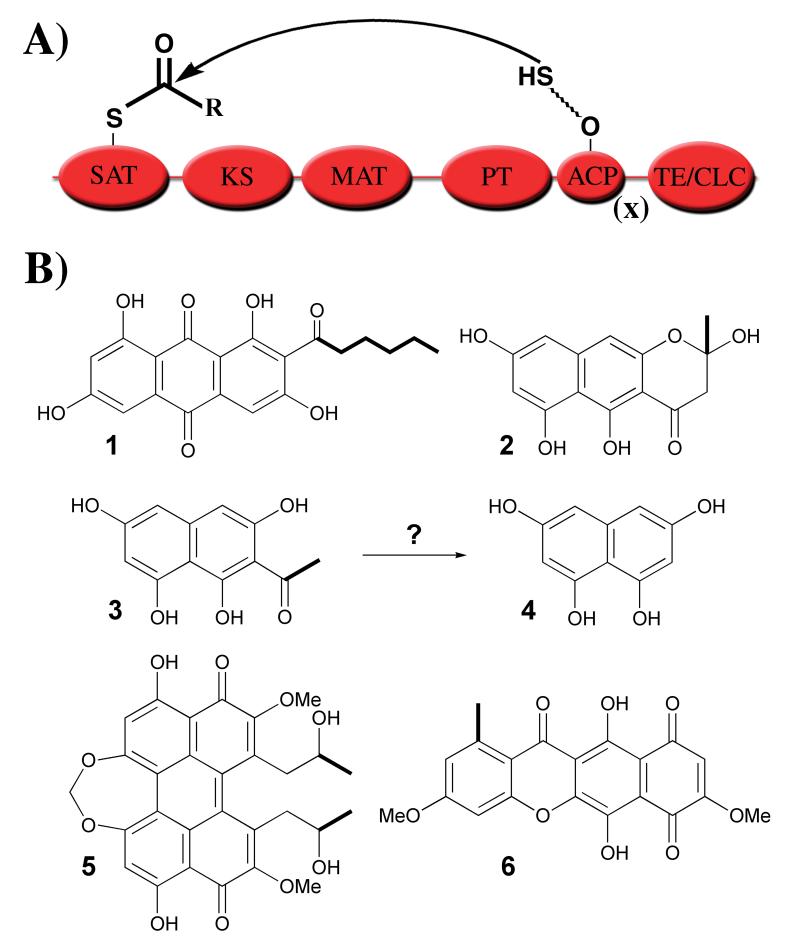
Figure 1.
A. Domain architecture. Domains include: starter unit:ACP transacylase (SAT), ketosynthase (KS), malonyl-CoA:ACP transacylase (MAT), putative product template (PT) [8], acyl-carrier protein (ACP), and thioesterase/Claisen cyclase (TE/CLC). (x) = 1 ACP for PksA, CTB1, and PKS4. (x) = 2 ACPs for WA and PKS1. (R) = methyl or pentyl. B. Selected fungal polyketide products. Compounds include norsolorinic acid (1), naphthopyrone YWA1 (2), acetyl-tetrahydroxynaphthalene (3), 1,3,6,8-tetrahydroxynaphthalene (THN, 4), cercosporin (5), and bikaverin (6). Starter units are indicated in bold.
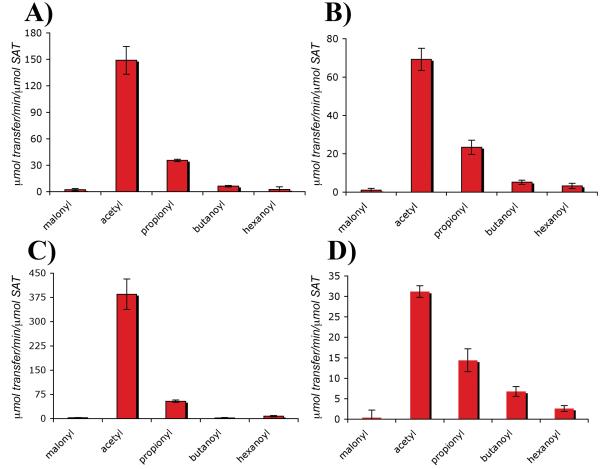
Employing the chromatographic assay previously used to examine acyl transfer in PksA [9], we found that the SAT domains from WA, PKS1, CTB1, and the revised PKS4 preferentially accepted acetyl and transferred it onto pantetheine, the biochemically relevant part of the holo-ACP arm. All of the SAT domains transferred acetyl faster (Figure 2) than the PksA SAT domain transferred hexanoyl (30.4 ± 0.6 μmol transfer/min/μmol SAT, hexanoyl-CoA [9]), the biosynthetically required starter unit for norsolorinic acid (1) biosynthesis, under identical conditions. Malonyl, however, was not accepted or transferred. Propionyl-CoA, while rarely, if ever, seen as a starter in fungal systems, was weakly accepted over any longer chain fatty-acyl CoAs (butanoyl- or hexanoyl-CoA) demonstrating minimal tolerance for alternative chain lengths. In a relevant precedent, Mosbach carried out the classic perturbation of the acetyl starter unit effect by administering [1-14C]propionate (0.24 mM) to Penicillium baarnense [18]. Even in this whole-cell experiment a new metabolite was formed, homoorsellinate, in a 1:5 ratio with orsellinate, which is normally produced. Stepwise degradation showed the former arose specifically from incorporation of a propionyl starter. Controls lacking SAT domains gave no detectable acyl transfer under the conditions of the experiment. However, low extents of spontaneous acyl transfer can take place only after extended periods of time. The SAT monodomains from WA, PKS1, CTB1, and PKS4 were shown by autoradiography to be able to covalently accept and transfer an acetyl group from [14C]-acetyl-CoA onto the PksA ACP monodomain, further supporting their roles in PKS initiation (Supporting Information).
Figure 2. SAT domain selectivity.
Rates are reported in μmol transfer/min/μmol SAT. A. WA SAT transfer B. PKS1 SAT transfer C. CTB1 SAT transfer D. PKS4 SAT transfer. Malonyl-CoA: WA, 2.2 ± 1.2; PKS1, 1.0 ± 1.0; CTB1, 2.6 ± 0.8; and PKS4, 0.4 ± 1.8. Acetyl-CoA: WA, 149 ± 15.7; PKS1, 69.2, ± 5.8; CTB1, 385 ± 47; and PKS4, 31.2 ± 1.4. Propionyl-CoA: WA 35.4 ± 1.3; PKS1, 23.4 ± 3.7; CTB1, 53.8 ± 3.8; and PKS4, 14.4 ± 2.8. Butanoyl-CoA: WA, 6.2 ± 0.8; PKS1 5.2 ± 1.1; CTB1, 2.2 ± 1.3; and PKS4, 6.8 ± 1.2. Hexanoyl-CoA: WA, 2.6 ± 3.0; PKS1, 3.2 ± 1.4; CTB1, 7.6 ± 2.1; and PKS4, 2.6 ± 0.7.
Acetyl specificity for the SAT domain in PKS1 was unexpected and conflicted with conclusions drawn from earlier biochemical experiments. Previously, PKS1 from C. lagenarium was heterologously expressed in Aspergillus oryzae, and the major product, THN (4), a precursor in fungal melanin biosynthesis, was proposed to be the direct PKS product [13]. Radiolabeled acetyl-CoA was employed in vitro with partially purified PKS1 to monitor incorporation into 4. An acetyl starter unit was not detected, and the authors proposed that malonyl-CoA was the starter as well as the extender unit [19]. In keeping with this study, we initially proposed that the SAT domain in PKS1 could either facilitate loading of the malonyl starter unit, act in tandem with the MAT, or be a non-functional domain. In the present study, however, the clear selectivity for acetyl-CoA suggests that an alternative mechanism could occur.
The dissected MAT monodomain from PksA was previously shown to load the ACP with the malonyl extender unit, but its expression was problematic [20]. Production of the corresponding MAT domains from WA, PKS1, CTB1, and PKS4 was similarly fraught with difficulties in expression, solubility, and activity. To circumvent these complications, it was resolved to express the PKS1 SAT-KS-MAT tridomain as a relevant representative example. The PKS1 SAT monodomain with the active site Cys mutated, C119A, exhibited a significantly reduced rate for acetyl transfer (8.0 ± 2.0 μmol/min/μmol SAT). Both the SAT and C119A mutant exhibited very similar spectral signatures when analyzed by CD in accord with retaining similar structures (Supporting Information). The mutated PksA SAT domain similarly showed a small amount of turnover for hexanoyl [9]. We rationalize this low extent of apparent activity to residual substrate binding to give effective concentrations sufficient for acyl transfer to occur. To monitor MAT selectivity, the PKS1 SAT-KS-MAT tridomain, with the SAT domain mutated (C119A), transferred malonyl (18,100 ± 2155) approximately 2,500-fold faster than acetyl (7.2 ± 1.9) suggesting that the MAT domain is highly selective for malonyl transfer. For PKS1, however, unlike PksA, which initiates synthesis specifically with a hexanoyl unit introduced via its SAT domain, decarboxylation (through hydration and loss of bicarbonate [21]) of malonyl-ACP to acetyl-ACP would provide a second avenue for the synthesis of acetyl-THN (3). This could be the primary route for PKSs that lack an active SAT domain [22]. For example, in vitro reconstitution of intact PKS4 with an inactivated SAT domain (CAB92399) did not result in detectable acetyl incorporation in the bikaverin (6) precursor, SMA76a [23], and decarboxylation of malonyl-ACP becomes the acting process in vitro. Notwithstanding, the evolutionary maintenance of SAT domains suggests that they play an important role in vivo. Further experimentation is required to determine the relative SAT contribution for acetyl starter unit introduction.
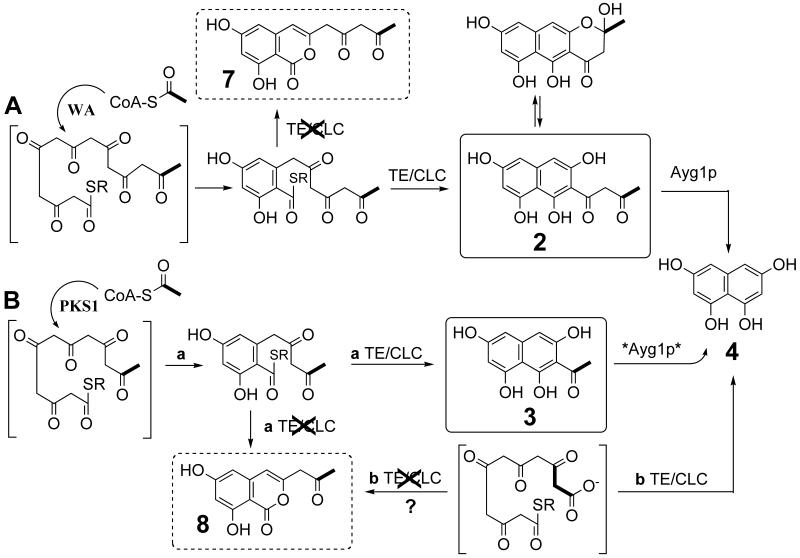
An alternative mechanism, however, can be envisioned that is in agreement with the loading of an acetyl starter unit by either initiation route in PKS1. In a parallel example, WA produces the heptaketide naphthopyrone YWA1 (2) when heterologously expressed in A. oryzae [12] (Scheme 1.A). Initially, a mutated copy of the wA gene from A. nidulans resulted in heterologous production of WA lacking the C-terminal domain and isolation of the heptaketide citreoisocoumarin 7 from the incorrect product release step [24]. Ebizuka and co-workers examined point mutants within this C-terminal domain, which has sequence similarity to thioesterases (TEs), and demonstrated that this TE-like domain acts as a Claisen cyclase (CLC) rather than a thioester hydrolase [25]. YWA1 (2) is then converted to THN (4) through a hydrolytic shortening reaction in Aspergillus fumigatus diverting this molecule into fungal melanin biosynthesis [26]. In vitro reactions enabled detailed mechanistic inquiry of the enzyme responsible for this reaction, Ayg1p, which also acts upon the hexaketide acetyl-THN (3), albeit with lower affinity, releasing the pentaketide THN [27]. Acetyl transfer by the SAT domain in PKS1 suggests initiation with an acetyl starter unit, which could account for the production of the hexaketide acetyl-THN (3) (Scheme 1.B). Supporting this alternative pathway, organic extracts from an inducible pks1 transformant in A. oryzae included 3 and 4 as well as several shorter chain derailment products [28]. It is important to note that by direct analogy to citreoisocoumarin 7 formation above, when the catalytic Ser in the TE/CLC domain was mutated, the incorrectly cyclized hexaketide isocoumarin 8 comprised 95% of the organic extract as opposed to the pentaketide isocoumarin. To account for this contradiction, the authors proposed that PKS1 accepted a malonyl starter unit and that the TE/CLC domain was responsible for chain length determination and cyclization control for the production of THN (Scheme 1.B.b). However, the present findings suggest that the hexaketide acetyl-THN (3) could be the direct polyketide product still bearing an acetyl starter unit (Scheme 1.B.a). A phenomenon similar to that catalyzed by Ayg1P could occur where 3 is converted to 4. As a consequence, both polyketide chain length control, and cyclization control of the first aromatic ring in fungal nonreducing PKSs would be carried out prior to the TE/CLC catalyzed product release.
Scheme 1.
A. Biosynthesis of the naphthopyrone YWA1 (2). The heptaketide is specifically cyclized into the naphthopyrone YWA1 (2) with a competent TE/CLC and into the citreoisocoumarin 7 without a competent TE/CLC. The partially cyclized open chain intermediate, which is common to the biosynthesis of both 2 and 7, is shown. Cyclization/aromatization of the first ring is carried out prior to the TE/CLC catalyzed product release. R = ACP. B. Biosynthesis of THN (3). a. Alternative proposal: acetyl-CoA incorporation into a hexaketide precursor accounts for the production of 3 and 4 with a competent TE/CLC, 8 without a competent TE/CLC, and an acetyl-starter. b. Current proposal: malonyl-CoA incorporation into a pentaketide precursor, giving rise to 4 with a competent TE/CLC and 8 without a competent TE/CLC. The latter is a contradictory situation and would require either a normal hexaketide precursor, or one relying on a malonate starter followed by decarboxylation to the pyrone 8. Proposal b does not account for SAT domain acetyl specificity.
Genome sequencing projects have revealed fungal species that have genes predicted to encode as many as 30 PKSs [29]. Only a small fraction of these have been correlated to structurally characterized natural products. We suggest a general pattern for iterative systems containing SAT domains that these typically activate acetyl-CoA to initiate polyketide biosynthesis and account for the observation of the classical “starter unit effect.” For the case of norsolorinic acid and other less common examples, dedicated FASs appear associated with the PKS to synthesize longer fatty acid chains to prime polyketide synthesis. Complementary to this role for the SAT domain, the existing experimental data suggest that MAT domains selectively activate malonyl units predominantly for chain extension, but decarboxylation of intermediary malonyl-ACPs can occur (as is known in Type II PKSs) to afford an alternate source of acetyl starter units. As data is accumulated in future experiments, the partitioning between SAT-derived acetyl and indirect decarboxylation from MAT-derived malonyl units, which may vary widely from case to case, will become clear.
Experimental Section
gDNA from C. orbiculare NRRL 28842 (pks1), A. nidulans RLMH67 (wA), C. nicotianae ATCC18366 (ctb1), and the imperfect form of G. fujikuroi, Fusarium fujikuroi M6884 (+) (pks4), were used as PCR templates. The predicted protein sequence data (NCBI) encoded by exons 1 and 2 in wA (Q03149), pks1 (BAA18956), ctb1 (AAT69682), and pks4 (CAB92399) were codon-optimized for E. coli using the GeMS software [17]. The synthons (synthetic gene fragments) with a 5′ NdeI site for cloning were assembled from standard 40-mer oligonucleotides using polymerase cycling assembly (PCA) [16] followed by PCR amplification (Supporting Information). The remaining portion of the SAT or SAT-KS-MAT within exon 3 was amplified from the gDNA templates including an engineered 5′ overlap to the codon-optimized fragment and a 3′ NotI site. The synthons were fused to the gDNA-amplified fragments by ligation or overlap extension PCR and inserted into pET24a (Novagen, Madison, WI) for expression. Upon discovery of the alternative coding sequence for PKS4, the gene was fixed by both gene synthesis and overlap extension PCR. The active site Cys119 codon in the PKS1 SAT monodomain and the PKS1 SAT-KS-MAT tridomain was mutated to Ala using overlap extension PCR. Protein production was carried out in BL21(DE3) E. coli and purified using nickel-nitrilotriacetic acid resin (Qiagen, Valencia, CA). The purified protein was dialyzed against potassium phosphate (100 mM, pH 7.0) for assay, and enzyme concentrations were determined with the Bradford assay in triplicate using bovine albumin as a standard. Chromatographic transacylase assays were carried out as previously described [9]. Methods and retention times are listed in the Supporting Information.
Supplementary Material
Acknowledgements
We thank Drs. K. O’Donnell, N. P. Keller, M. E. Daub, and R. H. Proctor for providing either spores or gDNA from C. orbiculare, A. nidulans, C. nicotianae, and G. fujikuroi, respectively. This work was supported by the National Institutes of Health Grant ES 001670.
References
- [1].Staunton J, Weissman KJ. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- [2].Rawlings BJ. Nat. Prod. Rep. 1999;16:425–484. doi: 10.1039/a900566h. [DOI] [PubMed] [Google Scholar]
- [3].Smith S, Tsai S-C. Nat. Prod. Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wakil SJ. Biochemistry. 1989;28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- [5].Tehlivets O, Scheuringer K, Kohlwein SD. Biochim. Biophys. Acta. 2007;1771:255–270. doi: 10.1016/j.bbalip.2006.07.004. [DOI] [PubMed] [Google Scholar]
- [6].Bentley R. Crit. Rev. Biotechnol. 1999;19:1–40. doi: 10.1080/0738-859991229189. [DOI] [PubMed] [Google Scholar]
- [7].Turner WB. Fungal Metabolites. Academic Press; London and New York: 1971. [Google Scholar]
- [8].Udwary DW, Merski M, Townsend CA. J. Mol. Biol. 2002;323:585–598. doi: 10.1016/s0022-2836(02)00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Crawford JM, Dancy BCR, Hill EA, Udwary DW, Townsend CA. Proc. Natl. Acad. Sci. USA. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Brown DW, Adams TH, Keller NP. Proc. Natl. Acad. Sci. USA. 1996;93:14873–14877. doi: 10.1073/pnas.93.25.14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hitchman T, Schmidt E, Trail F, Rarick M, Linz J, Townsend CA. Bioorg. Chem. 2001;29:293–307. doi: 10.1006/bioo.2001.1216. [DOI] [PubMed] [Google Scholar]
- [12].Watanabe A, Fujii I, Sankawa U, Mayorga M, Timberlake W, Ebizuka Y. Tetrahedron Lett. 1999;40:91–94. [Google Scholar]
- [13].Fujii I, Mori Y, Watanabe A, Kubo Y, Tsuji G, Ebizuka Y. Biosci. Biotechnol. Biochem. 1999;63:1445–1452. doi: 10.1271/bbb.63.1445. [DOI] [PubMed] [Google Scholar]
- [14].Choquer M, Dekkers KL, Chen HQ, Cao L, Ueng PP, Daub ME, Chung KR. Mol. Plant Microbe Interact. 2005;18:468–476. doi: 10.1094/MPMI-18-0468. [DOI] [PubMed] [Google Scholar]
- [15].Linnemannstons P, Schulte J, del Mar Prado M, Proctor RH, Avalos J, Tudzynski B. Fungal Genet. Biol. 2002;37:134–148. doi: 10.1016/s1087-1845(02)00501-7. [DOI] [PubMed] [Google Scholar]
- [16].Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Proc. Natl. Acad. Sci. USA. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jayaraj S, Reid R, Santi DV. Nucleic Acids Res. 2005;33:3011–3016. doi: 10.1093/nar/gki614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mosbach K. Acta. Chem. Scand. 1964;18:1591–1595. [Google Scholar]
- [19].Fujii I, Mori Y, Watanabe A, Kubo Y, Tsuji G, Ebizuka Y. Biochemistry. 2000;39:8853–8858. doi: 10.1021/bi000644j. [DOI] [PubMed] [Google Scholar]
- [20].Ma Y, Smith L, Cox RJ, Beltran-Alvarez P, Arthur CJ, Simpson TJ. ChemBioChem. 2006;7:1951–1958. doi: 10.1002/cbic.200600341. [DOI] [PubMed] [Google Scholar]
- [21].Witkowski A, Joshi AK, Smith S. Biochemistry. 2002;41:10877–10887. doi: 10.1021/bi0259047. [DOI] [PubMed] [Google Scholar]
- [22].Crawford JM, Vagstad AL, Ehrlich KC, Townsend CA. Bioorganic Chemistry. 2008 doi: 10.1016/j.bioorg.2007.11.002. doi:10.1016/j.bioorg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ma SM, Zhan J, Watanabe K, Xie X, Zhang W, Wang CC, Tang Y. J. Am. Chem. Soc. 2007;129:10642–10643. doi: 10.1021/ja074865p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Watanabe A, Ono Y, Fujii I, Sankawa U, Mayorga M, Timberlake W, Ebizuka Y. Tetrahedron Lett. 1998;39:7733–7736. [Google Scholar]
- [25].Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Chem. Biol. 2001;8:189–197. doi: 10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- [26].Tsai HF, Fujii I, Watanabe A, Wheeler MH, Chang YC, Yasuoka Y, Ebizuka Y, Kwon-Chung KJ. J. Biol. Chem. 2001;276:29292–29298. doi: 10.1074/jbc.M101998200. [DOI] [PubMed] [Google Scholar]
- [27].Fujii I, Yasuoka Y, Tsai HF, Chang YC, Kwon-Chung KJ, Ebizuka Y. J. Biol. Chem. 2004;279:44613–44620. doi: 10.1074/jbc.M406758200. [DOI] [PubMed] [Google Scholar]
- [28].Watanabe A, Ebizuka Y. Chem. Biol. 2004;11:1101–1106. doi: 10.1016/j.chembiol.2004.05.015. [DOI] [PubMed] [Google Scholar]
- [29].Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.