Abstract
Flavivirus gene expression is modulated by RNA secondary structure elements at the terminal ends of the viral RNA molecule. For tick-borne encephalitis virus (TBEV), four stem-loop (SL) elements have been predicted in the first 180 nucleotides of the viral genome: 5′-SL1, 5′-SL2, 5′-SL3 and 5′-SL4. The last three of these appear to be unique to tick-borne flaviviruses. Here, we report their characterization by mutagenesis in a TBEV luciferase reporter system. By manipulating their thermodynamic properties, we found that an optimal stability of the 5′-SL2 is required for efficient RNA replication. 5′-SL3 formation is also important for viral RNA replication, but although it contains the viral start codon, its formation is dispensable for RNA translation. 5′-SL4 appears to facilitate both RNA translation and replication. Our data suggest that maintenance of the balanced thermodynamic stability of these SL elements is important for temporal regulation of its different functions.
Keywords: Flavivirus, TBEV, RNA replication and translation, Replicon, 5′noncoding region
Introduction
Flaviviruses are small enveloped viruses with a positive-stranded RNA genome of about 11 kb, and they comprise one of the three genera within the family Flaviviridae (Lindenbach et al., 2007). The genus Flavivirus is further divided into three groups, based on their mode of transmission: mosquito-borne, tick-borne, and no known vector. The two arthropod-borne groups include important human pathogens such as West Nile virus (WNV), dengue virus (DENV), yellow fever virus (YFV), Japanese encephalitis virus (JEV) (all of them mosquito-borne) and tick-borne encephalitis virus (TBEV), the most important tick-transmitted flavivirus. TBEV is endemic in parts of Asia and Europe and causes thousands of cases of severe neurological illness every year (Gubler et al., 2007; Lindenbach et al., 2007).
The flavivirus genome is a multifunctional RNA molecule that serves as the sole mRNA for the translation of the viral polyprotein, the template for minus-strand synthesis, and the genomic RNA for incorporation into viral particles (Lindenbach et al., 2007; Wengler and Gross, 1978). To meet these diverse demands, it is likely that the flaviviral RNA molecule must change its overall secondary structure at specific stages of the viral life cycle. Specific RNA sequence motifs in the terminal regions of the flavivirus genome may mediate these essential rearrangements (Hahn et al., 1987; Markoff, 2003). A well-characterized aspect of these processes involves “cyclization” of the linear RNA molecule, in which base pairing of a set of complementary RNA stretches called cyclization sequences present in the terminal regions at opposite ends of the genome induces the formation of a panhandle-like structure (reviewed in Villordo and Gamarnik, 2009).
Genome cyclization is an important prerequisite for RNA replication because the promoter for minus-strand synthesis, i.e., the specific site where the replicase complex initially binds, has been shown to reside within a specific region of RNA in the 5′ noncoding region (NCR) that folds into a stable Y-shaped stem-loop (Filomatori et al., 2006; Li et al., 2010; Lodeiro et al., 2009). This element, called stem-loop A (SL-A) or 5′ stem-loop 1 (5′-SL1), is predicted to be formed in all flavivirus genomes (Brinton and Dispoto, 1988; Gritsun and Gould, 2007; Leyssen et al., 2002; Mandl et al., 1993; Thurner et al., 2004) and has recently been confirmed by RNAse mapping and chemical probing in DENV (Lodeiro et al., 2009).
Despite the fact that all flaviviruses have a similar genome structure and make use of RNA structural elements to mediate cyclization, minus-strand synthesis, and translation, other functional RNA elements at the distal ends of the genome with important roles in regulation of these processes exhibit a surprising lack of similarity between mosquito-borne and tick-borne flaviviruses (Markoff, 2003; Thurner et al., 2004), suggesting that there might be significant mechanistic differences in the way that RNA structural elements of mosquito-borne and tick-borne flaviviruses regulate translation and RNA replication. In addition to 5′-SL1, the tick-borne flaviviruses contain three other predicted stem-loop structures (5′-SL-2, 5′-SL3, and 5′-SL4) near their 5′ end, downstream of 5′-SL1 (Fig. 1A). 5′-SL2 contains the 5′ half of the TBEV cyclization element and is therefore expected to play a role in the regulation of cyclization and minus-strand synthesis. 5′-SL3 contains the AUG start codon of the viral polyprotein and thus has the potential to affect initiation of viral protein synthesis. 5′-SL4, which lies within the part of the translated region encoding the capsid (C) protein, is a potential analog of the capsid coding region hairpin element, cHP, which has been shown to participate in start codon selection as well as RNA synthesis in the mosquito-borne flaviviruses DENV and WNV (Clyde et al., 2008; Clyde and Harris, 2006).
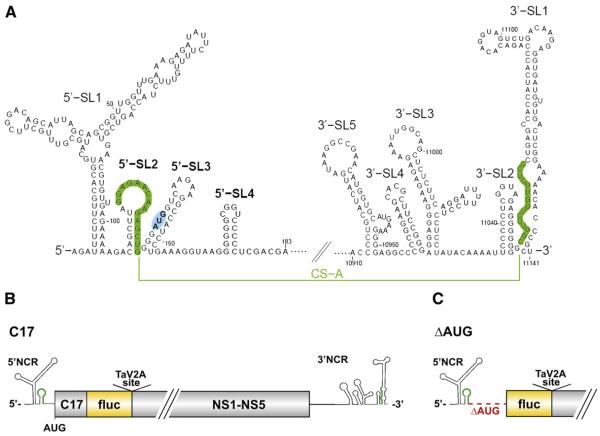
Fig. 1.
Genome secondary structure and organization of TBEV and derived replicon constructs. (A) The 5′ and 3′ regions of the TBEV genome. The stem-loop structures are shown as predicted for the linear form, without consideration of the long-range interaction of the cyclization sequences (CS) (depicted in green) in the viral5′-SL2 and 3′-SL1. The CS element was characterized in an earlier study (Kofler et al., 2006) and was originally called CS-A. The viral start codon AUG within the 5′-SL3 structure is highlighted in blue. (B) Schematic diagram of the parental replicon C17 (wt), in which the structural protein region of TBEV was replaced by an in-frame insertion of the firefly luciferase gene (fluc) (Rouha et al., 2010). C17, truncated capsid protein gene; NS1–NS5, the coding region for the non-structural proteins 1–5; NCR, non-coding region; TaV2A site, Thosea asigna virus 2A site. (C) Schematic diagram of mutant replicon ΔAUG, which has a deletion starting from nucleotide position 133, causing it to lack the entire region encoding the capsid protein, the 5′-SL3 element, and the viral start codon. Diagrams A–C are not drawn to scale.
In this study, we have carried out site-directed mutagenesis using a recently established TBEV reporter system to gain information about the possible functional roles of these structures. We provide evidence that increasing and decreasing the thermodynamic stability of each of these stem-loop structures can have both positive and negative effects on viral RNA replication efficiency. By looking at the kinetics of reporter expression, we were able to distinguish effects occurring at the level of translation and RNA replication, thus providing clues about the temporal regulation of translation and RNA replication in tick-borne flaviviruses.
Results
To analyze the roles of 5′-SL2, 5′-SL3 and 5′-SL4 of TBEV (Fig. 1A) in viral replication and translation, we made and tested a set of mutants in which each of these elements was modified individually in the reporter replicon C17 (Rouha et al., 2010), a modified genomic clone in which the genes for the viral structural proteins had been replaced by an in-frame insertion of the firefly luciferase gene.
In this system, in vitro-transcribed, capped RNA is used to transfect BHK-21 cells by electroporation, and when luciferase activity is measured, a biphasic kinetic profile is observed (Hoenninger et al., 2008). The first peak, which occurs at approximately 3 h posttransfection, represents translation of the original plus-strand RNA that was used for transfection. This is followed by a temporary decrease in luciferase activity, presumably due to degradation of a large proportion of the input RNA, which is then followed at later time points by a strong increase in activity due to replication of the original RNA and translation of the newly synthesized plus strand. Using appropriate controls to normalize for difference in transfection efficiency (see Materials and methods), the effects of mutations on the ability of the replicon to translate the input RNA (3 h peak) and on the kinetics of RNA replication (later time points) can be evaluated separately. This system has been applied successfully in earlier studies (Hoenninger et al., 2008; Rouha et al., 2010).
As negative controls for translation and RNA replication, two replicons were used throughout this study. The first one, called ΔAUG (Fig. 1C), is a construct lacking the entire region encoding the capsid protein and the AUG start codon (and therefore also lacking 5′-SL3 and 5′-SL4). This RNA construct cannot be translated and was therefore used as a negative control for examining translation of input RNA. The second one, NS5-GAA, is a translation-competent construct that is identical to the wild-type replicon except that it contains a mutated active-site motif (GDD to GAA) in the viral RNA-dependent RNA polymerase (NS5), making it unable to replicate (Hoenninger et al., 2008).
Analysis of 5′ stem-loop structure 2 (5′-SL2)
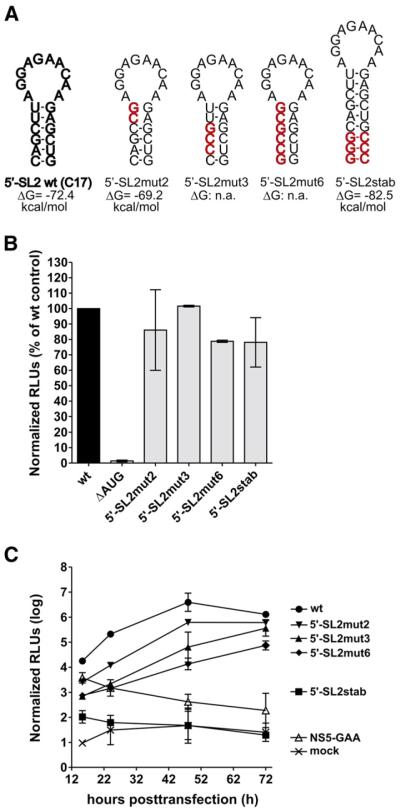
The 5′-SL2 element contains the 5′ cyclization sequence (5′-CS, in green in Fig. 1A). A long-range RNA interaction between the 5′-CS and a complementary sequence at the 3′ end (3′-CS) has been shown to be essential for TBEV RNA replication (Kofler et al., 2006), and the formation of 5′-SL2 would therefore be expected to interfere with this interaction. To investigate the significance of 5′-SL2 formation, we engineered a set of mutants in which the stability of this stem-loop structure was altered without changing the cyclization motif. In the mutants 5′-SL2mut3 and 5′-SL2mut6, three and six nucleotide mismatches, respectively, were introduced into the stem of 5′-SL2 to lower its stability (Fig. 2A, upper panel). Secondary structure predictions using the Vienna RNA Package (Hofacker, 2003) indicated that the original 5′-SL2 structure would no longer be formed in either of these mutants (not shown). Thermodynamic analysis of full-length genome sequences of both mutants, however, indicated that they retained their potential to establish long-range interactions with the 3′ cyclization element. Another mutant, 5′-SL2mut2, had two mismatches adjacent to the loop that were predicted to enlarge the loop but maintain the stem in a shorter and less stable form (Fig. 2A). In addition to the destabilizing mutations, we also made a construct to augment the stability of 5′-SL2 by extending its stem by three additional G-C base pairs at its bottom end (construct 5′-SL2stab, Fig. 2A). The calculated stability of 5′-SL2 in 5′-SL2stab was increased by 10.1 kcal/mol compared to the parental C17 replicon (Fig. 2A). As a consequence, long-range interaction with the 3′ cyclization element would be expected to be less thermodynamically favorable due to the increased energy required for this stem to open up.
Fig. 2.
Characterization of 5′-SL2. (A) Schematic drawing and thermodynamic analysis of the predicted secondary structure of wild-type 5′-SL2 and those of the engineered mutant replicons. Nucleotide changes are depicted in red. ΔG, Gibbs free energy of the secondary structure of the first 194 nucleotides of the viral genome. A decrease in the ΔG value relative to the wild-type indicates stabilization of the secondary structure, whereas an increase indicates destabilization. (B) Translation level of viral input RNA in BHK-21 cells 3 h posttransfection. Normalized luciferase levels are shown as percentages relative to the parental control C17 (wt). RNA of construct ΔAUG, which lacks the viral translational AUG start codon, was used as a negative control. Error bars represent standard deviation of at least two independent experiments. (C) Viral RNA replication efficiencies of the parental C17 replicon (wt) and mutants derived from it, monitored by comparing normalized firefly luciferase activity in BHK-21 cells 15.5–72 h posttransfection. Capped NS5-GAA RNA, which can be translated but cannot replicate due to a GDD-to-GAA mutation in the viral RNA polymerase gene NS5, was used as a control. Error bars represent the standard deviation of two independent experiments, each measured in triplicate.
To determine the effects of stabilization and destabilization of 5′-SL2 on translation and replication of the replicon RNA, the parental C17 construct and the mutant replicon RNAs described above were transcribed in vitro, and BHK-21 cells were transfected with equimolar amounts of each RNA together with the appropriate Renilla luciferase control construct (in vitro-transcribed RNA for early time points and plasmid DNA for later time points; see Materials and methods for details) to normalize for differences in transfection efficiency.
When normalized firefly luciferase activity was measured at 3 h after transfection, all four of the 5′-SL2 mutants exhibited the same level of activity as the parental wild-type construct (Fig. 2B), indicating that neither stabilization nor destabilization of this stem-loop had an effect on initial translation of the input RNA after transfection. However, the kinetics of luciferase activity at later time points (from 15.5 to 72 h posttransfection) revealed very clear differences between the mutants. Destabilization of 5′-SL2 led to impairment of RNA replication in an order corresponding to the degree of destabilization (mut6>mut3>mut2). Mutants 5′-SL2mut3 and 5′-SL2mut6, in which 5′-SL2 is predicted to not form at all, exhibited more than twentyfold less luciferase activity than the parental replicon C17 at 15.5 to 72 h after transfection (Fig. 2C). Strikingly, 5′-SL2stab was completely negative for RNA replication even though its ability to be translated was not impaired. These data suggest that the secondary structure of 5′-SL2 does not play a role in translation of the viral RNA but is important for RNA replication. The observation that both stabilization and destabilization of this element impair RNA replication is consistent with the idea that it is a transient structure that needs to be able to undergo dynamic changes in the course of the viral replication cycle, presumably to allow interaction of the cyclization sequences at the appropriate time.
Analysis of 5′ stem-loop structure 3 (5′-SL3)
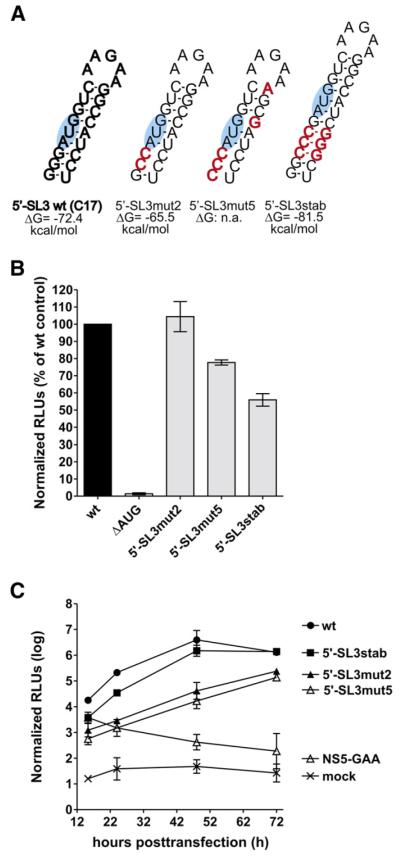
An obvious and important feature of the TBEV 5′-SL3 is the presence of the translational AUG start codon in the 5′ half of its stem (Fig. 1A), suggesting a possible role in translation of the viral polyprotein. To analyze the 5′-SL3 element, we designed mutants that would stabilize or destabilize its stem structure. The destabilizing mutants, called 5′-SL3mut2 and 5′-SL3mut5, had two and five silent nucleotide substitutions, respectively, leading to the mismatches in the stem shown in Fig. 3A. The mutations in 5′-SL3mut2 were predicted to cause a decrease in stability of 6.9 kcal/mol, and those in 5′-SL3mut5 were predicted to prevent stem-loop formation altogether. Another mutant, 5′-SL3stab, contained an insertion of three cytosines two nucleotides before the start codon and three guanines 16 nucelotides after the start codon to lengthen the stem and stabilize it by creating three additional base pairs (resulting in an additional glycine in the truncated capsid protein sequence preceding the reporter gene) (Fig. 3A). The stem-loop structure in 5′-SL3stab was predicted to be 9.1 kcal/mol more stable than its wild-type counterpart.
Fig. 3.
Characterization of 5′-SL3. (A) Schematic drawing and thermodynamic analysis of the predicted secondary structures of wild-type 5′-SL3 (wt, C17, bold letters) and engineered mutant replicons. Nucleotide changes are depicted in red. The viral start codon AUG within the 5′-SL3 structure is highlighted in blue. ΔG, Gibbs free energy of the secondary structure of the first 194 nucleotides of the viral genome. A decrease in the ΔG value relative to the wild-type indicates stabilization of the secondary structure, whereas an increase indicates destabilization. (B) Translation level of viral input RNA in BHK-21 cells 3 h posttransfection. Normalized luciferase levels are shown as percentages relative to the parental control C17 (wt). RNA of construct ΔAUG, lacking the viral translational start codon AUG, was used as a negative control. Error bars represent standard deviation of at least two independent experiments. (C) Viral RNA replication efficiencies of the parental C17 replicon and mutants derived from it, monitored by comparing normalized firefly luciferase activity in BHK-21 cells 15.5–72 h posttransfection. Capped NS5-GAA RNA, which can be translated but cannot replicate due to a GDD-to-GAA mutation in the viral RNA polymerase gene NS5, was used as a control. Error bars represent the standard deviation of two independent experiments, each measured in triplicate.
As shown in Fig. 3B, transfection of BHK-21 with each of these mutants resulted in luciferase activity at 3 h posttransfection, indicating that all of them were capable of being translated. However, quantitative differences were observed. While translation of the 5′-SL3mut2 construct was not impaired, the levels of luciferase expression with 5′-SL3mut5 and 5′-SL3stab were only about 80% and 55%, respectively, of that of the wild-type construct. Although this suggests a role of 5′-SL3 stability in achieving optimal translation, the differences in translation levels could also be caused by the sequence changes immediately preceding the AUG.
At the later time points (15.5–72 h; Fig. 3C), striking differences in the luciferase kinetic profiles were observed. While stabilizing 5′-SL3 appeared to have little effect on RNA replication (5′-SL3stab), destabilization of this element (5′-SL3mut2 and 5′-SL3mut5) resulted in a more than twenty-fold reduction in luciferase activity. It therefore appears that the ability to form 5′-SL3 is necessary for efficient RNA replication.
Analysis of 5′ stem-loop structure 4 (5′-SL4)
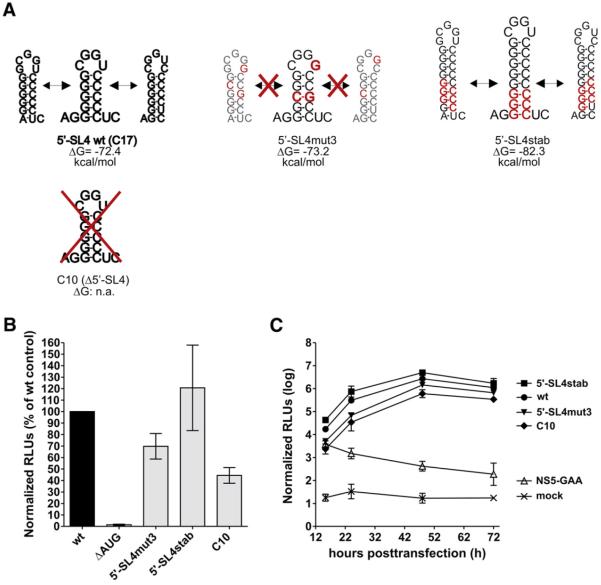
5′-SL4 occupies a position just within the beginning of the viral coding region. To study this element, we first performed a computational analysis of its thermodynamic properties. This revealed an exceptionally high potential of the stem-loop to form several slightly different variants, i.e., unlike the 5′-SL2 and 5′-SL3 structures, there was not a single strongly favored secondary structure. At least three different hairpins are likely to occur at this position in a pool of viral RNAs (Fig. 4A, upper left panel). This “secondary-structure ensemble” is caused by a series of six consecutive Gs in the 5′ half of the stem and a series of five consecutive Cs in the 3′ half, which allows the base pairing to shift for one or two nucleotides in 5′ or 3′ direction without a significant change in thermodynamic stability. To investigate whether this slippage to form an ensemble of structures is itself important in viral RNA replication or translation, we generated three different mutants (Fig. 4A). In 5′-SL4mut3, three silent nucleotide changes were introduced that were predicted by thermodynamic analysis to prevent the hairpin element from forming alternative base pairings. This was done by introducing a G-C anchor in the middle of the stem region, thus favoring only one of the possible structures. The thermodynamic stability of the resulting hairpin, however, was largely unchanged compared to the wild-type structure ensemble (Fig. 4A). An opposite outcome was achieved with mutant 5′-SL4stab. Three additional G-C pairs were introduced at the base of the stem, resulting in an elongation and strong stabilization of the resulting hairpin structures, but in this case, the potential of these stabilized hairpins to slide like those in the wild-type structure was retained. In mutant C10, the entire 5′-SL4 sequence element was deleted (the mutant designation C10 indicates that it encodes only the first 10 amino acids of the capsid protein, whereas the parental C17 encodes the first 17 residues).
Fig. 4.
Characterization of 5′-SL4. (A) Schematic drawing of the predicted secondary structure for wild-type 5′-SL4 (C17 wt) and engineered mutant replicons including thermodynamic analysis. Nucleotide changes are depicted in red. In mutant C10 only the codons for the first 10 (instead of 17) amino acid residues were maintained. It therefore lacks the entire 5′-SL4 element. ΔG, Gibbs free energy of the secondary structure of a segment of the first 194 nucleotides of the viral genome. A decrease in the ΔG value relative to the wild-type indicates stabilization of the secondary structure, whereas an increase reflects destabilization. (B) Translation level of viral input RNA in BHK-21 cells 3 h posttransfection. Normalized luciferase levels are shown as percentages relative to the parental control C17. RNA of construct ΔAUG, which lacks the viral translational AUG start codon, was used as a negative control. Error bars represent the standard deviation of at least two independent experiments. (C) Viral RNA replication efficiencies of the parental C17 replicon and mutants derived from it, monitored by comparing normalized firefly luciferase activity in BHK-21 cells 15.5–72 h posttransfection. Capped NS5-GAA RNA, which can be translated but cannot replicate due to a GDD-to-GAA mutation in the viral RNA polymerase gene NS5, was used as a control. Error bars represent the standard deviation of two independent experiments, each measured in triplicate.
All three replicons were tested for translation and replication as described above. As shown in Fig. 4B, deletion of the entire 5′-SL4 element (C10) reduced translation of the input RNA by approximately 50%. Interestingly, the mutant in which slippage was prevented (5′-SL4mut3) also seemed to be impaired in translation efficiency. Although this mutant is predicted to maintain one structure of the original ensemble, the luciferase assay revealed a more than 30% diminished translation efficiency compared to the parental C17 replicon (Fig. 4B). In contrast, the elongation and stabilization of 5′-SL4 did not negatively affect the efficiency of translation of input RNA, and possibly even increased it (Fig. 4B). This supports the hypothesis that the structural flexibility of 5′-SL4 has a positive effect on translation. However, the fact that the C10 deletion mutant was also functional means that 5′-SL4 is not absolutely essential.
Monitoring luciferase activity from 15.5 to 72 h posttransfection revealed that all three mutant replicons were capable of replicating efficiently. Their kinetic curves (Fig. 4C) were generally parallel in form, but the absolute levels of luciferase activity differed by as much as 30 fold. However, since the relative luciferase levels at each time point reflected those initially seen after transfection (5′-SL4stab>wt>5′-SL4mut3>C10), this might also be a combinatory or indirect effect due to the differences in translation efficiency.
Discussion
RNA secondary structures located in both terminal regions of the viral genome play decisive roles in the flavivirus life cycle. Although this is a common characteristic of all flaviviruses, notable differences exist between tick-borne and mosquito-borne members of the genus regarding the functional interplay, specific genomic location and exact sequence composition of these cis-acting elements.
In general, tick-borne flaviviruses, including the Far Eastern and the Western subtypes of TBEV and Powassan virus, tend to have longer 5′ NCRs than the mosquito-borne members of the genus (Markoff, 2003). Although the 5′-terminal regions of mosquito-borne and tick-borne flaviviruses otherwise have little in common, they nevertheless share at least two important features: (i) one or more cyclization elements that interact with a complementary sequence in the 3′ NCR during replication and (ii) a Y-shaped stem-loop structure (SL-A or 5′-SL1) at the beginning of the genome that serves as a promoter for minus-strand synthesis.
Recently, the predicted Y-shaped SL-A structure of dengue virus was confirmed experimentally, and specific structural elements within SL-A that are important for RNA synthesis were identified (Filomatori et al., 2011, 2006; Lodeiro et al., 2009; Polacek et al., 2009). It has been shown that specific binding of the viral replicase to this structure brings it into close proximity to the 3′-terminus, where it initiates synthesis of the negative-strand RNA molecule (Filomatori et al., 2006). Similar results could also be obtained with West Nile virus (Dong et al., 2008) and the presence of the 5′-SL1/SL-A structure in all flavivirus genomes suggests that its functional role is conserved throughout the genus.
In addition to the SL-A element, two additional stem-loops have been predicted in the 5′ part of the genomes of DENV types 1–4: the SL-B element within the 5′ NCR (which contains the 5′ half of one of the mosquito-borne cyclization sequences) and the cHP element, which lies just within the capsid coding region and is involved in RNA translation and replication (for a review see Paranjape and Harris, 2010). Recently, both of these stem-loops were confirmed by solution structure probing (Polacek et al., 2009).
Although tick-borne and mosquito-borne flaviviruses both use cyclization sequences, the specific sequences, their locations, and even the number of elements involved differ significantly. In mosquito-borne flaviviruses, a 5′ cyclization sequence (5′-CS) within the capsid coding region binds to a complementary and highly conserved 3′-CS element in the 3′ NCR (Alvarez et al., 2005; Hahn et al., 1987; Khromykh et al., 2001; Lo et al., 2003). In addition, a second, separate interaction involved in cyclization has been reported (Alvarez et al., 2008, 2005; Zhang et al., 2008), termed 5′–3′ UAR (upstream AUG region because the 5′ element is located upstream of the translation initiator AUG in the 5′ UTR). A third pair of complementary interacting sequence elements has been proposed recently, the 5′–3′-DAR (downstream AUG region) elements (Friebe and Harris, 2010). In tick-borne flaviviruses, a different set of sequence motifs mediates this process. Using a series of disruptive and compensatory mutations, it has been demonstrated that the tick-borne 5′ cyclization element (5′-CS) is located upstream rather than downstream of the viral start codon and that the complementary 3′ part (3′-CS) is located at the bottom of the terminal stem-loop 3′-SL1 (Kofler et al., 2006)(Fig. 1A). Therefore, this part of the 3′-SL1 cannot form when the genome has assumed the circularized conformation. Although another set of inverted sequence motifs located in the capsid coding region and the 3-NCR had been identified as potential cyclization elements by computational analysis (Khromykh et al., 2001; Kofler et al., 2006; Mandl et al., 1993), experimental evidence suggests that these sequences do not contribute to cyclization, and TBEV therefore appears to possess only a single pair of cyclization sequences (Kofler et al., 2006).
Since both halves of the TBEV cyclization element are predicted to be sequestered within stem-loop elements (5′-SL2 and 3′-SL1), it is very likely that these structures have to be disrupted at the appropriate time to allow cyclization to occur. In this study, we introduced mutations to both stabilize and destabilize 5′-SL2 without altering the cyclization sequence. These mutations did not affect translation of the viral input RNA, indicating that 5′-SL2 is not directly involved in RNA translation, and this is consistent with previous reports that flavivirus translation does not require genome cyclization (Alvarez et al., 2005; Chiu et al., 2005; Lo et al., 2003). However, both the ability of 5′-SL2 to form and its ability to open up were required for efficient RNA replication. In fact, stabilization of this element by approximately 10 kcal/mol abolished replication completely. Not only does this indicate that the 5′-SL2 element plays an important role in the process of RNA replication, but it also provides indirect but compelling evidence that the viral RNA molecule assumes different conformations during its replication cycle. As long as the cyclization sequence is internally base-paired within the 5′-SL2 structure, interaction with the corresponding 3′ sequence at the bottom of the terminal stem-loop 3′-SL1 is prevented (Fig. 1A). Therefore, the ability of the viral RNA to cyclize probably depends on the stability of 5′-SL2 remaining within an optimal range. If the stability of 5′-SL2 is too high (as seen with 5′-SL2stab), the 5′ half of the cyclization sequence is not free to undergo a long-range interaction with the 3′ half. On the other hand, if the stability is too low or if no stem-loop is formed at all, this might result in a permanent cyclized state, which could disrupt the timing of the replication cycle. These observations are in excellent agreement with recently reported data on alternative conformations of the dengue virus genome. Filomatori et al. (2011) demonstrated that efficient initiation of DENV RNA synthesis depends on the presence of unpaired 3′-terminal nucleotides of the template RNA. This implies that the secondary structure element at the 3′-terminus, which is highly conserved among all flaviviruses, has to partially open up to lift the repression of minus-strand synthesis. A mechanistic explanation for this is provided by the 3D structure of the RNA polymerase protein NS5 (Yap et al., 2007), which shows the template channel of this enzyme to be unable to accommodate double-stranded RNA. The notion of an important role of achieving a balanced equilibrium between linear and cyclic confirmations of the flavivirus genome is further supported by another publication from the same group on dengue virus (Villordo et al., 2010). In that study, a new small hairpin element (sHP) that is present only in the linear genomic conformation was identified and shown to have an important role in replication and infectivity. Our results suggest that balancing the linear and cyclic genome conformations plays a similarly important role in the replication of tick-borne flaviviruses. Future investigations should focus on the relationship between alternative RNA secondary structures and temporal regulation of the virus life cycle and clarify whether a functional analog to the DENV sHP motif is also present in the TBEV genome.
Although the mechanism is less apparent than with 5′-SL2, 5′-SL3 also appears to have a potential regulatory function. This element contains the start codon for the viral polyprotein, but earlier studies performed in our laboratory with mutant viruses that lack this element have already established that there is no absolute requirement for this stem-loop element in TBEV replication. In the absence of 5′-SL3, translation can be initiated from an alternative start codon, and this is still sufficient to generate a detectable signal in immunofluorescence staining (Kofler et al., 2006). The more sophisticated experimental system used here has now allowed us to test whether stabilization or destabilization of 5′-SL3 influences RNA translation or replication levels. As expected, neither translation nor replication was fully abolished by any of our mutations. Interestingly, however, we observed a 10- to 20-fold reduction in RNA replication levels with our destabilized mutants. We therefore propose that the formation of 5′-SL3 is necessary for optimal RNA replication.
The TBEV 5′-SL4 is a putative analog of the cHP element found in DENV and WNV, which is believed to cause stalling of the ribosome during translation initiation, and this has been proposed to favor the recognition of the correct start codon (Clyde and Harris, 2006). Its presence at the appropriate position has also been shown to be required for RNA replication, although its specific sequence does not appear to be important (Clyde et al., 2008). It has been speculated that this hairpin might have been maintained in different flaviviruses by sequence covariation (Clyde and Harris, 2006). Considering that the TBEV virus start codon is – in contrast to the AUG of most mosquito-borne flaviviruses (exceptions JEV and YFV) – embedded in a strong Kozak initiation context and that the 5′-SL4 is not situated at distance from the start codon that would correspond to a ribosomal footprint if the ribosome were paused over the start codon (26–28 nt in case of the 5′-SL4 instead of 12–15 nt), (Clyde and Harris, 2006; Kozak, 1991), a contribution of the 5′-SL4 in TBEV RNA translation is questionable. Interestingly, our folding predictions suggested that the 5′-SL4 does not fold into a single dominant secondary structure but instead exists in an ensemble of hairpins that are able to shift towards the viral 5′ or 3′ end. According to our computational analysis, this is a specific feature of the tick-borne 5′-SL4 element but not the mosquito-borne cHP element. Our results indicate that the TBEV 5′-SL4 is a determinant of RNA replication efficiency, either directly or indirectly. Reducing its stability and eliminating its structural flexibility both had a negative effect on these processes, whereas no negative effect was observed upon stabilization of 5′-SL4. Therefore, unlike 5′-SL2 and 5′-SL3, there is no evidence that 5′-SL4 needs to be able to open up at a specific stage in the replication cycle. The different RNA stem-loop structures at the 5′ end of the TBEV genome thus appear to be used in different ways to modulate translation and RNA replication.
Materials and methods
Plasmids and cells
The replicon constructs used in this study were all derived from the cDNA clone of TBEV Western subtype prototypic strain Neudoerfl (GenBank accession no. U27495). All of them were constructed by modifying the previously described C17 replicon construct. Construction of the C17 replicon, which was generated by the deletion of the start codon at position 132–134 (numbering according to the full-length sequence of strain Neudoerfl) of C17fluc-TaV2A (Hoenninger et al., 2008) has been described elsewhere. Briefly, C17 lacks most of the coding region for the structural proteins, which is replaced by a firefly luciferase (fluc) gene with the start codon (ATG) deleted, followed by a 60-nucleotide-long 2A processing site of Thosea asigna virus (Fig. 1B) (Rouha et al., 2010). All of the constructs contained a T7 promoter, allowing the RNA replicon to be generated by in vitro transcription.
Cell culture
BHK-21 cells (ATCC CL10) were grown in Eagle’s minimal essential medium (EMEM) supplemented with 5% fetal calf serum (FCS). The cells were electroporated using a GenePulser apparatus (Bio-Rad) as described previously (Mandl et al., 1997). After transfection, the growth medium was replaced by a maintenance medium consisting of EMEM without phenol red (Cambrex) containing 1% FCS.
Mutant construction
Firefly luciferase reporter replicons ΔAUG and C10 were constructed by PCR amplification with the forward primer F-SalIreg (GCATCGGTCGACTTAATACGA) and the reverse primer NS1_SS_R (GCTCATGGACATTGTAGGGTTTCT) using the parental plasmid clones Δ5′-SL3-(Δ130–183) and C10-(Δ163–183), which do not contain a reporter gene and have been described in a previous study (Kofler et al., 2006). The PCR products were trimmed with SalI and PacI, replacing a fragment extending from a SalI site preceding the T7 promoter to a PacI site at nucleotide 184 with the corresponding region from firefly luciferase replicon C17. The replicon 5′-SL4mut3 was created by amplification using a partial cDNA clone, C17ΔCME (containing the region extending from a unique SalI restriction site to the unique ClaI site at position 3155 within the NS1 coding region), as the template, with primer SalIreg and mutagenic primers R-5′-SL4mut3 GGTCTTAATTAATCGTCGAGGCGGCCCGCCGC.
Plasmid 5′-SL2stab was created by three-step PCR. PCR I was performed using wild-type primers (F-SalIreg and R-E1) with the mutagenic primer R-3′-SL2stab 5′-GGGCAGCTCTTGTTCTCCTAAGCTGCCCTCTTTTTCTCAACACGTT-3′ and F-5′-SL2stab 5′-GGGCAGCTTAGGAGAACAAGAGCTGCCCGGGATGGTCAAGAAGGC-3′.
The mutants 5′-SL2mut2 and 5′-SL2mut3 were generated using a GeneTailor Site-Directed Mutagenesis System (Invitrogen) with mutagenic forward primers F-5′-SLmut2 (5′-CGTGTTGAGAAAAAGACAGCCGAGGAGAACAA-3′) and F-5′-SLmut3 (5′-ACGTGTTGAGAAAAAGACCCGTTAGGAGAA) and a wild-type reverse primer.
To generate all of the other mutants (SL2mut6, SL3mut2, SL2mut5, SL2stab, SL4mut6, SL4stab), the region from SalI and PacI was synthesized by the company “GeneArt”, and the synthetic DNA was inserted into the C17 replicon, taking advantage of the SalI and PacI restriction sites.
All plasmids were amplified in Escherichia coli HB101 and purified using commercially available plasmid purification systems (QIAGEN).
In vitro transcription, transfection and luciferase assay
RNA was transcribed from 1-μg aliquots of the plasmid DNA by T7 polymerase transcription, using commercially available reagents (Ambion) and conditions described in detail elsewhere (Mandl et al., 1997). Briefly, after the transcription reaction, template DNA was digested by incubation with DNAseI, purified, and separated from unincorporated nucleotides using an RNeasy Mini Kit (QIAGEN). RNA was then quantified spectrophotometrically, and equal amounts (corresponding to approximately 1.4 × 1012 copies) were introduced into BHK-21 cells by electroporation with a Bio-Rad Gene Pulser as described previously (Mandl et al., 1997). RNA or DNA containing the Renilla luciferase (Rluc) gene was used as internal control for the standardization of transfection efficiency. For translation experiments (analysis of input RNA, 3 h posttransfection) capped Rluc mRNA, in vitro transcribed from BamHI-linearized Rluc expression plasmid phRL-SV40 was used. For measuring RNA replication at later time points, the expression plasmid phRL-SV40 itself was co-electroporated as the control. Luciferase assays were performed as described elsewhere (Hoenninger et al., 2008).
Folding predictions
For all calculations of secondary structures, we used the Vienna RNA Package (Hofacker, 2003). Calculations using the complete sequence of the wild-type genome C17 as well as all of the mutant contructs used in this study revealed long-range interactions predicted to result to a panhandle-like overall genome structure. For calculation of 5′ and 3′ secondary structures as shown in Fig. 1, we recalculated the complete sequence with the constraints that nt 115–123, nt 11060–11063, and 11068–11071 were not allowed to participate in any secondary structure. We obtained the secondary structures shown in Fig. 1A for the 5′ and 3′ terminal regions of the genome. Because closer inspection of folding probabilities showed that there were no secondary structures starting in the first 194 nucleotides that folded together with sections of the genome farther downstream, we decided that it was feasible to use the first 194 nucleotides as the template for estimating the ΔG values of the 5′ stem loops SL2, SL3 and SL4. As a reference value for the Gibbs free energy, we calculated the difference between the Gibbs free energies of the 5′ 194 nucleotides with and without the constraint that the respective stem loops would be allowed to form. We repeated the same calculations using the mutant sequences and thus obtained the contribution of the mutated stem-loops to the overall Gibbs free energy of the structure.
Acknowledgments
We gratefully acknowledge Steven L. Allison for the helpful discussions in data evaluation and his invaluable assistance during the manuscript preparation, and Karen Pangerl for critically reading the manuscript. Funding for this research was provided by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (grants P19411-B11 and P20533-B03).
References
- Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA–RNA interactions circularize the dengue virus genome. J. Virol. 2005;79(11):6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez DE, Filomatori CV, Gamarnik AV. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology. 2008;375(1):223–235. doi: 10.1016/j.virol.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1988;162(2):290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- Chiu WW, Kinney RM, Dreher TW. Control of translation by the 5′- and 3′-terminal regions of the dengue virus genome. J. Virol. 2005;79(13):8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K, Harris E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J. Virol. 2006;80(5):2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K, Barrera J, Harris E. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology. 2008;379(2):314–323. doi: 10.1016/j.virol.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhang B, Shi PY. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology. 2008;381(1):123–135. doi: 10.1016/j.virol.2008.07.040. [DOI] [PubMed] [Google Scholar]
- Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20(16):2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J. Biol. Chem. 2011;286(9):6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Harris E. Interplay of RNA elements in the dengue virus 5′ and 3′ ends required for viral RNA replication. J. Virol. 2010;84(12):6103–6118. doi: 10.1128/JVI.02042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsun TS, Gould EA. Origin and evolution of flavivirus 5′UTRs and panhandles: trans-terminal duplications? Virology. 2007;366(1):8–15. doi: 10.1016/j.virol.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Kuno G, Markoff R. Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed Lippincott Williams & Wilkins Co.; Philadelphia, PA: 2007. pp. 1154–1252. [Google Scholar]
- Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 1987;198(1):33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Hoenninger VM, Rouha H, Orlinger KK, Miorin L, Marcello A, Kofler RM, Mandl CW. Analysis of the effects of alterations in the tick-borne encephalitis virus 3′-noncoding region on translation and RNA replication using reporter replicons. Virology. 2008;377(2):419–430. doi: 10.1016/j.virol.2008.04.035. [DOI] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31(13):3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001;75(14):6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler RM, Hoenninger VM, Thurner C, Mandl CW. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J. Virol. 2006;80(8):4099–4113. doi: 10.1128/JVI.80.8.4099-4113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266(30):19867–19870. [PubMed] [Google Scholar]
- Leyssen P, Charlier N, Lemey P, Billoir F, Vandamme AM, De Clercq E, de Lamballerie X, Neyts J. Complete genome sequence, taxonomic assignment, and comparative analysis of the untranslated regions of the Modoc virus, a flavivirus with no known vector. Virology. 2002;293(1):125–140. doi: 10.1006/viro.2001.1241. [DOI] [PubMed] [Google Scholar]
- Li XF, Jiang T, Yu XD, Deng YQ, Zhao H, Zhu QY, Qin ED, Qin CF. RNA elements within the 5′ untranslated region of the West Nile virus genome are critical for RNA synthesis and virus replication. J. Gen. Virol. 2010;91(5):1218–1223. doi: 10.1099/vir.0.013854-0. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel H-J, Rice CM. Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed Lippincott Williams & Wilkins Co.; Philadelphia, PA: 2007. pp. 1101–1152. [Google Scholar]
- Lo MK, Tilgner M, Bernard KA, Shi PY. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003;77(18):10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodeiro MF, Filomatori CV, Gamarnik AV. Structural and functional studies of the promoter element for dengue virus RNA replication. J. Virol. 2009;83(2):993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl CW, Holzmann H, Kunz C, Heinz FX. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology. 1993;194(1):173–184. doi: 10.1006/viro.1993.1247. [DOI] [PubMed] [Google Scholar]
- Mandl CW, Ecker M, Holzmann H, Kunz C, Heinz FX. Infectious cDNA clones of tick-borne encephalitis virus European subtype prototypic strain Neudoerfl and high virulence strain Hypr. J. Gen. Virol. 1997;78(5):1049–1057. doi: 10.1099/0022-1317-78-5-1049. [DOI] [PubMed] [Google Scholar]
- Markoff L. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 2003;59:177–228. doi: 10.1016/S0065-3527(03)59006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjape SM, Harris E. Control of dengue virus translation and replication. Curr. Top. Microbiol. Immunol. 2010;338:15–34. doi: 10.1007/978-3-642-02215-9_2. [DOI] [PubMed] [Google Scholar]
- Polacek C, Foley JE, Harris E. Conformational changes in the solution structure of the dengue virus 5′ end in the presence and absence of the 3′ untranslated region. J. Virol. 2009;83(2):1161–1166. doi: 10.1128/JVI.01362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouha H, Thurner C, Mandl CW. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 2010;38(22):8328–8337. doi: 10.1093/nar/gkq681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurner C, Witwer C, Hofacker IL, Stadler PF. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 2004;85(5):1113–1124. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- Villordo SM, Gamarnik AV. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139(2):230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villordo SM, Alvarez DE, Gamarnik AV. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA. 2010;16(12):2325–2335. doi: 10.1261/rna.2120410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G, Gross HJ. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology. 1978;89(2):423–437. doi: 10.1016/0042-6822(78)90185-x. [DOI] [PubMed] [Google Scholar]
- Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, Vasudevan SG, Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 2007;81(9):4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Dong H, Stein DA, Iversen PL, Shi PY. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA in-teractions. Virology. 2008;373(1):1–13. doi: 10.1016/j.virol.2008.01.016. [DOI] [PubMed] [Google Scholar]