Abstract
Three halophilic isolates, strains Halo-G*T, AUS-1 and Naxos II, were compared. Halo-G* was isolated from an evaporitic salt crystal from Baja California, Mexico, whereas AUS-1 and Naxos II were isolated from salt pools in Western Australia and the Greek island of Naxos, respectively. Halo-G*T had been exposed previously to conditions of outer space and survived 2 weeks on the Biopan facility. Chemotaxonomic and molecular comparisons suggested high similarity between the three strains. Phylogenetic analysis based on the 16S rRNA gene sequences revealed that the strains clustered with Halorubrum species, showing sequence similarities of 99.2–97.1 %. The DNA–DNA hybridization values of strain Halo-G*T and strains AUS-1 and Naxos II are 73 and 75 %, respectively, indicating that they constitute a single species. The DNA relatedness between strain Halo-G*T and the type strains of 13 closely related species of the genus Halorubrum ranged from 39 to 2 %, suggesting that the three isolates constitute a different genospecies. The G+C content of the DNA of the three strains was 65.5–66.5 mol%. All three strains contained C20C20 derivatives of diethers of phosphatidylglycerol, phosphatidylglyceromethylphosphate and phosphatidylglycerolsulfate, together with a sulfated glycolipid. On the basis of these results, a novel species that includes the three strains is proposed, with the name Halorubrum chaoviator sp. nov. The type strain is strain Halo-G*T (=DSM 19316T =NCIMB 14426T =ATCC BAA-1602T).
The current classification of halophilic archaea is based on phenotypic characteristics, chemical data (polar lipid composition) and genetic data (16S rRNA gene sequence information and DNA–DNA hybridization) (Oren et al., 1997; Grant et al., 2001). Strains of the genus Halorubrum are known to use carbohydrates as sources of carbon and energy, as was first described for Halorubrum saccharovorum (Tomlinson & Hochstein, 1976), the type species of the genus (McGenity & Grant, 2001). At the time of writing, the genus Halorubrum contains 19 species with validly published names: Hrr. saccharovorum (Tomlinson & Hochstein, 1976), Hrr. sodomense (Oren, 1983), Hrr. lacusprofundi (Franzmann et al., 1988), Hrr. trapanicum (McGenity & Grant, 1995), Hrr. coriense and Hrr. distributum (Kamekura & Dyall-Smith, 1995), Hrr. vacuolatum (Kamekura et al., 1997; Grant & Larsen, 1989), Hrr. tebenquichense (Lizama et al., 2002), Hrr. terrestre (Ventosa et al., 2004), Hrr. tibetense (Fan et al., 2004), Hrr. xinjiangense (Feng et al., 2004), Hrr. alkaliphilum (Feng et al., 2005), Hrr. lipolyticum and Hrr. aidingense (Cui et al., 2006), Hrr. orientale (Castillo et al., 2006), Hrr. ezzemoulense (Kharroub et al., 2006), Hrr. arcis (Xu et al., 2007), Hrr. litoreum (Cui et al., 2007) and Hrr. ejinorense (Castillo et al., 2007).
We describe here three halophilic archaeal strains that were isolated from a marine intertidal area along the coast of Baja California, Mexico (strain Halo-G*T; 28° N 114° W), natural salt-water pools on the Western Australian coast (strain AUS-1) and from a salt lake on the island of Naxos, Greece (strain Naxos II; 37° 04′ 35.77″ N 25° 20′ 52.21″ E). The three strains belong to the genus Halorubrum and proved to be very similar in their properties, suggesting a wide distribution of these haloarchaea. In addition, strain Halo-G*T is of special significance, because it had been dried onto quartz disks and flown on the Biopan facility, a small retrievable capsule developed by the European Space Agency for exposure of biological samples in low Earth orbit (ESA, 2005), and survived exposure to conditions of outer space for 2 weeks (Mancinelli et al., 1998).
All strains were isolated by enrichment in liquid medium and repeated streaking on agar medium as follows. For strain Halo-G*T, the medium contained (g l−1) casein hydrolysate (HyCase; Sigma), 5; yeast extract (Difco), 5; NaCl, 200; KCl, 2; MgCl2 .6H2O, 20; CaCl2 .2H2O, 0.2 (adjusted to pH 7.4). For strain AUS-1, the medium contained (g l−1) polypeptone (Daigo Eiyo), 3.3; trisodium citrate, 3; NaCl, 250; KCl, 2; MgSO4, 10; CaCl2 .2H2O, 0.2 (adjusted to pH 7.2 with NaOH). For strain Naxos II, M2 medium was used, containing (g l−1) HyCase, 5; yeast extract (Difco), 5; Tris, 12.1; NaCl, 200; KCl, 2; MgCl2 .6H2O, 20; CaCl2 .2H2O, 0.2 (adjusted to pH 7.4 with HCl). For solidification, 20 g agar l−1 was added to each medium. Routine cultivation was in M2 medium at 40 °C and pH 7.4. Growth ranges and optima of NaCl and MgCl2 were determined using the growth medium containing various concentrations of NaCl (0.9–5.2 M) and MgCl2 (0–0.5 M). Phenotypic tests were performed according to the proposed minimal standards for the description of new taxa in the order Halobacteriales (Oren et al., 1997). The methods used were described previously (Stan-Lotter et al., 2002; Gruber et al., 2004). All tests were performed at least in triplicate with the exception of utilization of amino acids, which was tested in duplicate. Unless otherwise indicated, tests were done in M2 medium at pH 7.2–7.4 with incubation at 37 °C. The utilization of carbohydrates or amino acids was tested in a semi-defined medium which contained (g l−1) yeast extract, 0.2; Tris, 6.05; NaCl, 233; KCl, 2; MgCl2 .6H2O, 20; CaCl2 .2H2O, 0.2 ; NH4Cl, 0.053; trace element solution (Malik, 1983), 0.1 ml; adjusted to pH 7.4 with HCl. Incubation was done in test tubes without shaking for 7 weeks and utilization of substrates was judged by cellular growth (Stan-Lotter et al., 2002). Susceptibility to antibiotics was tested by spreading cell suspensions on culture plates and applying discs impregnated with the following amounts of antibiotic: ampicillin (10 μg), anisomycin (10 μg), bacitracin (10 μg), chloramphenicol (10 μg), erythromycin (10 μg), kanamycin (10 μg), neomycin (10 μg), novobiocin (5 μg), rifampicin (10 μg) and tetracycline (10 μg).
Cell motility and morphology were observed under a phase-contrast light microscope and in dark field (Leica DM E). Gram staining of cells was performed according to Dussault (1955). Colony morphology was observed on agar medium under optimal growth conditions after incubation for 30 days.
Polar lipids were extracted with chloroform/methanol as described previously (Stan-Lotter et al., 2002). One- and two-dimensional TLC was performed with silica gel 60 plates (10×10 cm), using the solvent systems of Kamekura & Dyall-Smith (1995) and Stan-Lotter et al. (2002), respectively. Detection of phospholipids and functional groups was done as described previously (Stan-Lotter et al., 2002); in addition, sulfated lipids were detected by spraying with 0.016 % azure A (Sigma) in 1 mM H2SO4, according to Sprott et al. (2003).
The 16S rRNA genes of strains Halo-G*T and Naxos II were amplified by PCR using the primers Archae21F and 1525R, as described previously (Gruber et al., 2004). The nearly full-length nucleotide sequence (approx. 1400 bp) was determined for each strain. The 16S rRNA gene sequence of strain AUS-1 had been determined and deposited previously (Ihara et al., 1999). The web-based software mega 3 (http://www.megasoftware.net; Kumar et al., 2004) was used for sequence analysis and for construction of the phylogenetic tree. Comparison of the sequences with those of members of the family Halobacteriaceae was based on the neighbour-joining method (Saitou & Nei, 1987). In addition, maximum-parsimony and maximum-likelihood algorithms were used as described previously (Gruber et al., 2004) .
Chromosomal DNA for hybridization experiments was isolated and purified according to the methods described by Wilson (1987) and Marmur (1961). Determination of the G+C content was performed by the DSMZ Identification Service, following cell disruption with a French press and purification on hydroxyapatite (Cashion et al., 1977). Further details of the method have been described previously (Stan-Lotter et al., 2002). DNA–DNA hybridization studies were performed by the competition procedure of the membrane method (Johnson, 1994), described in detail by Arahal et al. (2001a, b). The hybridization temperature was 57.1 °C, which is within the limit of validity for the filter method (De Ley & Tijtgat, 1970), and the percentage of hybridization was calculated according to Johnson (1994). The experiments were carried out in triplicate. A few DNA–DNA hybridization experiments were performed by the DSMZ Identification Service (Stan-Lotter et al., 2002), using the thermal renaturation method of De Ley et al. (1970) with modifications by Huß et al. (1983).
The organisms are rods, 2–5 μm long. Liquid 96-h cultures of strains Halo-G*T, AUS-1 and Naxos II were motile and pleomorphic, although rod-shaped cells were most common (see Supplementary Fig. S1, available in IJSEM Online). All three strains were capable of growing over a range of NaCl concentrations from 2.0 M (12 %) to 5 M (30 %). They grew optimally in the presence of 4.3 M (25 %) NaCl, as has been shown for most extremely halophilic archaea. More details on phenotypic characteristics and results from nutritional tests are given in the species description.
TLC of polar lipids (Supplementary Fig. S2) suggested that all three strains contained phosphatidylglycerol, phosphatidylglyceromethylphosphate and phosphatidylglycerolsulfate derived from C20C20 glycerol diethers. A sulfated glycolipid S-DGD was also detected. This profile is similar to those reported for the neutrophilic species of Halorubrum (McGenity & Grant, 2001).
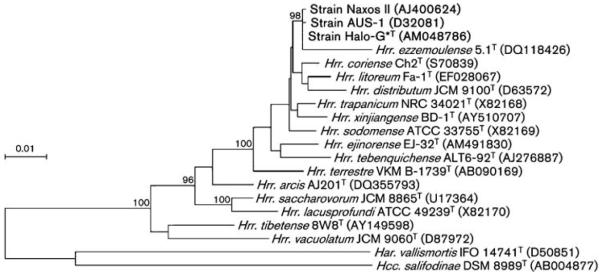
The 16S rRNA gene sequence of strain Halo-G*T was very similar to those of AUS-1 and Naxos II (99.8 % similarity to both); it was closely related to those of Hrr. coriense Ch2T (98.8 %), Hrr. trapanicum NRC 34021T (98.8 %), Hrr. xinjiangense BD-1T (98.8 %), Hrr. litoreum Fa-1T (98.7 %), Hrr. sodomense ATCC 33755T (98.7 %), Hrr. ejinorense EJ-32T (98.2 %), Hrr. distributum JCM 9100T (97.9 %) and Hrr. ezzemoulense 5.1T (97.3 %). Fig. 1 shows a phylogenetic tree that delineates the relationship of the three strains to each other and to the Halorubrum species. Similar topologies were obtained when other treeing methods were used (maximum-parsimony and maximum-likelihood; not shown). The signature sequences A, B and C for the genus Halorubrum (Grant et al., 2001) were present in all three strains without mismatches; Hrr. xingjiangense BD-1T had two mismatches in sequence C and Hrr. ezzemoulense 5.1T had two inserts in sequence B. In summary, it was concluded that strains Halo-G*T, AUS-1 and Naxos II formed a new distinct phylogenetic branch within the genus Halorubrum.
Fig. 1.
Phylogenetic tree based on the neighbour-joining algorithm, showing the relationships of strains Halo-G*T, AUS-1 and Naxos II and several Halorubrum type strains. The tree is based on an alignment of 16S rRNA gene sequences. Sequence accession numbers are given in parentheses. Bootstrap values higher than 80 out of 100 subreplicates are indicated at the respective bifurcations. The sequences of Haloarcula vallismortis IFO 14741T and Halococcus salifodinae DSM 8989T were used as the outgroup. Bar, 0.01 expected changes per site.
The DNA–DNA relatedness between strain Halo-G*T and strains Naxos II and AUS-1 was 75 and 73 %, respectively (determined in triplicate). In addition, the 13 Halorubrum type strains that showed 16S rRNA gene sequence similarities higher than 97 % with strain Halo-G*T [determined by using the fasta search and/or the EzTaxon 2.0 program; http://www.eztaxon.org (Chun et al., 2007)] were included in DNA–DNA hybridization experiments. The level of DNA–DNA relatedness between strain Halo-G*T and related Halorubrum species was as follows (three experiments each): 39 % with Hrr. ezzemoulense CECT 7099T, 35 % with Hrr. ejinorense EJ-32T, 32 % with Hrr. litoreum JCM 13561T, 31 % with Hrr. coriense JCM 9275T, 28 % with Hrr. distributum JCM 10118T, 25 % with Hrr. californensis SF3-213T, 23 % with Hrr. tebenquichense JCM 12290T, 21 % with Hrr. xinjiangense JCM 12388T, 20 % with Hrr. arcis JCM 13916T, 19 % with Hrr. terrestre VKM B-739T, 5 % with Hrr. sodomense JCM 8880T, 3 % with Hrr. saccharovorum ATCC 29252T and 2 % with Hrr. trapanicum JCM 10477T. These data indicated that strain Halo-G*T does not belong to any of these 13 other species, since DNA relatedness values <70 % have been suggested to justify designation to different species (Wayne et al., 1987); on the other hand, they showed that strains Halo-G*T, Naxos II and AUS-1 are members of the same species.
The phenotypic features, DNA–DNA hybridization values and phylogenetic data based on the 16S rRNA gene sequence comparison clearly supported the placement of strains Halo-G*T, AUS-1 and Naxos II in a novel species of Halorubrum, for which we propose the name Halorubrum chaoviator sp. nov. Table 1 shows features of all three strains that permit differentiation of the novel species from other related Halorubrum species.
Table 1. Differential characteristics of strains Halo-G*T, AUS-1 and Naxos II and type strains of related Halorubrum species.
Strains used for comparison: 1, Hrr. coriense DSM 10284T; 2, Hrr. trapanicum JCM 10477T; 3, Hrr. xinjiangense JCM 12388T; 4, Hrr. litoreum JCM 13561T, 5, Hrr. sodomense ATCC 33755T; 6, Hrr. ejinorense CECT 7194T; 7, Hrr. ezzemoulense CECT 7099T. Unless indicated, data for reference strains were taken from McGenity & Grant (2001), Feng et al. (2004), Kharroub et al. (2006), Castillo et al. (2007) and Cui et al. (2007). +, Positive; −, negative; PGS, phosphatidylglycerolsulfate; nd, no data available.
| Feature | Halo-G*T | AUS-1 | Naxos II | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|---|---|---|
| Morphology† | r/p | r/p | r/p | lr/p | r/p | sr | r | r/p | r | r/p |
| Cell width (μm) | 0.8 | 0.8 | 0.5–0.8 | 0.5 | 0.7–1.0 | 0.8–1.2 | 0.3–0.5 | 0.5 | 1.0–1.5 | 0.6 |
| Cell length (μm) | 2.0–4.0 | 2.0–4.0 | 2.0–5.0 | 5.0 | 1.5–3.0 | 1.8–2.6 | 2.0–5.0 | 2.5–5.0 | 5.0–8.0 | 1.5–3.0 |
| Pigmentation‡ | rd | rd | rd | rd–o | po | rd | rd | o–rd | rd | rd |
| Motility | + | + | + | + | − | + | + | + | − | + |
| Growth at 2 M NaCl | −/+§ | −/+§ | −/+§ | −/+∥§ | − | + | + | + | − | − |
| Temperature optimum (°C) |
37 | 37 | 37 | 50 | 37 | 40 | 37–42 | 40 | 37 | 37–40 |
| Growth at 10 °C | − | − | − | − | − | + | nd | − | nd | nd |
| Mg2+ requirement | 50 mM | None | 20 mM | 5 mM | nd | None | 30 mM | 5 mM | None | Required |
| Nitrite from nitrate | − | + | − | − | nd | − | + | nd | + | + |
| Hydrolysis of: | ||||||||||
| Starch | + | − | + | +∥ | − | − | − | + | − | − |
| Gelatin | − | − | − | − ∥ | − | nd | − | − | − | − |
| Utilization of: | ||||||||||
| (+)-d-Glucose | + | + | + | + | + | + | + | + | − | + |
| (−)-d-Fructose | + | + | + | + | + | + | − | + | − | − |
| (+)-d-Galactose | + | + | + | +∥ | + | − | + | + | − | − |
| Sucrose | − | − | − | + | + | + | + | − | − | + |
| Lactose | + | + | + | +∥ | − | − | − | + | − | − |
| Maltose | + | + | + | +∥ | + | + | + | + | − | + |
| Presence of: | ||||||||||
| PGS | + | + | + | + | + | + | + | + | − | + |
| S-DGD | + | + | + | + | + | + | + | + | − | + |
| G+C content (mol%) | 65.5 | 65.5 | 66.5 | 67.3∥ | 64.3 | 68.0 | 64.9 | 67.4 | 61.0 | 61.9 |
lr, Long rod; p, pleomorphic; r, rod; sr, short rod.
o, Orange; po, pale orange; prd, pale red; rd, red.
Very slow growth, after about 4 weeks.
Data from this study.
Description of Halorubrum chaoviator sp. nov.
Halorubrum chaoviator [cha.o.vi.a’tor. Gr. n. chaos empty space, the void; L. n. viator traveller; N.L. n. chaoviator (nominative in apposition) the traveller of the void, referring to the exposure of the type strain to conditions of outer space in the Biopan facility].
Cells stain Gram-negative. Cells are pleomorphic, although most are rod-shaped (Supplementary Fig. S1). Cells are approx. 2.0–5.0×0.5–0.8 μm. Colonies are circular and red pigmented, 1.5–2 mm in diameter following incubation for 30 days at 37 °C. Growth occurs at pH 7.0–8.5, 28–50 °C and NaCl concentrations of 2.0–5.0 M (12–30 %). No growth at 10 °C. Optimal growth occurs at pH 7.4, 37 °C and 4.3 M (25 %) NaCl. Extremely halophilic; cells lyse in water. The requirement for magnesium is variable among strains. Chemo-organotrophic, aerobic and oxidase- and catalase-positive. β-Galactosidase-positive; α-galactosidase activity is variable among strains. Anaerobic growth with nitrate or l-arginine does not occur. Tween 80, aesculin and gelatin are not hydrolysed. Starch hydrolysis and nitrate reduction to nitrite are variable among strains. Indole is not formed. Acid is produced from (+)-d-glucose, (+)-d-galactose, lactose and maltose, but not from (−)-d-fructose or sucrose. The following substrates are utilized as sole carbon and energy sources: (+)-d-galactose, (+)-d-glucose, (−)-d-fructose, maltose and lactose. No growth occurs on sucrose, l-arginine, l-glutamic acid or dl-phenylalanine. Polar lipids include phosphatidylglycerol, phosphatidylglyceromethylphosphate and phosphatidylglycerolsulfate derived from C20C20 glycerol diethers and the sulfated glycolipid S-DGD. Susceptible to bacitracin and novobiocin; not susceptible to ampicillin, anisomycin, chloramphenicol, erythromycin, kanamycin, neomycin, rifampicin or tetracycline. The DNA G+C content of the three known strains is 65.5–66.5 mol% (Tm).
The type strain is strain Halo-G*T (=DSM 19316T =NCIMB 14426T =ATCC BAA-1602T), which was isolated from an evaporitic salt crystal from Baja California, Mexico. Its G+C content is 65.5 mol%. Strains AUS-1 (=JCM 9573) and Naxos II, reference strains of the species, were isolated from Western Australia and the Greek island of Naxos, respectively.
Supplementary Material
Acknowledgements
This work was supported in part by the Austrian Science Funds (FWF), project P16260 (to H. S.-L.), and the NASA Astrobiology Institute Cooperative Agreement NNA04CC05A (to R. L. M.). We thank Michael Sulzner, Austrian Ministry of Health, for help with the isolation of strain Naxos II.
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequences of strains Halo-G*T, AUS-1 and Naxos II are AM048786, D32081 and AJ400624.
Phase-contrast micrographs of cells of strains Halo-G*T and Naxos II and 2D TLC of the polar lipids of strains Halo-G*T, AUS-1 and Naxos II are available as supplementary material with the online version of this paper.
References
- Arahal DR, García MT, Ludwig W, Schleifer KH, Ventosa A. Transfer of Halomonas canadensis and Halomonas israelensis to the genus Chromohalobacter, as Chromohalobacter canadensis comb. nov. and Chromohalobacter israelensis comb. nov. Int J Syst Evol Microbiol. 2001a;51:1443–1448. doi: 10.1099/00207713-51-4-1443. [DOI] [PubMed] [Google Scholar]
- Arahal DR, García MT, Vargas C, Cánovas D, Nieto JJ, Ventosa A. Chromohalobacter salexigens sp. nov., a moderately halophilic species that includes Halomonas elongata DSM 3043 and ATCC 33174. Int J Syst Evol Microbiol. 2001b;51:1457–1462. doi: 10.1099/00207713-51-4-1457. [DOI] [PubMed] [Google Scholar]
- Cashion P, Holder-Franklin MA, McCully J, Franklin M. A rapid method for the base ratio determination of bacterial DNA. Anal Biochem. 1977;81:461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- Castillo AM, Gutiérrez MC, Kamekura M, Xue Y, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A. Halorubrum orientale sp. nov., a halophilic archaeon isolated from Lake Ejinor, Inner Mongolia, China. Int J Syst Evol Microbiol. 2006;56:2559–2563. doi: 10.1099/ijs.0.64420-0. [DOI] [PubMed] [Google Scholar]
- Castillo AM, Gutiérrez MC, Kamekura M, Xue Y, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A. Halorubrum ejinorense sp. nov., isolated from Lake Ejinor, Inner Mongolia, China. Int J Syst Evol Microbiol. 2007;57:2538–2542. doi: 10.1099/ijs.0.65241-0. [DOI] [PubMed] [Google Scholar]
- Chun J, Lee J-H, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57:2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- Cui HL, Tohty D, Zhou PJ, Liu SJ. Halorubrum lipolyticum sp. nov. and Halorubrum aidingense sp. nov., isolated from two salt lakes in Xin-Jiang, China. Int J Syst Evol Microbiol. 2006;56:1631–1634. doi: 10.1099/ijs.0.64305-0. [DOI] [PubMed] [Google Scholar]
- Cui H-L, Lin Z-Y, Dong Y, Zhou P-J, Liu S-J. Halorubrum litoreum sp. nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol. 2007;57:2204–2206. doi: 10.1099/ijs.0.65268-0. [DOI] [PubMed] [Google Scholar]
- De Ley J, Tijtgat R. Evaluation of membrane filter methods for DNA-DNA hybridization. Antonie van Leeuwenhoek. 1970;36:461–474. doi: 10.1007/BF02069048. [DOI] [PubMed] [Google Scholar]
- De Ley J, Cattoir H, Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970;12:133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Dussault HP. An improved technique for staining red halophilic bacteria. J Bacteriol. 1955;70:484–485. doi: 10.1128/jb.70.4.484-485.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESA . European Users Guide to Low Gravity Platforms. European Space Agency; Noordwijk, Netherlands: 2005. FOTON retrievable capsules. chapter 6. http://www.spaceflight.esa.int/users/downloads/userguides/chapter_6_foton.pdf. [Google Scholar]
- Fan H, Xue Y, Ma Y, Ventosa A, Grant WD. Halorubrum tibetense sp. nov., a novel haloalkaliphilic archaeon from Lake Zabuye in Tibet, China. Int J Syst Evol Microbiol. 2004;54:1213–1216. doi: 10.1099/ijs.0.03032-0. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou PJ, Liu SJ. Halorubrum xinjiangense sp. nov., a novel halophile isolated from saline lakes in China. Int J Syst Evol Microbiol. 2004;54:1789–1791. doi: 10.1099/ijs.0.63209-0. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou P, Zhou YG, Liu SJ, Warren-Rhodes K. Halorubrum alkaliphilum sp. nov., a novel haloalkaliphile isolated from a soda lake in Xinjiang, China. Int J Syst Evol Microbiol. 2005;55:149–152. doi: 10.1099/ijs.0.63320-0. [DOI] [PubMed] [Google Scholar]
- Franzmann PD, Stackebrandt E, Sanderson K, Volkman JK, Cameron DE, Stevenson PL, McMeekin TA, Burton HR. Halobacterium lacusprofundi sp. nov., a halophilic bacterium isolated from Deep Lake, Antarctica. Syst Appl Microbiol. 1988;11:20–27. [Google Scholar]
- Grant WD, Larsen H. Group III. Extremely halophilic archaebacteria. Order Halobacteriales ord. nov. In: Staley JT, Bryant MP, Pfennig N, Holt JG, editors. Bergey’s Manual of Systematic Bacteriology. vol. 3. Williams & Wilkins; Baltimore: 1989. pp. 2216–2219. [Google Scholar]
- Grant WD, Kamekura M, McGenity TJ, Ventosa A. Order I. Halobacteriales Grant and Larsen 1989b, 495VP (Effective publication: Grant and Larsen 1989a, 2216) In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology. 2nd edn vol. 1. Springer; New York: 2001. pp. 294–299. [Google Scholar]
- Gruber C, Legat A, Pfaffenhuemer M, Radax C, Weidler G, Busse HJ, Stan-Lotter H. Halobacterium noricense sp. nov., an archaeal isolate from a bore core of an alpine Permian salt deposit, classification of Halobacterium sp. NRC-1 as a strain of H. salinarum and emended description of H. salinarum. Extremophiles. 2004;8:431–439. doi: 10.1007/s00792-004-0403-6. [DOI] [PubMed] [Google Scholar]
- Huß VAR, Festl H, Schleifer KH. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol. 1983;4:184–192. doi: 10.1016/S0723-2020(83)80048-4. [DOI] [PubMed] [Google Scholar]
- Ihara K, Umemura T, Katagiri I, Kitajima-Ihara T, Sugiyama Y, Kimura Y, Mukohata Y. Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation. J Mol Biol. 1999;285:163–174. doi: 10.1006/jmbi.1998.2286. [DOI] [PubMed] [Google Scholar]
- Johnson JL. Similarity analysis of DNAs. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR, editors. Methods for General and Molecular Bacteriology. American Society for Microbiology; Washington, DC: 1994. pp. 655–681. [Google Scholar]
- Kamekura M, Dyall-Smith ML. Taxonomy of the family Halobacteriaceae and the description of two genera Halorubrobacterium and Natrialba. J Gen Appl Microbiol. 1995;41:333–350. [Google Scholar]
- Kamekura M, Dyall-Smith ML, Upasani V, Ventosa A, Kates M. Diversity of alkaliphilic halobacteria: proposals for transfer of Natronobacterium vacuolatum, Natronobacterium magadii, and Natronobacterium pharaonis to Halorubrum, Natrialba, and Natronomonas gen. nov., respectively, as Halorubrum vacuolatum comb. nov., Natrialba magadii comb. nov., and Natronomonas pharaonis comb. nov., respectively. Int J Syst Bacteriol. 1997;47:853–857. doi: 10.1099/00207713-47-3-853. [DOI] [PubMed] [Google Scholar]
- Kharroub K, Quesada T, Ferrer R, Fuentes S, Aguilera M, Boulahrouf A, Ramos-Cormenzana A, Monteoliva-Sanchez M. Halorubrum ezzemoulense sp. nov., a halophilic archaeon isolated from Ezzemoul sabkha, Algeria. Int J Syst Evol Microbiol. 2006;56:1583–1588. doi: 10.1099/ijs.0.64272-0. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lizama C, Monteoliva-Sánchez M, Suárez-García A, Rosselló-Mora R, Aguilera M, Campos V, Ramos-Cormenzana A. Halorubrum tebenquichense sp. nov, a novel halophilic archaeon isolated from the Atacama Saltern, Chile. Int J Syst Evol Microbiol. 2002;52:149–155. doi: 10.1099/00207713-52-1-149. [DOI] [PubMed] [Google Scholar]
- Malik KA. A modified method for the cultivation of phototrophic bacteria under anaerobic conditions. J Microbiol Methods. 1983;1:343–352. [Google Scholar]
- Mancinelli RL, White MR, Rothschild LJ. Biopan-survival I: exposure of the osmophiles Synechococcus sp. (Nägeli) and Haloarcula sp. to the space environment. Adv Space Res. 1998;22:327–334. [Google Scholar]
- Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- McGenity TJ, Grant WD. Transfer of Halobacterium saccharovorum, Halobacterium sodomense, Halobacterium trapanicum NRC 34021 and Halobacterium lacusprofundi to the genus Halorubrum gen. nov., as Halorubrum saccharovorum comb. nov., Halorubrum sodomense comb. nov., Halorubrum trapanicum comb. nov., and Halorubrum lacusprofundi comb. nov. Syst Appl Microbiol. 1995;18:237–243. [Google Scholar]
- McGenity TJ, Grant WD. Genus VII. Halorubrum McGenity and Grant 1996, 362VP (Effective publication: McGenity and Grant 1995, 241) In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology. 2nd edn vol. 1. Springer; New York: 2001. pp. 320–324. [Google Scholar]
- Oren A. Halobacterium sodomense sp. nov., a Dead Sea halobacterium with an extremely high magnesium requirement. Int J Syst Bacteriol. 1983;33:381–386. [Google Scholar]
- Oren A, Ventosa A, Grant WD. Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol. 1997;47:233–238. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sprott GD, Sad S, Fleming LP, Dicaire CJ, Patel GB, Krishnan L. Archaeosomes varying in lipid composition differ in receptor-mediated endocytosis and differentially adjuvant immune responses to entrapped antigen. Archaea. 2003;1:151–164. doi: 10.1155/2003/569283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan-Lotter H, Pfaffenhuemer M, Legat A, Busse HJ, Radax C, Gruber C. Halococcus dombrowskii sp. nov., an archaeal isolate from a Permo-Triassic alpine salt deposit. Int J Syst Evol Microbiol. 2002;52:1807–1814. doi: 10.1099/00207713-52-5-1807. [DOI] [PubMed] [Google Scholar]
- Tomlinson GA, Hochstein LI. Halobacterium saccharovorum sp. nov., a carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol. 1976;22:587–591. doi: 10.1139/m76-087. [DOI] [PubMed] [Google Scholar]
- Ventosa A, Gutierrez MC, Kamekura M, Zvyagintseva IS, Oren A. Taxonomic study of Halorubrum distributum and proposal of Halorubrum terrestre sp. nov. Int J Syst Evol Microbiol. 2004;54:389–392. doi: 10.1099/ijs.0.02621-0. [DOI] [PubMed] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, et al. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Green Publishing & Wiley-Interscience; New York: 1987. pp. 2.4.1–2.4.5. [Google Scholar]
- Xu X-W, Wu Y-H, Zhang HB, Wu M. Halorubrum arcis sp. nov., an extremely halophilic archaeon isolated from a saline lake on the Qinghai–Tibet Plateau, China. Int J Syst Evol Microbiol. 2007;57:1069–1072. doi: 10.1099/ijs.0.64921-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.