Abstract
The nonclassic class I human leukocyte antigen E (HLA-E) molecule engages the inhibitory NKG2A receptor on several cytotoxic effectors, including natural killer (NK) cells. Its tissue distribution was claimed to be wider in normal than in neoplastic tissues, and surface HLA-E was undetectable in most tumor cell lines. Herein, these issues were reinvestigated taking advantage of HLA-E-specific antibodies, immunohistochemistry, and biochemical methods detecting intracellular and surface HLA-E regardless of conformation. Contrary to published evidence, HLA-E was detected in a few normal epithelia and in a large fraction (approximately 1/3) of solid tumors, including those derived from HLA-E-negative/low-normal counterparts. Remarkably, HLA-E was detected in 30 of 30 tumor cell lines representative of major lymphoid and nonlymphoid lineages, and in 11 of 11, it was surface-expressed, although in a conformation poorly reactive with commonly used antibodies. Coexpression of HLA-E and HLA class I ligand donors was not required for surface expression but was associated with NKG2A-mediated protection from lysis by the cytotoxic cell line NKL and polyclonal NK cells from healthy donors, as demonstrated by antibody-mediated relief of protection in 10% to 20% of the tested target-effector combinations. NKG2A-mediated protection of additional targets became evident on NK effector blocking with antibodies to activating receptors (DNAM-1, natural cytotoxicity receptors, and NKG2D). Thus, initial evidence that the long-elusive HLA-E molecule is enhanced by malignant transformation and is functional in tumor cells is presented here, although its importance and precise functional role remain to be addressed in the context of a general understanding of the NK ligand-receptor network.
Introduction
Human leukocyte antigen E (HLA-E) is a nonclassic class I molecule recognized by natural killer (NK) cells, CD8 cytotoxic T lymphocytes (CTLs), and a more recently described subset of CD8 effectors with memory phenotype, called by some authors NK-CTLs [1–4]. NK cells and certain CTL subsets engage cell surface HLA-E through heterodimeric lectin-like receptors, both inhibitory (CD94/NKG2A) and activating (i.e., CD94/NKG2C). NK-CTLs primarily engage HLA-E through (oligo)clonally rearranged T-cell receptor (TcR) and lyse target cells, but they may also express NKG2A [1–4].
Inhibition through NKG2A is possibly the most thoroughly understood function of HLA-E. It requires the stabilization of the HLA-E heavy chain through association with its light-chain subunit (β2m) and short peptide ligands cleaved from the signal sequences of “permissive” class I alleles (the classic HLA-A, -B, and -C and the nonclassic HLA-G heavy chains), with the aid of class I-dedicated chaperones such as TAP and tapasin (reviewed in Rodgers and Cook [5]).
Coexpression of HLA-E and permissive alleles, crucial to this mechanism of ligand donation/stabilization, is thought to protect the conceptus from a maternal hemiallogeneic response [6] and prevent the inappropriate recognition of somatic self [7], but HLA-E may also favor immunoevasion. For instance, some viral genomes encode proteins acting as surrogate donors of HLA-E ligands [8,9], and ovarian carcinoma cells were shown to express increased levels of the ligand donor HLA-G as a result of interferon γ (IFN-γ) treatment [10]. However, because IFN-γ also upregulates antigen-presenting HLA-A, -B, and -C molecules, that is, a full set of major activating T-cell ligands, it is difficult to predict the final outcome (evasion or tumor control) in this and in similar [11] situations.
Unfortunately, the critical issue of whether HLA-E levels differ between normal and neoplastic tissues remains largely not addressed. For instance, immunohistochemistry detected HLA-E at several extra-placental locations, including normal white blood cells, liver, skin, and lung, but the reactive cell types were not specified [12]. Expression in the skin was subsequently confirmed [11], and HLA-E was also detected in certain endothelia but not in the few tested glandular epithelia [13]. As to biochemistry and flow cytometry studies, the commonly used 3D12 and MEM antibodies detected HLA-E polypeptides in the soluble extracts and/or on the surface of only 10 of 37 [14] and 4 of 31 [15] tumor cell lines. To complicate interpretations, HLA-E transcripts could be detected in the absence of HLA-E polypeptides [15], and HLA-E polypeptides were detected at an intracellular location but not on the cell surface [11].
On the basis of the available data, one might conclude that HLA-E is expressed in an undefined, possibly wide, range of normal tissues, but only in a few tumor cells in culture, either constitutively (seldom) or (possibly more often) following IFN-γ treatment, providing a weak rationale to investigate its function in tumor cells. Possibly for this reason, there are, to our knowledge, few published studies on this topic [11,16].
A more recent study of ours may help to reinterpret some of these results. In this study [17], it was shown that 3D12 and the MEM antibodies [11,13–15,18] selectively bind a subpopulation of unfolded HLA-E molecules free of β2m, whereas biochemical approaches, among which the most effective is a reverse biotin labeling method, detect surface HLA-E regardless of conformation.
Using the HLA-E-specific [17] MEM-E02 antibody, we report herein the tissue distribution of HLA-E in normal nonlymphoid tissues and their malignant counterparts. Through biotin labeling and cytotoxicity assays, we measure surface HLA-E expression and assess for the first time the NK-inhibitory function of HLA-E constitutively expressed under the control of its own promoter in untransfected neoplastic cell lines, in the presence and absence of permissive alleles. The findings reported herein reconcile previous conflicting results, alleviate some theoretical inconsistencies, and bear several implications in tumor immunology.
Materials and Methods
Immunohistochemistry
Neoplastic tissues from patients (free from therapy) undergoing surgery were obtained upon written consent following the recommendations of the latest (March 1, 2006) Regina Elena Institute Ethical Committee Official Guideline. For further details, see Supplemental Materials and Methods.
Cell Lines
The 221B lymphoblastoid cell line and its transfectants, namely, 221.AEH [19], 221.G1, 221.B15 [20], and 221.B*0702 [21], were obtained through the courtesy of different investigators and/or the collaborative efforts of the HLA-G and -E Workshops (see acknowledgments). Epstein-Barr virus-immortalized B lymphocytes (EBV-B) and tumor cell lines are listed in the Supplemental Materials and Methods along with their HLA-A, -B, and -C typing. Some of these are early passage (<10 subcultures) tumor cells previously established [22]. Normal human epidermal melanocytes (NHEMs) were purchased from Lonza (Walkersville, MD). HLA-E genotyping was obtained by direct sequencing of genomic DNA, as described [17].
Biochemical Methods
The mouse monoclonal antibodies MEM-E/02, MEM-E/06, MEM-E/07, and MEM-E/08 [13,17,18] are all from Exbio, Prague, Czech Republic; 3D12, 4D12 [14,23], W6/32 [24], Namb-1 [25], L31 [26], and a polyclonal to ERp57 were used in previous publications of ours [22,26–29]. The reverse surface biotin labeling method is described [17].
Flow Cytometry
Tumor cells and peripheral blood mononuclear cells (PBMCs)/purified NK cells were stained on ice with either fluorochrome-labeled antibodies or with a predetermined optimal (10 µg/ml) concentration of primary antibody/chimeric immunoglobulin (Ig). In the latter case, primary antibody binding was revealed by fluorescence isothiocyanate-labeled rabbit antibodies to either mouse or human Ig (Dako, Glostrup, Denmark). Isotype-matched control antibodies, or a chimeric Ig of irrelevant specificity, were included as negative controls. Specifically bound fluorescence was immediately analyzed without fixation by a FACScan flow cytometer (Becton, Dickinson & Co, Mountain View, CA). Antibodies to MICA(159227), MICB (MAB), ULBP1 (170818), ULBP2 (165903), ULBP3 (166510), NKG2A (131411), NKG2C (134591), NKG2D (149810), DNAX accessory molecule 1 (DNAM-1/CD226) (102511), NKp30 (210845), NKp44 (253415), NKp46/CD335 (195314), and recombinant human Fc chimeras to activating immune receptors DNAM-1-Fc, NKp30-Fc, NKp44-Fc, and NKp46-Fc were from R&D Systems (Minneapolis, MN). SKII.4 to polyovirus receptor (CD155) was from Dr Marco Colonna. Antibody to Nectin-2 was from BD Pharmingen (San Jose, CA). Antibodies to CD3 (UCHT1) and CD56 (MOC-1) were from Dako. Antibodies to killer Ig-like receptor (KIR) KIR2DL2/DL3 (DX27), KIR3DL1 (DX9), and KIR2DL1/DS1 (11PB6) were from Miltenyi Biotec GmbH (Bergisch Gladbach, Germany). Antibodies to Ig-like transcript (ILT) ILT2 (GHI/75) were from Biolegend (San Diego, CA).
NK Cell-Mediated Cytotoxicity
The continuous NK cell line NKL [30] expresses NKG2A and ILT2 [31]. Polyclonal NK cells (typically 60% to 75% CD56+/CD3-, and 60% to 80% NKG2A+) were established by culturing healthy donor PBMCs in vitro for 10 to 12 days on feeder layers of RPMI 8866 cells, as described [32]. They were further purified (>98% CD56+/CD3-) by negative immunomagnetic selection (Miltenyi). Cytotoxicity was measured by a standard 4-hour 51Cr release assay, at the indicated effector-target (E/T) ratios, using as targets 5 x 103 cells per microplate well in triplicate. Antibodies (10 µg/ml) or Ig fragment antigen binding 2 fragments (7.5 µg/ml) to HLA molecules and NK cells were separately incubated at room temperature for 15 minutes with target and effector cells, respectively, before dispensing into the 96-well microplate.
Results
Tissue Distribution of HLA-E in Normal Adult Tissues and Neoplastic Lesions
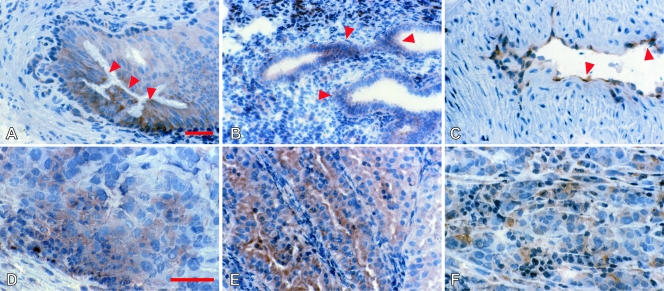
Immunohistochemical staining with MEM-E/02 gave a fine ground-glass pattern sparing the nuclei in a few normal tissues (Figure 1, A–C, and Table W1) and in a much wider spectrum of tumor lesions (Figure 1, D–F). Frequency and intensities of positive reactions were different in different histotypes (Table W2). For instance, ovarian and testicular tumors, non-small cell lung carcinomas, soft tissue tumors (Figure 1D), as well as cutaneous melanoma (Figure 1F), most of which derive from a tested counterpart in which HLA-E was undetectable, displayed from intermediate to high frequencies of MEM-E/02 reactivity. Unlike endothelia from normal tissues, some tumor endothelia were clearly reactive (Figure 1C). HLA-E down-regulation was exclusively observed in 50% approximately of endometrial carcinoma lesions (Table W2; Figure 1E displays a positive case). In summary, one third of the tumor lesions were stained by MEM-E/02, most often because enhancement or de novo appearance associated with malignant transformation.
Figure 1.
Immunohistochemical analysis of HLA-E. Normal (A–C) and neoplastic (D–F) nonlymphoid human tissues were stained with MEM-E/02 and nuclear counterstained with Mayer hematoxylin. A ground-glass pattern was detected in the principal but not the basal cells in the epididymis (A, arrows), in endometrial cells (B, arrows), and in the vascular endothelium of the myometrium (C, arrows). In tumors, variable expression was seen in a case of osteosarcoma (D), a well-differentiated endometrial carcinoma (E), and an in transit metastatic melanoma (F). Original magnifications, x160 (A–C, E); x250 (D and F). Scale bars, 100 µm.
Widespread Accumulation and Surface Expression of HLA-E Polypeptides in Tumor Cells
Western blot analysis with MEM-E/02 of 24 tumor cell lines from representative lineages (Figure W1) detected HLA-E polypeptides of the correct 42-kDa size at extremely variable levels. All the tumor cells were clearly positive, including the K562 cell line that was found to express the lowest HLA-E levels, only detectable on long filter exposure (see below).
Next, 33 tumor cell lines were assayed by flow cytometry with the HLA-E-restricted MEM-E/02,MEM-E/06, MEM-E/07, and MEM-E/08 antibodies, and 15 were also tested with the 3D12 and 4D12 antibodies. Although four of these cells (U937, Jurkat, Raji, and K562) had previously been shown to differ in 3D12 surface binding [14], all the antibodies including 3D12 and 4D12 gave a dull fluorescence pattern in our hands, with differences among antibodies and cell lines being too small to represent a reliable ranking method (representative results are shown in Figure W2 and Table W3).
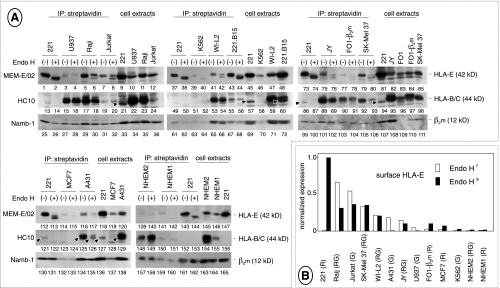
To detect surface HLA-E molecules regardless of antibody reactivity, 11 tumor cell lines and two cultures of NHEMs (NHEM1 and NHEM2) were assessed for surface HLA-E expression by reverse biotin labeling [17]. Because there are two nonsynonymous HLA-E alleles [33] with high (HLA-E107G) and low (HLA-E107R) surface expression [34], all the cells were HLA-E typed.
As opposed to the similar, dull staining detected by flow cytometry, biotin labeling (Figure 2A) detected widely different levels of surface HLA-E. Cells lacking coexpression of permissive alleles (221, K562, and the 221.B15 transfectant) only expressed immature (Endo H-sensitive) glycoforms (lanes 1–2, 39–40, and 43–44), as described [17]. In contrast, U937, Raji, Jurkat, WI-L2, JY, FO-1-β2m, SKMEL-37, A431, and NHEM2 cells expressing detectable levels of HC10-reactive class I molecules (lanes 15–20, 53–54, 88–93, 125–126, and 148–149), namely, potential donors of HLA-E ligands, expressed both mature (Endo H-insensitive) and immature (Endo H-sensitive) HLA-E glycoforms (lanes 3–8, 41–42, 75–80, 116–117, and 139–140). NHEM1 melanocytes and MCF7 cells were the only exceptions, in that the former did react with HC10 (lanes 150–151) but only expressed immature glycoforms (lanes 141–142), whereas the latter reacted barely, if at all, with HC10 (lanes 123–124) but did express some mature HLA-E glycoforms (lanes 114–115). Endo H-sensitivity of surface HLA-E was not due to biotin labeling of intracellular class I heavy chains because HC10-reactive heavy chains recovered from the same soluble extracts were insensitive to Endo H digestion (lanes 16, 18, 20, 54, 56, 89, 91, 93, 126, 149, and 151), as expected.
Figure 2.
HLA-E expression on the tumor cell surface. (A) NP40 extracts (1 mg) from biotin-labeled cells were immunoprecipitated by streptavidin-conjugated beads and either digested with Endoglycosidase H [41] or mock-incubated. Surface proteins were resolved by SDS-PAGE, electroblotted, and identified by sequential staining/stripping of the filters with the indicated antibodies. Extracts from biotinlabeled cells (100 µg) were also run side by side. Arrowheads mark residual staining by MEM-E/02 not removed by stripping. Five experiments are shown, each including 221 cells as an internal control. Each cell line was tested at least twice with similar results, except FO-1-β2m cells in which HLA-E expression was slightly underestimated in the experiment shown in the figure, as assessed by two additional experiments. (B) Values of densitometric scans of the Endo H-resistant and Endo H-sensitive HLA-E bands from lanes 2, 4, 6, 8, 40, 42, 76, 78, 80, 115, and 117, sorted by intensity. HLA-E typing, shown in parentheses, was either reported previously [14,15] or assessed by direct DNA sequencing as described [17]. G indicates HLA-E107G; R, HLA-E107R; RG, heterozygous condition.
In summary, all the cells were positive and could be ranked by densitometry according to the expression of the two surface HLA-E glycoforms (Figure 2B). Reminiscent of immunohistochemistry (see previous paragraph), normal cells (melanocytes) clustered at the low end of the rank, whereas tumor cell lines were distributed across the whole range of HLA-E levels. HLA-E levels showed no obvious correlation with either HLA-E typing or coexpression with ligand donors. For instance, 221 and K562 cells both lack permissive alleles but lie at the opposite ends of the rank, and none of the HC10-reactive cell lines expressed as much HLA-E as 221.
Finally, widespread expression of HLA-E is not an artifact of in vitro passaging because reverse biotin-labeling confirmed its presence on the surface of three early passage (<10 subcultures) tumor cell lines (Figure W3).
Surface HLA-E and Lysis by the Cytotoxic Cell Line NKL
Because biotin labeling revealed surface HLA-E molecules on 221 and K562 cells, previously described to be unreactive with antibodies to HLA-E [14,15,19], we wanted to rule out a low-level baseline NKG2A-mediated protection that may have gone unnoticed in previous studies [12,31]. To this end, two experiments were performed in which 221 and K562 cells were tested as targets of the widely used NK cytotoxic cell line NKL.
In the first experiment (Figure W4), whole Ig and Ig fragment antigen binding 2 fragments of W6/32 (to HLA-E and most class I HLA molecules) and Z199 (to NKG2A) did not appreciably affect NKL lysis of parental 221 cells, but they efficiently restored, as expected, lysis of 221.AEH cells expressing a hybrid HLA-E molecule (AEH) capable of self ligand donation [19].
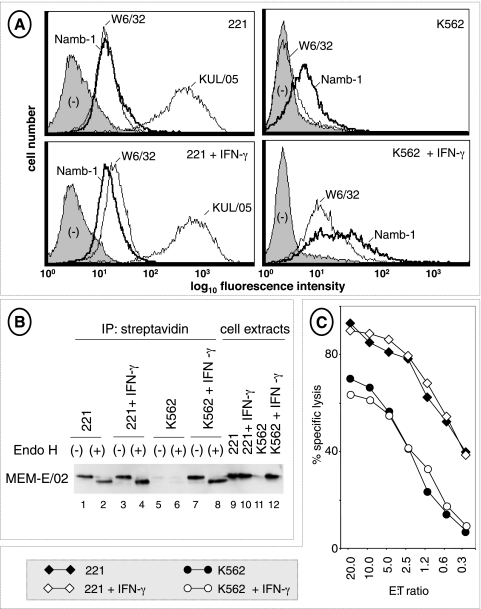
In the second experiment, 221 and K562 cells were treated with IFN-γ, a potent enhancer of class I expression also capable of modifying the pool of peptide ligands generated by the proteasome [35]. Flow cytometry with antibodies to conformed HLA class I molecules (W6/32) and β2m (Namb-1) revealed slight (in 221) and drastic (in K562) increases in surface class I expression (Figure 3A). These increases were in agreement with those detected by surface biotin labeling (Figure 3B, lanes 1–8), conclusively demonstrating IFN-γ-dependent surface enhancement of HLA-E in these cells. However, NKL-mediated lysis remained unaffected by IFN-γ over a wide range of E/T ratios (Figure 3C).
Figure 3.
Up-regulation of HLA-E heavy chains by IFN-γ and NKL lysis. (A) 221 and K562 cells, either untreated or treated for 72 hours with 200 U/ml of recombinant IFN-γ, were tested by flow cytometry with antibodies to class I heavy chains (W6/32) and β2m (Namb-1). An antibody to HLA class II molecules (KUL/05) was used as a control for IFN-γ responsiveness of 221 cells. (B) Control and IFN-γ-treated (as previously mentioned) 221 and K562 cells were surface-labeled with biotin and HLA-E was revealed as described in the legend to Figure 2. (C) Control and IFN-γ-treated (as previously mentioned) 221 and K562 cells were tested for their susceptibility to NKL lysis at the indicated E/T ratios. The experiment was repeated three times.
Altogether, these results (Figures 2, 3, and W4) demonstrate that surface-expressed HLA-E does not appreciably protect 221 and K562 cells from NKL lysis.
Next, we looked for a protective effect of HLA-E coexpressed with variable levels of its ligand donors. NKL cells were used as cytolytic effectors on 11 tumor cell lines (Jurkat, WI-L2, Raji, U937, M10, MNT-1, SK-MEL-37, SK-MEL-93, FO-1-β2m, A431, and MCF7). In these cells, HLA-A, -B, and -C molecules are expected to contribute peptide ligands enabling HLA-E inhibition through NKG2A and, in addition, to act as direct inhibitory ligands of the KIR and ILT2 receptors. Because they express ILT2 and NKG2A, but not KIR ([31] and our own unpublished data), NKL cells provide a simplified readout for NK lysis.
As expected, the 11 target tumor cell lines were lysed to a different extent. FO-1-β2m cells were consistently most sensitive to lysis (representative results shown in Figure W5) and displayed the largest repertoire of activating NK ligands among the five selected cell lines (Table W4). No other obvious correlations were apparent between susceptibility to lysis and expression of NK ligands, including HLA-E. For instance, FO-1-β2m, A431, and U937 cells were lysed to a different extent (Figure 4) in spite they express similar (intermediate) surface HLA-E levels (compared with Figure 2B). As also shown by representative results in Figure W5, NKL lysis of 7 of 11 cell lines was unaffected by antibodies to ILT2 (GHI/75) and NKG2A (Z199), but GHI/75 enhanced lysis in the remaining 4 cell lines (FO-1-β2m, A431, M10, and SK-Mel 37), 2 of which (FO-1-β2m and A431) became more susceptible on incubation with antibodies to NKG2A. We conclude that a fraction of tumor targets can be demonstrated to be protected through ILT2 and, possibly to a lesser extent, through NKG2A.
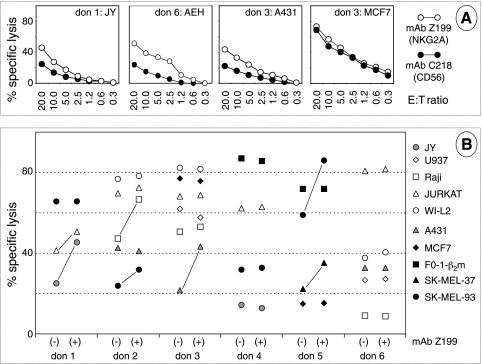
Figure 4.
Cytotoxic activity of polyclonal NK cells against tumor cells. (A) CD56+/CD3- effector cells were tested in a 51Cr release assay, in the presence of antibodies to CD56 (C218) and to NKG2A (Z199) on the indicated targets. (B) Summary of cytotoxicity results (at an E/T ratio of 20:1) with NK cells from 6 donors and 10 tumor cell targets in presence of either C218 (-) or Z199 (+). A line connects statistically significant lytic enhancements by Z199, that is, differences in average values of triplicates more than three times the SD.
Surface HLA-E and Lysis by Polyclonal NK Cells
One may wonder whether the role of the HLA-E/NKG2A axis might be similarly appreciated using a panel of nonimmortalized effector NK cells. Polyclonal NK cells (>98% CD56+/CD3-) were then obtained by immunomagnetic sorting from the PBMCs of six different healthy donors cultured on feeder layers without interleukin 2, as described [32]. Like NKL, these NK cell populations expressed high levels of inhibitory receptors, not only NKG2A, NKG2D, and ILT2 but also KIRs, and in addition they also expressed activating receptors such as DNAM-1 and the natural cytotoxicity receptors (NCRs) NKp30, NKp44, and NKp46 (for additional details, see Figure W6). Polyclonal NK cells were used as effectors in a cytotoxicity assay using as targets the immortalized JY B lymphoid line and nine tumor cell lines (Jurkat, WI-L2, Raji, U937, SK-MEL-37, SK-MEL-93, FO-1-β2m, A431, and MCF7). The protective role of NKG2A from NK lysis was assessed as previously mentioned in the presence of the NKG2A-specific antibody Z199.
NKG2A-mediated protection (i.e., protection in donor 1:JY and donor 3:A431 but not in donor 3:MCF7; Figure 4A) was detected in a minority of E/T combinations. A complete synopsis is provided in Figure 4B in which the highest E/T ratios are shown for simplicity. Altogether, NKG2A-mediated protection was observed in 7 (23% approximately) of 27 cases (see connecting bars). As expected, six distinct patterns were observed, that is, the six polyclonal NK cell populations differed from one another and from NKL cells in their ability to kill a given target and to display NKG2A-mediated protection. Overall, four of six NK cell cultures could be demonstrated to be inhibited through NKG2A with at least one target. Similar experiments performed with bulk NK cells (not submitted to immunomagnetic purification) from four donors resulted in 5 (11% approximately) of 44 Z199-mediated lysis enhancements (not shown).
Functional Role of NKG2A in the Context of Activating NK Cell Circuitry
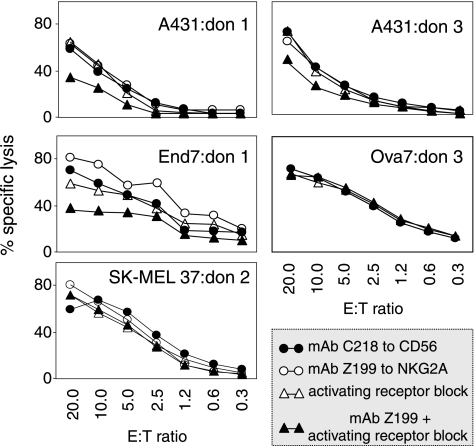
In the attempt to account for the low percentage of E/T combinations in which the inhibitory role of NKG2A is appreciable, we hypothesized that a strong overriding activation might obscure NKG2A-mediated inhibition. To obtain evidence in favor of this hypothesis, a panel of three long-term (FO-1-β2m, SK-MEL-37, and A431) and four early passage (Mel20, Br7, End7, and Ova7) tumor cell lines expressing one or more major NK-activating ligands (Table W4) were tested in cytotoxicity as targets of polyclonal NK cells from three distinct donors expressing essentially all the cognate activating receptors (Figure W6). Z199-mediated lysis enhancement was assessed not only in the absence but also in the presence of mixtures of antibodies blocking those activating receptors known to bind the triggering ligands detectable (Table W4) on the target.
Representative results (Figure 5) confirmed that Z199 by itself enhances NK lysis in a minority (5/21) of E/T combinations, but on blocking of activating receptors and the consequent reduction in NK lysis (observed in 14/21 E/T combinations), antibodies to NKG2A become capable of restoring susceptibility to lysis of three additional targets. It may be concluded that overriding activation (by DNAM-1 and/or NCR and/or NKG2D) may prevent appreciation of the inhibitory HLA-E/NKG2A axis.
Figure 5.
Block of activating receptors reveals NKG2A-mediated protection from NK lysis. The indicated tumor cell lines were tested as targets in a 51Cr release assay using CD56+/CD3- cells as effectors, in the presence of the indicated antibodies. Blocking antibodies to activating receptors were selected on the basis of the ligands expressed by the target: A431 (DNAM-1), End7 (NKG2D + DNAM-1), SK-MEL 37 (DNAM-1), and Ova7 (NKG2D+DNAM-1). Differences in 51Cr release values were statistically significant when the average values of triplicates differed from control more than three times the SD, that is, at E/T ratios from 20:1 to 2.5:1 in A431: donor 1 as well as End7:donor 1 and at E/T ratios of 20:1 and 10:1 in A431:donor 3. The top two panels display two E/T combinations in which the inhibitory effect of NKG2A becomes evident only when activating receptors are antibody-blocked. End7:donor 1 exemplifies a case in which NKG2A similarly protects in the presence and absence of antibody block. In the remaining cases, activating receptors and NKG2A cannot be demonstrated to influence susceptibility to lysis.
Discussion
Consistent with their primary and specialized role at the fetal-maternal interface, the expression of several nonclassic class I HLA molecules is believed to be highly restricted [5,6]. The extensive immunohistochemical testing performed herein is consistent with this view and, in addition, shows that the few normal epithelia and tissues that constitutively express HLA-E are from the genitourinary/reproductive tract and the colonic mucosa. Perhaps, HLA-E contributes to safeguarding somatic self integrity in these normal tissues.
In contrast, HLA-E expression is wider than previously appreciated in malignant tissues, one third of which (including those derived from an HLA-E-negative counterpart) are reactive with MEM-E/02 (Figure 1 and Tables W1 and W2). Herein, we present evidence supporting a functional role of HLA-E expression in tumors.
Widespread Surface Expression of HLA-E in Tumor Cells: Comparison with Previous Studies
Two previous flow cytometry studies with either 4D12 or MEM antibodies detected weak surface expression of HLA-E in a minority of tumor cell lines [14,15]. In our hands, however, the same antibodies resulted in a dull and substantially invariant surface fluorescence pattern on 33 tumor cell lines, 11 of which also tested in the two previously mentioned studies (Figures W2 and 3 and Table W3). Only by reverse biotin labeling (Figures 2 and 3) were we able to semiquantitatively detect a wide range of HLA-E levels. HLA-E was invariably surface-expressed under the species of two distinct glycoforms. Surprisingly, tumor cells previously [14] ranked by flow cytometry in the order U937 > Jurkat > Raji and K562 (Raji and K562 both negative) were ranked by reverse biotin labeling in the order Raji ≠ Jurkat >> U937 > K562 (all positive), regardless of the glycoform being considered.
A detailed comparison among flow cytometry studies is difficult because few staining profiles are published, and/or the results are charted on the basis of semiquantitative, subjective evaluation scales. Regardless of protocols and interpretations, it may be concluded that HLA-E molecules, although poorly reactive (at least in our hands) with the available antibodies, are invariably present on the surface of tumor cell lines.
HLA-E Contributes to Protect Tumors from NK Cell Lysis
As a first step to assess the function of HLA-E on tumor cells, so far supported by limited evidence [10,16], we focused on its well-known ability to protect from NK cell lysis through NKG2A expressed by the cytotoxic cell line NKL and resting polyclonal NK cells from healthy donors. Specific antibodies to NKG2A relieved protection in the case of some tumor cell lines coexpressing permissive alleles but not in the case of parental 221 (B lymphoblastoid) and K562 (erythromyeloid) cells lacking permissive alleles (Figures 3–5 and W3). We conclude that surface HLA-E molecules are functional in at least some tumor cell lines but not in 221 and K562 despite that these cells do express HLA-E, both constitutively and because of IFN-γ treatment.
The observation that protection occurred in a minority (10%–20%) of the tested E/T combinations is reminiscent of the bare minority of HLA-A2-restricted CTL clones to melanoma cells in which NKG2A-mediated inhibition is detectable [16] and is not surprising, given the many differences in lineage, genetic background, and intrinsic susceptibility to lysis among tumor targets. Most likely, a redundancy in activating and inhibitory ligand-receptor interactions masks or quenches the specific contribution of HLA-E in certain tumor-NK and tumor-CTL lytic combinations. Experiments (Figure 5) in which the inhibitory role of NKG2A could be appreciated only upon block of activating NK receptors (DNAM-1 and/or NCRs and/or NKG2D) provide evidence in support of this interpretation.
A Model for the Regulation and Function of HLA-E in Tumors
There is evidence that HLA-E may favor both tumor escape and tumor immune surveillance. Tumor escape is supported by a negative prognostic association of HLA-E expression in colorectal and breast carcinomas [36,37]. A possible mechanism has been suggested by at least two groups [14,36]. In this model, selective loss of HLA-A, -B, and -C alleles (a phenotype proposed by some to be frequently associated with immune evasion) alleviates competition for β2m, allowing ligand donation from the residual class I allele(s) in amounts sufficient to stabilize HLA-E, enhance its surface expression, engage the inhibitory NKG2A receptor, and further promote immune escape.
However, three lines of evidence argue against the previously mentioned model and a purely negative role of HLA-E on survival: 1) although HLA-E may have a low affinity for β2m in defined model systems [34], there is a subset of HLA-E molecules that efficiently binds β2m [17]; 2) HLA-E is widely expressed in human tumors, as shown herein; and 3) high HLA-E seems to correlate with good, and not poor, prognosis in melanoma and glioblastoma [38–40].
By providing evidence that HLA-E is a widespread tumor recognition structure enabling NK cell recognition and monitoring, the present study suggests that HLA-E might represent in at least some tumors a very efficient checkpoint and fail safe mechanism against all kinds of HLA losses and not a vulnerable soft spot toward immune evasion in tumor mutants that have already accumulated multiple HLA defects.
It will be of interest to determine whether HLA-E on the tumor cell surface engages rearranging and nonrearranging receptors, either activating or inhibitory, expressed on different populations of NK, CTL, and NK-CTL effectors, and whether it modulates immune lysis. The applicable importance of HLA-E in tumor immunology remains to be precisely addressed on the clonal level and in the context of a global appreciation of the immune ligand-receptor network.
Supplementary Material
Supplemental Materials and Methods
Immunohistochemistry
Samples were snap frozen in liquid nitrogen and stored at -80°C. Four-micrometer cryostat sections were fixed in cold absolute acetone for 10 minutes and either immediately used for indirect immunoperoxidase or stored at -20°C up to 6 months with no appreciable changes in reactivity. Slides were incubated overnight with antibodies (50 µg/ml) at 4°C in a moistened chamber. Control sections were incubated with isotype-matched IgG. An indirect avidin-biotin immunoperoxidase staining was performed with commercially available reagents (VECTASTAIN Elite, Mountain View, CA), and the enzymatic activity was developed using 3-amino-9-ethylcarbazole as the chromogenic substrate for 8 minutes. Slices were then rinsed with phosphate-buffered saline, counterstained with Mayer hematoxylin, and mounted with buffered glycerol. At least three nonconsecutive sections from each specimen were analyzed. Normal tissues were from at least two patients and were collected distal to transformed tissues.
Cell Lines
JY and BSM are EBV-immortalized cell lines. Tumor cell lines are listed along with their lineage and HLA-A, -B, and -C typing [1–8]: Molt 4 (T lymphoblastoid; A1, 25; B18, 57; Cw6, 12), Jurkat (T lymphoblastoid; A3; B35), WI-L2 (B lymphoblastoid; A1, 2; B51, 17), Raji (B lymphoblastoid; A3), U937 (myelomonocytic; A3, 19; B51, 18; Cw1, 3), HL-60 (myelomonocytic; A10; B57; Cw6), K562 (erythromyeloid; A11, 31; B18, 40; Cw3), Colo 38 (melanoma; A24, 11; B35, 15; Cw3, 4), SK-MEL-37 (melanoma; A2, 11; B15, 55; Cw1), SK-MEL-93 (melanoma; A2, 31; B21, 35; Cw4), FO-1 and its β2m transfectant FO-1-β2m (melanoma; A25; B8; Cw7), A549 (carcinoma; A25, 30; B18, 44; Cw12, 16), BT20 (carcinoma; A24; B15; Cw3, 12), Calu-1 (carcinoma; A2, B40, 41; Cw2, 17), End 9 (carcinoma; A24, 25; B15, 52; Cw1, 3), HT-29 (carcinoma; A1, 24; B35, 44; Cw4), MCF7 (carcinoma; A2, B18, 44; Cw5), T24 (carcinoma; A1; B18; Cw5), HeLa (carcinoma; A68; B15; Cw16), KJ29 (carcinoma; A2; B27; Cw1), and JAR (choriocarcinoma). Early passage cell lines are melanoma Mel20 (A1, 31; B14, 18), ovarian carcinoma Ova7 (A1, 11; B14, 51), endometrial carcinoma End7, and breast carcinoma Br7.
As shown in Table W3 (representative results are shown in Figure W2), all the antibodies with nominal HLA-E specificity detected HLA-E on the surface of 221.AEH transfectants overexpressing a hybrid HLA-E heavy chain capable of self (in cis) ligand donation [9], but they failed to detect levels of HLA-E exceeding twice the background in all the other tested cell lines. These include parental 221 cells (expressing HLA-E but lacking HLA-A, -B, and -C and HLA-G), the 221.G1 and 221.B7 transfectants coexpressing “permissive” alleles, and four EBV-immortalized, nontransformed B lymphoid cell lines previously shown to synthesize from intermediate to high levels of HLA-E [10].
Some of the listed tumor cell lines have previously been tested by others [7,11]: U937 and Jurkat (3D12-positive), Raji (3D12-negative), BT-20 and HT-29 (negative for MEM-E/06, MEM-E/07, and MEM-E/08), Calu 1, A549, and K562 (negative for 3D12 and the three MEM antibodies as well), and MCF-7 (negative for the MEM antibodies except MEM-E/07).
Acknowledgments
The authors thank Marco Colonna for antibodies to polyovirus receptor; Daniel E. Geraghty for 221.AEH, 221.G1, 3D12 and 4D12; and Michael J. Robertson for NKL cells. Rocco Fraioli and Dina Milana are gratefully acknowledged for skillful technical assistance. Maria Vincenza Sarcone and Paula Franke provided secretarial support and revised the English text.
Abbreviations
- CTL
cytotoxic Tlymphocyte
- DNAM-1
DNAX accessory molecule 1
- E/T
effector-target
- HLA
human leukocyte antigen(s)
- IFN
interferon
- ILT
immunoglobulin-like transcript
- KIR
killer immunoglobulin-like receptor
- NCR
natural cytotoxicity receptor
- NHEM
normal human epidermal melanocyte
- NK
natural killer
- PBMC
peripheral blood mononuclear cell
- TcR
T-cell receptor
- β2m
β2-microglobulin
Footnotes
Work supported by the Italian Ministries of Public Health (C.C., A.Z., and P.G.), University and Research (MIUR) (C.C. and A.Z.), AIRC (C.C., A.Z., P.G.N., and P.G.), and the Center of Excellence (BEMM) Rome, Italy (C.C. and A.Z.). The authors have no conflict of interest.
This article refers to supplementary materials, which are designated by Tables W1 to W4 and Figures W1 to W6 and are available online at www.neoplasia.com.
References
- 1.Braud VM, Allan DSJ, O'Callaghan CA, Söderström K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 2.Garcia P, Llano M, Heredia AB, Willberg CB, Caparros E, Aparicio P, Braud VM, Lopez-Botet M. Human T cell receptor-mediated recognition of HLA-E. Eur J Immunol. 2002;32:936–944. doi: 10.1002/1521-4141(200204)32:4<936::AID-IMMU936>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 3.Pietra G, Romagnani C, Falco M, Vitale M, Castriconi R, Pende D, Millo E, Anfossi S, Biassoni R, Moretta L, et al. The analysis of the natural killer-like activity of human cytolytic T lymphocytes revealed HLA-E as a novel target for TCRα/β-mediated recognition. Eur J Immunol. 2002;31:3687–3693. doi: 10.1002/1521-4141(200112)31:12<3687::aid-immu3687>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 4.Moretta L, Romagnani C, Pietra G, Moretta A, Mingari MC. NK-CTLs, a novel HLA-E-restricted T-cell subset. Trends Immunol. 2003;24:136–143. doi: 10.1016/s1471-4906(03)00031-0. [DOI] [PubMed] [Google Scholar]
- 5.Rodgers J, Cook R. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- 6.Houlihan JM, Biro PA, Harper HM, Jenkinson HJ, Holmes CH. The human amnion is a site of MHC class Ib expression: evidence for the expression of HLA-E and HLA-G. J Immunol. 1995;154:5665–5674. [PubMed] [Google Scholar]
- 7.Carosella ED, Paul P, Moreau P, Rouas-Freiss N. HLA-G and HLA-E: fundamental and pathophysiological aspects. Immunol Today. 2000;21:532–534. [PubMed] [Google Scholar]
- 8.Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, Cerundolo V, Borysiewicz LK, McMichael AJ, Wilkinson GWG. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031–1033. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 9.Ulbrecht M, Martinozzi S, Grzeschik M, Hengel H, Ellwart JW, Pla M, Weiss EH. The human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J Immunol. 2000;164:5019–5022. doi: 10.4049/jimmunol.164.10.5019. [DOI] [PubMed] [Google Scholar]
- 10.Malmberg K, Levitsky V, Norell H, de Matos CT, Carlsten M, Schedvins K, Rabbani H, Moretta A, Soderstrom K, Levitskaya J, et al. IFN-γ protects short-term ovarian carcinoma cell lines from CTL lysis via a CD94/NKG2A-dependent mechanism. J Clin Invest. 2002;110:1515–1523. doi: 10.1172/JCI15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derré L, Corvaisier M, Charreau B, Moreau A, Godefroy E, Moreau-Aubry A, Jotereau F, Gervois N. Expression and release of HLA-E by melanoma cells and melanocytes: potential impact on the response of cytotoxic effector cells. J Immunol. 2006;177:3100–3107. doi: 10.4049/jimmunol.177.5.3100. [DOI] [PubMed] [Google Scholar]
- 12.Lee N, Llano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty D. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coupel S, Moreau A, Hamidou M, Horejsi V, Soulillou JP, Charreau B. Expression and release of soluble HLA-E is an immunoregulatory feature of endothelial cell activation. Blood. 2007;109:2806–2814. doi: 10.1182/blood-2006-06-030213. [DOI] [PubMed] [Google Scholar]
- 14.Marìn R, Ruiz-Cabello F, Pedrinaci S, Mendez R, Jimenez P, Geraghty DE, Garrido F. Analysis of HLA-E expression in human tumors. Immunogenetics. 2003;54:767–775. doi: 10.1007/s00251-002-0526-9. [DOI] [PubMed] [Google Scholar]
- 15.Palmisano GL, Contardi E, Morabito A, Gargaglione V, Ferrara GB, Pistillo MP. HLA-E surface expression is independent of the availability of HLA class I signal sequence-derived peptides in human tumor cell lines. Hum Immunol. 2005;66:1–12. doi: 10.1016/j.humimm.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Speiser DE, Pittet MJ, Valmori D, Dunbar R, Rimoldi D, Liénard D, MacDonald HR, Cerottini JC, Cerundolo V, Romero P. In vivo expression of natural killer cell inhibitory receptors by human melanoma-specific cytolytic T lymphocytes. J Exp Med. 1999;190:775–782. doi: 10.1084/jem.190.6.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo Monaco E, Sibilio L, Melucci E, Tremante E, Suchànek M, Horejsi V, Martayan A, Giacomini P. HLA-E: strong association with β2m and surface expression in the absence of HLA class I signal sequence-derived peptides. J Immunol. 2008;181:5442–5450. doi: 10.4049/jimmunol.181.8.5442. [DOI] [PubMed] [Google Scholar]
- 18.Menier C, Saez B, Horejsi V, Martinozzi S, Krawice-Radanne I, Bruel S, Le Danff C, Reboul M, Hilgert I, Rabreau M, et al. Characterization of monoclonal antibodies recognizing HLA-G or HLA-E: new tools to analyze the expression of nonclassical HLA class I molecules. Hum Immunol. 2003;64:315–326. doi: 10.1016/s0198-8859(02)00821-2. [DOI] [PubMed] [Google Scholar]
- 19.Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol. 1998;160:4951–4960. [PubMed] [Google Scholar]
- 20.Lee N, Malacko AR, Ishitani A, Chen MC, Bajorath J, Marquardt H, Geraghty DE. The membrane-bound and soluble forms of HLA-G bind identical sets of endogenous peptides but differ with respect to TAP association. Immunity. 1995;3:591–600. doi: 10.1016/1074-7613(95)90130-2. [DOI] [PubMed] [Google Scholar]
- 21.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. Specificity of HLA class I antigen recognition by human NK clones: evidence for clonal heterogeneity, protection by self and non-self alleles, and influence of the target cell type. J Exp Med. 1993;178:1321–1336. doi: 10.1084/jem.178.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giacomini P, Giorda E, Fraioli R, Nicotra MR, Vitale N, Setini A, Delfino L, Morabito A, Benevolo M, Venturo I, et al. Low prevalence of selective human leukocyte antigen (HLA)-A and HLA-B epitope losses in early-passage tumor cell lines. Cancer Res. 1999;59:2657–2667. [PubMed] [Google Scholar]
- 23.Strong RK, Holmes MA, Li P, Braun L, Lee N, Geraghty DE. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J Biol Chem. 2003;278:5082–5090. doi: 10.1074/jbc.M208268200. [DOI] [PubMed] [Google Scholar]
- 24.Parham P, Barnstable CJ, Bodmer WF. Use of monoclonal antibody (W6/32) in structural studies of HLA-A,B,C antigens. J Immunol. 1979;23:342–349. [PubMed] [Google Scholar]
- 25.Pellegrino MA, Ng AK, Russo C, Ferrone S. Heterogenous distribution of determinants defined by monoclonal antibodies on HLA-A, -B antigen bearing molecules. Transplantation. 1982;34:18–23. doi: 10.1097/00007890-198207000-00004. [DOI] [PubMed] [Google Scholar]
- 26.Setini A, Beretta A, De Santis C, Meneveri R, Martayan A, Mazzilli MC, Appella E, Siccardi AG, Natali PG, Giacomini P. Distinctive features of the α1 domain α helix of HLA-C heavy chains free of β2-microglobulin. Hum Immunol. 1996;46:69–81. doi: 10.1016/0198-8859(96)00011-0. [DOI] [PubMed] [Google Scholar]
- 27.Giorda E, Sibilio L, Martayan A, Moretti S, Venturo I, Mottolese M, Ferrara GB, Cappellacci S, Eibenschutz L, Catricalà C, et al. The antigen processing machinery of human leukocyte antigens: linked patterns of gene expression in neoplastic cells. Cancer Res. 2003;63:4119–4127. [PubMed] [Google Scholar]
- 28.Martayan A, Fraioli R, Giorda E, Setini A, Ciccarelli G, Delfino L, Ferrara GB, Giacomini P. Biosynthesis of HLA-C heavy chains in melanoma cells with multiple defects in the expression of HLA-A, -B, -C molecules. Br J Cancer. 1999;80:639–649. doi: 10.1038/sj.bjc.6690405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delfino L, Ciccarelli G, Bini D, Morabito A, Pozzi S, Martayan A, Giorda E, Setini A, Fraioli R, Giacomini P, et al. HLA-A, -B, -C genotyping and expression in human nonlymphoid tumor cell lines. J Immunother. 1999;22:7–15. doi: 10.1097/00002371-199901000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Robertson MJ, Cochran KJ, Cameron C, Le JM, Tantravahi R, Ritz J. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp Hematol. 1996;24:406–415. [PubMed] [Google Scholar]
- 31.Navarro F, Llano M, Bellón T, Colonna M, Geraghty DE, López-Botet M. The ILT2(LIR1) and CD94/NKG2A NK cell receptors respectively recognize HLA-G1 and HLA-E molecules co-expressed on target cells. Eur J Immunol. 1999;29:277–283. doi: 10.1002/(SICI)1521-4141(199901)29:01<277::AID-IMMU277>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Perussia B, Ramoni C, Anegon I, Cuturi MC, Faust J, Trinchieri G. Preferential proliferation of natural killer cells among peripheral blood mononuclear cells cocultured with B lymphoblastoid cell lines. Nat Immun Cell Growth Regul. 1987;6:171–188. [PubMed] [Google Scholar]
- 33.Grimsley C, Kawasaki A, Gassner C, Sageshima N, Nose Y, Hatake K, Geraghty DE, Ishitani A. Definitive high resolution typing of HLA-E allelic polymorphisms: identifying potential errors in existing allele data. Tissue Antigens. 2002;60:206–212. doi: 10.1034/j.1399-0039.2002.600302.x. [DOI] [PubMed] [Google Scholar]
- 34.Ulbrecht M, Couturier A, Martinozzi S, Pla M, Srivastava R, Peterson PA, Weiss EH. Cell surface expression of HLA-E: interaction with human β2m and allelic differences. Eur J Immunol. 1999;29:537–547. doi: 10.1002/(SICI)1521-4141(199902)29:02<537::AID-IMMU537>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Benham AM, Neefjes JJ. Proteasome activity limits the assembly of MHC class I molecules after IFN-gamma stimulation. J Immunol. 1997;159:5896–5904. [PubMed] [Google Scholar]
- 36.Levy EM, Bianchini M, Von Euw EM, Barrio MM, Bravo AI, Furman D, Domenichini E, Macagno C, Pinsky V, Zucchini C, et al. Human leukocyte antigen-E protein is overexpressed in primary human colorectal cancer. Int J Oncol. 2008;32:633–641. [PubMed] [Google Scholar]
- 37.de Kruijf EM, Sajet A, van Nes JG, Natanov R, Putter H, Smit VT, Liefers GJ, van den Elsen PJ, van de Velde CJ, Kuppen PJ. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol. 2010;185:7452–7459. doi: 10.4049/jimmunol.1002629. [DOI] [PubMed] [Google Scholar]
- 38.John T, Black MA, Toro TT, Leader D, Gedye CA, Davis ID, Guilford PJ, Cebon JS. Predicting clinical outcome through molecular profiling in stage III melanoma. Clin Cancer Res. 2008;14:5173–5180. doi: 10.1158/1078-0432.CCR-07-4170. [DOI] [PubMed] [Google Scholar]
- 39.Kren L, Slaby O, Muckova K, Lzicarova E, Sova M, Vybihal V, Svoboda T, Fadrus P, Lakomy R, Vanhara P, et al. Expression of immune-modulatory molecules HLA-G and HLA-E by tumor cells in glioblastomas: an unexpected prognostic significance? Neuropathology. 2011;31:129–134. doi: 10.1111/j.1440-1789.2010.01149.x. [DOI] [PubMed] [Google Scholar]
- 40.Mandruzzato S, Callegaro A, Turcatel G, Francescato S, Montesco MC, Chiarion-Sileni V, Mocellin S, Rossi CR, Bicciato S, Wang E, et al. A gene expression signature associated with survival in metastatic melanoma. J TranslMed. 2006;4:50. doi: 10.1186/1479-5876-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sibilio L, Martayan A, Setini A, Fraioli R, Fruci D, Shabanowitz J, Hunt DF, Giacomini P. Impaired assembly results in the accumulation of multiple HLA-C heavy chain folding intermediates. J Immunol. 2005;175:6651–6658. doi: 10.4049/jimmunol.175.10.6651. [DOI] [PubMed] [Google Scholar]
Supplementary References
- 1.Sarkar S, Glassy MC, Ferrone S, Jones OW. Cell cycle and the differential expression of HLA-A,B and HLA-DR antigens on human B lymphoid cells. Proc Natl Acad Sci USA. 1980;77:7297–7301. doi: 10.1073/pnas.77.12.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hakem R, Le Bouteiller P, Barad M, Trujillo M, Mercier P, Wietzerbin J, Lemonnier FA. IFN-mediated differential regulation of the expression of HLA-B7 and HLA-A3 class I genes. J Immunol. 1989;142:297–305. [PubMed] [Google Scholar]
- 3.Martayan A, Fiscella M, Setini A, Ciccarelli G, Gambari R, Feriotto G, Beretta A, Siccardi AG, Appella E, Giacomini P. Conformation and surface expression of free HLA-CW1 heavy chains in the absence of β2-microglobulin. Hum Immunol. 1997;53:23–33. doi: 10.1016/S0198-8859(96)00256-X. [DOI] [PubMed] [Google Scholar]
- 4.Martayan A, Fraioli R, Giorda E, Setini A, Ciccarelli G, Delfino L, Ferrara GB, Giacomini P. Biosynthesis of HLA-C heavy chains in melanoma cells with multiple defects in the expression of HLA-A, -B, -C molecules. Br J Cancer. 1999;80:639–649. doi: 10.1038/sj.bjc.6690405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delfino L, Ciccarelli G, Bini D, Morabito A, Pozzi S, Martayan A, Giorda E, Setini A, Fraioli R, Giacomini P, et al. HLA-A, -B, -C genotyping and expression in human nonlymphoid tumor cell lines. J Immunother. 1999;22:7–15. doi: 10.1097/00002371-199901000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Giacomini P, Giorda E, Fraioli R, Nicotra MR, Vitale N, Setini A, Delfino L, Morabito A, Benevolo M, Venturo I, et al. Low prevalence of selective human leukocyte antigen (HLA)-A and HLA-B epitope losses in early-passage tumor cell lines. Cancer Res. 1999;59:2657–2667. [PubMed] [Google Scholar]
- 7.Palmisano GL, Contardi E, Morabito A, Gargaglione V, Ferrara GB, Pistillo MP. HLA-E surface expression is independent of the availability of HLA class I signal sequence-derived peptides in human tumor cell lines. Hum Immunol. 2005;66:1–12. doi: 10.1016/j.humimm.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Adams S, Robbins FM, Chen D, Wagage D, Holbeck SL, Morse HC, Stroncek D, Marincola FM. HLA class I and II genotype of the NCI-60 cell lines. J Transl Med. 2005;3:1–11. doi: 10.1186/1479-5876-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol. 1998;160:4951–4960. [PubMed] [Google Scholar]
- 10.Lo Monaco E, Sibilio L, Melucci E, Tremante E, Suchànek M, Horejsi V, Martayan A, Giacomini P. HLA-E: strong association with β2m and surface expression in the absence of HLA class I signal sequence-derived peptides. J Immunol. 2008;181:5442–5450. doi: 10.4049/jimmunol.181.8.5442. [DOI] [PubMed] [Google Scholar]
- 11.Marìn R, Ruiz-Cabello F, Pedrinaci S, Mendez R, Jimenez P, Geraghty DE, Garrido F. Analysis of HLA-E expression in human tumors. Immunogenetics. 2003;54:767–775. doi: 10.1007/s00251-002-0526-9. [DOI] [PubMed] [Google Scholar]
- 12.Fruci D, Ferracuti S, Limongi MZ, Cunsolo V, Giorda E, Fraioli R, Sibilio L, Carroll O, Hattori A, van Endert PM, et al. Expression of endoplasmic reticulum aminopeptidases in EBV-B cell lines from healthy donors and in leukemia/lymphoma, carcinoma, and melanoma cell lines. J Immunol. 2006;176:4869–4879. doi: 10.4049/jimmunol.176.8.4869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.