Abstract
Mycobacterium tuberculosis is one of the world's most deadly human pathogens; an integrated understanding of how it successfully survives in its host is crucial to developing new treatment strategies. One notable characteristic of infection with M. tuberculosis is the formation of granulomas, aggregates of immune cells whose structure and function may reflect success or failure of the host to contain infection. One central regulator of host responses to infection, including granuloma formation, is the pleiotropic cytokine tumor necrosis factor-α (TNF). Experimental work has characterized roles for TNF in macrophage activation; regulation of apoptosis; chemokine and cytokine production; and regulation of cellular recruitment via trans-endothelial migration. Separating the effects of these functions is presently difficult or impossible in vivo. To this end, we applied a computational model to understand specific roles of TNF in control of tuberculosis in a single granuloma. In the model, cells are represented as discrete entities on a spatial grid responding to environmental stimuli by following programmed rules determined from published experimental studies. Simulated granulomas emerge as a result of these rules. After confirming the importance of TNF in this model, we assessed the effects of individual TNF functions. The model predicts that multiple TNF activities contribute to control of infection within the granuloma, with macrophage activation as a key effector mechanism for controlling bacterial growth. Results suggest that bacterial numbers are a strong contributing factor to granuloma structure with TNF. Finally, TNF-dependent apoptosis may reduce inflammation at the cost of impairing mycobacterial clearance.
Keywords: Cytokines, Bacterial Infections, Chemotaxis, Inflammation
Introduction
Tuberculosis (TB) kills more people per year than any other single infectious disease. Infection by its causative agent, Mycobacterium tuberculosis (Mtb), results in active disease in only a minority of cases (∼10%); the majority of infections are controlled and clinically silent, although the host often remains infected (reviewed in 1).
The classic feature of pulmonary Mtb infection arises as a result of the immune response where aggregates of immune cells and bacteria, called granulomas, form in the lungs. In humans and non-human primates with latent pulmonary infection, granulomas form as well-circumscribed masses in the lung parenchyma comprised of resting, infected and activated macrophages with a characteristic cuff of T cells on the periphery (e.g. 2, 3) and a caseous necrotic center (4). Macrophages within a granuloma have dual roles in Mtb infection: they are the primary mechanism for Mtb containment and the preferred location for bacterial growth. During infection, there are often multiple and different types of granulomas within the lung; each could have a different outcome depending on local environment. At the level of a single granuloma, macrophages may fail to control infection, leading to necrotic granulomas harboring large numbers of bacteria within macrophages (3). However, the relationship between bacterial control in a single granuloma and the outcome of infection at the level of the entire host is not well established (3).
Type-1 adaptive immunity is required to control infection at the host level (5). Activated T cells migrate to the site of infection and act as immune effectors. We distinguish three primary T cell types based on their effector function (c.f. 6). Pro-inflammatory T cells (CD4+ or CD8+) provide macrophage-activating cytokines (e.g. IFN-γ) while cytotoxic T cells (predominantly CD8+) provide cytolytic functions to control infection (reviewed in 1). A third T cell class, regulatory T cells (Treg; reviewed in 7), are present in mouse (8) and human (9) Mtb infections, and may prevent efficient Mtb clearance by immune responses (10, 11), or may modulate local responses to control pathology. Treg are CD4+Foxp3+ cells that comprise approximately 5-10% of all CD4+ T cells (12, 13). They suppress the action of pro-inflammatory T cells (14) through poorly understood mechanisms that may occur by cell-contact, secretion of immunosuppressive cytokines (15), or both.
The pro-inflammatory cytokine tumor necrosis factor-α (TNF) is a central, multi-faceted contributor to the immune response in Mtb infection produced by activated macrophages and pro-inflammatory T cells (16-20). The role of TNF is of clinical interest due to the association of anti-inflammatory TNF-blocking drugs with reactivation of latent TB in humans (21, 22). TNF is also necessary for Mtb containment in mouse models (16). TNF gene-disrupted or neutralized mice have disorganized granulomas in Mtb infections (17), underscoring the link between granuloma structure and effective containment of infection.
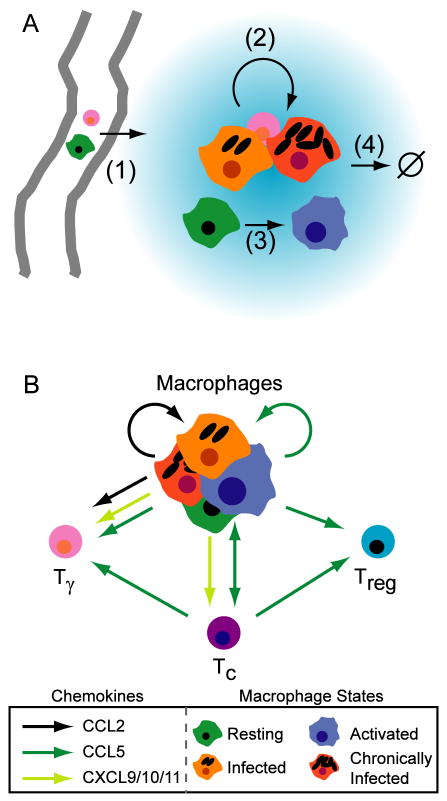
TNF has multiple immunological functions during infection with Mtb (Figure 1A). TNF has a direct role in immune cell recruitment via up-regulation of endothelial adhesion molecules (23), facilitating trans-endothelial migration of immune cells to the site of infection. TNF regulates production of chemokines by macrophages (24, 25); chemokines can further induce trans-endothelial migration (reviewed in 26) and coordinate recruitment (reviewed in 27) of immune cells within the tissues. TNF activates macrophages in conjunction with the cytokine IFN-γ (28-30); such activated macrophages can kill intracellular mycobacteria. TNF can also induce necrotic or apoptotic cell death in macrophages (31) that is promoted by Mtb infection (32). Figure 1A summarizes these effects.
Figure 1.
Models of molecular signaling networks that affect granuloma formation during infection with Mycobacterium tuberculosis. A. TNF (blue gradient) is an immunological effector with multiple roles. (1) TNF-dependent enhancement of transendothelial migration of monocytes and T cells to the lung parenchyma occurs via upregulation of endothelial adhesion molecules. (2) TNF-dependent activation of macrophages in concert with IFN-γ stimulates chemokine production and bacterial killing. (3) TNF-dependent apoptosis, a second pathway for mycobacterial killing. B. Model of the chemokine network induced during infection with Mycobacterium tuberculosis. CXCL9/10/11 are α-chemoattractants that bind the same chemokine receptor (CXCR3); CCL2 binds CCR2; CCL5 binds CCR5. Tγ are pro-inflammatory Th1 cells. Tc are cytotoxic T cells. Treg are regulatory T cells.
The effects of TNF in Mtb granuloma formation are likely related to the chemokine network induced during infection. We have identified a simplified model of chemokines based on three classes that affect recruitment of macrophages and T cells to the granuloma via binding of appropriate chemokine receptors on the cell surface (Figure 1B). The α-chemoattractant class (CXCL9,10, and 11; formerly Mig, IP-10 and I-TAC, respectively) binds chemokine receptor CXCR3 on pro-inflammatory CD4+ and CD8+ T cells (33), but not regulatory T cells (34). CCL2 (formerly MCP-1) binds CCR2 on macrophages (35) and proportions of pro-inflammatory T cell populations (36). CCL5 (formerly RANTES) binds CCR5 on macrophages and T cells, and is involved in migration of regulatory T cells to the site of other infections (37), although this has not been demonstrated for Mtb.
Each of the four roles of TNF (cellular migration, induction of chemokine/TNF secretion, macrophage activation and apoptosis) may contribute separately to establishing and maintaining control of Mtb infection at the level of a single granuloma. Currently, it is impossible to study these separate TNF functions using in vitro or in vivo models. Here we use a specific type of computational model known as an agent-based model (ABM) to study the contributions of these immune effectors on granuloma formation (illustrated in Figure 2). The power of this approach lies in the emergence of behaviors that arise from interactions between agents that would otherwise be impossible to know a priori. We build on our previously described ABM (38), which predicted the emergence of a two dimensional spherical structures (granulomas) without any rules specifying such spatial behavior to occur. We now incorporate various functions of TNF, different T cell classes and a simple chemokine network. This type of model, which combines spatial and temporal behaviors, is particularly suited to the investigation of granulomas due to the representation of cells, cytokines and bacteria, allowing tractable analysis of each factor independently or in combination. Analysis of an ABM reveals the specific contributions of TNF functions, alone and in combination, to control of Mtb infection and granuloma structure at the level of a single granuloma.
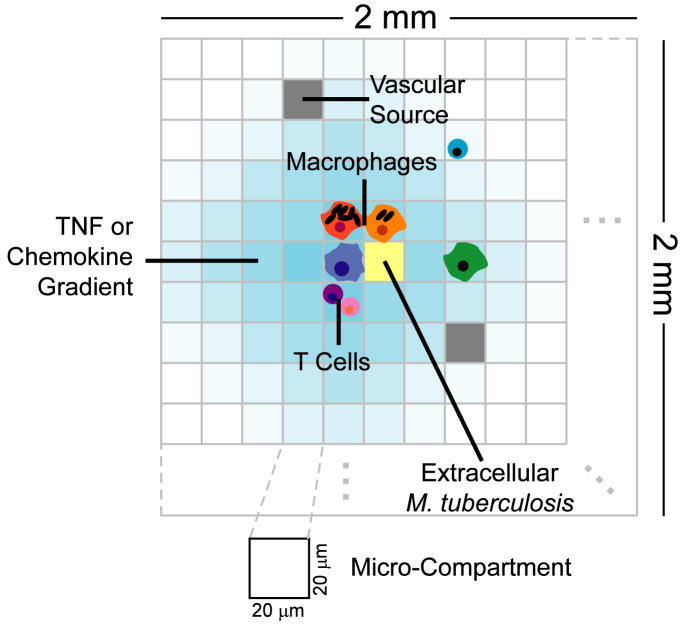
Figure 2.
Structure of the agent-based model environment. A 100 × 100 grid of micro-compartments represents a 2 mm × 2 mm section of lung tissue. Discrete entities include macrophages and T cells. TNF, chemokines and extracellular M. tuberculosis are represented as continuous entities. Each micro-compartments can contain either one macrophage or up to two T cells along with extracellular bacteria, TNF and chemokines. A percentage of randomly chosen micro-compartments are designated as vascular sources that allow new macrophages and T cells to be recruited to the grid by chemokines and TNF.
Methods
Supplememental methods, results and time-lapse movies are available at http://malthus.micro.med.umich.edu/lab/movies/Granuloma2009/.
Hybrid agent-based model
The model presented here is an extension of a previous ABM that captured cellular interactions leading to granuloma formation during infection with Mtb (38). The model is considered hybrid since we incorporated both discrete entities (cells) and continuous entities (chemokines, TNF and Mtb) that interact simultaneously. ABMs are developed based on four considerations: an environment, agents that reside there, the rules that describe the agents and their interactions, and the timescales on which events are defined.
The environment represents a 2 mm × 2 mm section of lung parenchyma as a 100 × 100 square 2-dimensional lattice with individual micro-compartments scaled to the approximate size of a single macrophage: 20 μm in diameter (39). Discrete agents move on the lattice and respond to their environment based on rules reflecting known biological activities. Bacteria and effector molecules can reside anywhere on the lattice and undergo diffusion when appropriate.
Caseation represents inflammation of, and damage to, the lung parenchyma from macrophage cell death. We note a change of terminology to “caseation” from “necrosis” in previous work (38), as strict necrosis within the granuloma is now believed to be caused by substantial neutrophil infiltration and death, while caseation is likely initiated by macrophage death (unpublished data, JLF). In the ABM, caseation is defined to occur when a threshold number of activated or infected macrophage deaths take place in a micro-compartment. A final environmental feature is designation of spaces as vascular sources.
We include two types of discrete agents in the model: macrophages and T cells. As previously (20, 38), macrophage agents are either resting (Mr, uninfected), infected (Mi; have taken up bacteria), chronically infected (Mci; are unable to clear their intracellular bacterial load), or activated (Ma; can effectively kill bacteria). In contrast to our previous study (38), where a single T cell class captured all cell behaviors, here we represent three distinct T cell subpopulations based on function: the Tγ class captures CD4+ and CD8+ pro-inflammatory T cells; Tc represent cytotoxic T cells; and Treg represent regulatory T cells. In this representation, all T cells in a particular class have identical function; this is simpler than in vivo, but we capture enough detail in this representation for a qualitative representation of known T cell effects.
In addition to the discrete entities, extracellular bacteria, diffusing effector molecules (CCL2, CCL5, CXCL9/10/11 and TNF) are agents (concentrations) that are tracked continuously over time. The chemokine model used here is a simplification that was chosen to include a ligand for each key chemokine receptor, while minimizing the distinct chemokine classes represented to save on computation.
Cells respond to signals in the surrounding environment according to rules that represent known activities in vivo. During simulations, each agent responds depending on its state. Examples of rules include uptake of bacteria, macrophage activation by T cells, secretion of cytokines and chemokines, etc. For a full list of rules, see Supplement 1.
Computer Simulations
At the beginning of a simulation, the grid has 105 randomly placed resident resting macrophages (Mr) that move randomly with no chemokine or cytokine present. Infection is initiated with one infected macrophage (Mi), containing a single bacterium, placed at the center. We chose this inoculum since we assume a single inhalation event. We also compared our results using a single bacteriurm to one using a larger inoculum (15 bacteria within an infected macrophage). In that scenario, clearance of infection occurred less often, with different kinetics at early timepoints, but after 20 days the two infection trajectories, and bacterial loads become similar from that point forward. Every 10 minutes of simulation time, positions and interactions between T cells and macrophages are updated, including recruitment of cells from vascular sources and secretion of TNF and chemokines. The landscape of molecular concentrations serves as the starting point for computing cytokine and chemokine diffusion for 10 minutes of simulation time. Cell states and interactions are then updated again, in the beginning of the next 10-minute timestep in an asynchronous fashion, and the algorithm continues in this way for 200 days (2,880,000 6s timesteps) of simulation time.
Parameter estimates and uncertainty and sensitivity analyses
It is difficult to estimate the rates and probabilities (parameters) of events occurring within the lung environment. We approximate parameter values in three ways: directly from available data, using uncertainty analysis, and model calibration with other data. Relative macrophage and T cell production were estimated from the literature (40-44). The macrophage CCL2 saturation point and detection threshold were estimated based on published data (45); T cell saturations are inferred from this. Tregs are more sensitive to CCL5 than Tγ or Tc cells (42, 46). The resting macrophage lifespan (Mrls) is based on (47); we assume a lifespan 10-fold shorter for activated macrophages (Mals). The lifespan of a T cell is based on (48). The delay of T cell infiltration (Tdelay) is based on (49).
Since absolute levels of chemokine production, saturation, etc were not known, we estimated the relative effects using calibration. Chemokine diffusion is based on (50). The initial number of macrophages (Minit) represents approximately 1% coverage of the grid (38). The number of bacteria engulfed or killed by resting macrophages (Nrk) was chosen to be low, but non-zero. The killing of ingested bacteria by activated macrophages (Nphag) is related to the maximum number of intracellular Mtb that can exist in an infected macrophage before it is designated as chronically infected. Ntact is a guess based on physical interactions of several T cells around a single macrophage. Other parameter value estimates are discussed in supplemental material
All other parameters not discussed above that are listed in Tables I and III were estimated using uncertainty analysis, with various parameters calibrated to ensure distinct outcomes between control and TNF-deletion cases. Parameters in Table II, including production, sensitivity and saturation thresholds, and effects on recruitment for all other chemokines, were defined to preserve a relative relationship with the CCL5 parameters (dependent parameters). Thus, when Table I parameters (independent parameters) vary, Table II parameters also vary since they are proportional.
Table I. Parameters varied for Latin hypercube sampling.
| Parameter | Description1 | Default | Range | Distribution | Varied in focused analysis? |
|---|---|---|---|---|---|
| αBi | Intracellular Mtb growth rate (per 10 minutes) | 0.0015 | [0.0002, 0.002] | Uniform | No |
| αBe | Extracellular Mtb growth rate (per 10 minutes) | 0.0007 | [0.00015, 0.015] | Log-Uniform | No |
| pk | Probability of Mr killing bacteria | 0.0187 | [0.01, 0.1] | Uniform | No |
| Tactm | Probability of Mi activation by Tγ | 0.03 | [0.0001, 0.1] | Log-Uniform | No |
| Mrecr | Probability of macrophage recruitment | 0.04 | [0.01, 0.1] | Uniform | No |
| Trecr | Probability of T cell recruitment | 0.09 | [0.01, 0.1] | Uniform | No |
| Tmove | Prob of a T cell moving onto an occupied micro-compartment | 0.02 | [0.00001, 0.1] | Log-Uniform | No |
| Trrecr | Proportion of Treg cells out of all T cells recruited | 0.1 | [0.01, 0.2] | Uniform | No |
| λc | Chemokine diffusion rate (cm2 per 0.1 minutes) | 6.27×10−7 | [1,7]×10−7 | Uniform | No |
| δc | Chemokine degradation rate (per 0.1 minutes) | 0.000916 | [0.0005, 0.0015] | Uniform | No |
| rT | Combined TNF/chemokine threshold for T cell recruitment at a vascular source2 | 1,767 | [0.1,10]×l04 | Log-Uniform | No |
| rM | Combined TNF/chemokine threshold for Mr recruitment at a vascular source2 | 6,507 | [0.1,10]×l04 | Log-Uniform | No |
| sc5 | CCL5 production rate (molecules per 10 minutes) | 3.55×106 | [1,10]×106 | Uniform | No |
| s5m | Macrophage CCL5 saturation threshold (molecules) | 5.79×106 | [1,100]×l06 | Log-Uniform | No |
| τ5m | Macrophage CCL5 threshold (molecules) | 2×l04 | [1,100]×104 | Log-Uniform | No |
| λTNF | TNF diffusion rate (cm2 per 0.1 minutes) | 6.56×10−7 | [1,7]×10−7 | Uniform | Yes |
| δTNF | TNF degradation rate (per 0.1 minutes) | 0.0005 | [0.0001, 0.001] | Uniform | Yes |
| sTNF | TNF production rate (molecules per 10 minutes) | 1.80×105 | [1,100]×l04 | Log-Uniform | Yes |
| papopt | Probability of TNF-induced apopt per 10 minute interval | 0.04 | [0.001, 0.2] | Uniform | Yes |
| τTNF3 | Macrophage TNF detection threshold (molecules) | 10×105 | [1,15]×105 | Uniform | Yes |
| rMTNF | Effect of TNF on Mr recruitment2 | 676 | [10,1000] | Log-Uniform | Yes |
All probabilities are per 10 minute interval.
Non-dimensional; c.f. II.3.iv-v. of the Agent-Based Model Rules (Supplement 1).
τTNF was divided into separate thresholds for activation (τTNFact) and apoptosis (τTNFapopt) in the focused analysis.
Table III. Parameters not varied in uncertainty analysis.
| Parameter | Description | Value | Reasoning |
|---|---|---|---|
| Minit | Number of resident macrophages | 105 | 1 |
| Kbe | Carrying capacity of micro-compartment for extracellular Mtb | 220 | 2, 3 |
| Nrk | Number of Mtb engulfed/killed by Mr | 2 | 3 |
| Nphag | Number of Mtb killed by Ma every 10 minutes | 10 | 3 |
| Ntact | Maximum Tγ number in Moore of Mi having effect | 4 | 3 |
| Ncaseum | Number of Ma, Mi and Mci deaths for caseation | 6 | 3 |
| tregTγ | Tγ inactivity time after Treg interactions (min) | 110 | 3 |
| Nc | Number of Mtb for Mi → Mci transition | 10 | 3, 4 |
| Kbi | Number of bacteria causing bursting | 20 | 3, 4 |
| Mals | Lifespan of Ma in days | 10 | 5 |
| Tls | Lifespan of T cells in days | 3 | 5 |
| Mrls | Lifespan of Mr in days | 100 | 5 |
| Tdelay | T cell recruitment delay in days | 20 | 6 |
| Pkill | Fraction Mtb killed by Fas/FasL apoptosis | 0.5 | 7 |
| pTk | Prob of Fas/FasL (TNF-independent) apoptosis by T cells | 0.006 | 7 |
| Tckmtb | Probability of Tc killing Mtb in Mci death | 0.75 | 7 |
| Tckmci | Probability of Tc killing Mci | 0.95 | 7 |
1. Set to the reference number for containment to have identical initial conditions.
2. Set ∼10-fold larger than the amount causing macrophage bursting. There is physical space for approximately 450 bacilli in one micro-compartment (tightly packed), but lack of nutrients for growth limits this.
3. These parameters have integer values that cannot be continuously varied over at least 250 different values.
4. The same effect as varying this is captured by changing intracellular growth rate: the faster Mtb grow, the sooner the transition to chronic infection. If Nc is varied in say [5, 25] we should set Kbi = 2*Nc.
5. Relative lifespans are well known. Vary cell age between 0 and the maximum age, so changing these would have questionable relevance.
6: Many parameter sensitivities change before and after this time, so it was held constant but multiple uncertainty analyses were performed to show the effect of this parameter.
7. Preliminary analysis revealed little effect for reasonable ranges. Thus, this was not varied to reduce the number of parameters varied.
Table II. Parameter relationships constrained for analyses.
| Parameter | Description | Value |
|---|---|---|
| sc2 | CCL2 production rate (molecules per 10 minutes) | sc5 |
| sc9 | CXCL9/10/11 production rate (molecules per 10 minutes) | 2*sc5 |
| s2m | Macrophage CCL2 saturation (molecules) | 10*s5m |
| τ2m | Macrophage CCL2 threshold (molecules) | 0.1*τ5m |
| s2Tγ | Tγ CCL2 saturation (molecules) | 10*s5m |
| τ2Tγ | Tγ CCL2 threshold (molecules) | 0.1*τ5m |
| s5Tγ | Tγ CCL5 saturation (molecules) | s5m |
| τ5Tγ | Tγ CCL5 threshold (molecules) | τ5m |
| s9Tγ | Tγ CXCL9 saturation (molecules) | 10*s5m |
| τ9Tγ | Tγ CXCL9 threshold (molecules) | τ5m |
| s5Tc | Tc CCL5 saturation (molecules) | s5m |
| τ5Tc | Tc CCL5 threshold (molecules) | τ5m |
| s9Tc | Tc CXCL9 saturation (molecules) | 10*s5m |
| τ9Tc | Tc CXCL9 threshold (molecules) | τ5m |
| s5Tr | Treg CCL5 saturation (molecules) | s5m |
| τ5Tr | Treg CCL5 threshold (molecules) | 0.1*τ5m |
| rM2 | Effect of CCL2 on Mr recr1,2 | rMTNF |
| rM5 | Effect of CCL5 on Mr recr1,2 | 0.1*rMTNF |
| rTTNF | Effect of TNF on T cell recr1 | rMTNF |
| rT9 | Effect of CXCL9 on Tγ, Tc cell recr1,2 | 0.1*rMTNF |
| rTγ2 | Effect of CCL2 on Tγ cell recr1,2 | rMTNF |
| rT5 | Effect of CCL5 on Tγ, Tc cell recr1,2 | 0.1*rMTNF |
| rTr5 | Effect of CCL5 on Treg cell recr1,2 | rMTNF |
| Tγrecr | Proportion of Tγ cells recruited | 0.6*(1 – Trrecr) |
| Tcrecr | Proportion of Tc cells recruited | 0.4*(1 – Trrecr) |
Non-dimensional; c.f. II.3.iv-v. of the Agent-Based Model Rules (Supplement 1).
These parameters were held constant in the focused sensitivity analysis at the default value of rMTNF given in Table 1.
Since not all parameters can be estimated from available data, it is necessary to use uncertainty and sensitivity methods to explore possible model outcomes using different parameter values. We have developed extensive methods for performing this analysis on ABMs and applied them here [see (51); for more details see Supplement 2]. Simulations using uncertainty analyses yielded a wide range of different granuloma structures and bacterial numbers within the granulomas (Supplementary Figure 1). Data from humans and non-human primates with tuberculosis strongly supports the existence of a diverse array of granuloma types even within a single host (4, 52).
Simulated deletion and depletion of TNF activities
To examine the effect of individual TNF activities on granuloma formation and maintenance, we performed virtual deletions and depletions of relevant parameters using a baseline parameter set that leads to control of infection (Tables I-III). Loss of activity was brought about by setting relevant probabilities to zero and/or raising relevant thresholds to an unattainable level. Virtual deletion refers to loss of the activity from the beginning of the simulation at the onset of infection. Virtual depletion refers to the loss of the activity after the establishment of a stable granuloma, 100 days post-infection. The timing of the depletion was determined by examining the results of sensitivity analysis and the baseline control scenario. Parameter sensitivities in the model stabilize by day 50 (c.f. Results), suggesting that 100 days post-infection represents a reasonable time for an established, stable granuloma. Significant differences between outcome variables were determined with a mean difference test (Welch's approximate t test) for 15 repeated simulations of each single deletion or depletion, and for 10 repeated simulations in deletion or depletion of two or more TNF activities simultaneously (for details, see Supplement 2).
Simulations with bacterial levels fixed after 100 days post-infection
To control for the effects of bacterial growth on granuloma structure, we fixed total bacterial levels (Be + Bi). Specifically, we simulated a baseline granuloma for 100 days and then fixed the total bacterial numbers to a constant level (2.75 +/- 0.51 log10 Mtb) for the next 100 simulation days. This allowed exploration of the effects of TNF depletions without the confounding effects of bacterial growth. To attain a fixed Mtb population, all bacterial growth and death was prevented, with the effects of bacteria on host macrophages allowed to otherwise proceed normally. Upon exit of intracellular Mtb from macrophages due to cell lysis or death, the bacteria were forced to deposit on the single micro-compartment where the macrophage resided after day 100 (unlike other simulations, where bacteria were distributed in the entire (Moore) neighborhood of a macrophage). This step prevents a biologically unrealistic scenario that could occur where the bacteria are “shuffled” to the outside of the granuloma. Note that intracellular and extracellular Mtb quantities still changed in the simulations, but the total bacterial number was conserved.
Results
Agent based model for dissection of granuloma structure and function
Computational models of the immune response to a pathogen can be deterministic (based on ordinary differential equations representing rates of change) or stochastic (based on probabilities of discrete agents and the rules governing their behavior). For the current investigation examining the contribution of various functions of TNF to granuloma structure and bacterial control in M. tuberculosis infection, we chose to use an ABM that is a hybrid model. This approach is appropriate because both spatial and temporal events must be considered when investigating the tuberculous granuloma. This ABM predicts the dynamics of Mtb infection at the level of a single granuloma.
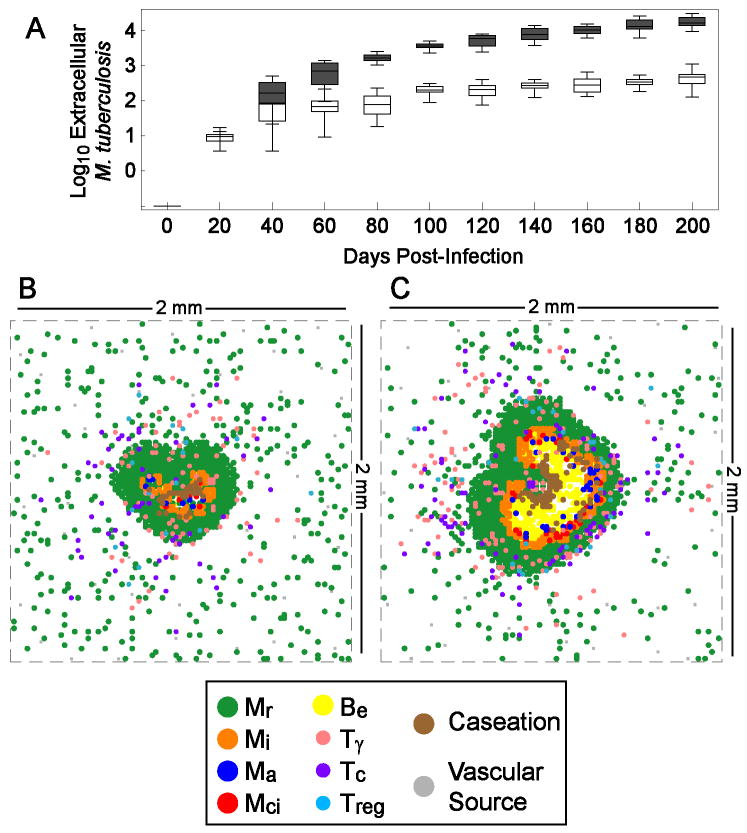
To first identify variables that determine initial containment or maintenance of an established infection at the scale of a single granuloma, we established a reference parameter set (Tables I-III) using uncertainty and sensitivity analyses as outlined in Methods and Supplement 2. The baseline simulations lead to stable bacterial numbers contained within the granuloma (∼103 total bacteria) up to 200 days post-infection (Figure 3A, white bars). Repeated simulations show that the granuloma is organized with relatively tightly packed cells, predominantly uninfected macrophages (green agents in Figure 3B), with T cell localization at the periphery of the granuloma (pink, purple, and light blue agents in Figure 3B). The model is robust: for 15 repeated runs that established stable bacterial levels, all showed controlled infection with variable outcomes of granuloma structure (Supplement 3 Figure S2A presents repeat simulations of the baseline scenario illustrating extent of variability).
Figure 3.
Simulated kinetics of extracellular M. tuberculosis and typical granuloma structures at 200 days post-infection in baseline control and TNF deletion scenarios. A. Box-whisker plots represent minimum, median, maximum and interquartile range of bacterial numbers for 15 simulations each for the containment scenario (white bars) and lacking TNF (gray bars). B. Containment granuloma using the baseline set of parameters (Tables 1-3). C. Irregular granuloma with uncontrolled bacterial growth resulting from lack of TNF in the simulation. Mr, Mi, Ma, and Mci are resting, infected, activated and chronically infected macrophages, respectively. Be is extracellular mycobacteria. Tγ, Tc, and Treg are pro-inflammatory, cytotoxic and regulatory T cells, respectively. Parameters are as in Tables 1-3 except for TNF deletion (where parameter sTNF = 0).
Simulated infection with all parameters set to the control scenario but lacking TNF (virtual TNF deletion) results in a granuloma that is irregular in shape, increased in size, and with wide-spread caseation (Figure 3C). Bacterial numbers are significantly higher than in the baseline scenario (Figure 3A, gray bars: ∼104 compared to 103; p < 0.01). Numbers of all macrophage and T cell populations in the model are significantly elevated in comparison to the baseline scenario within the first 20 days after infection (not shown). Therefore, loss of TNF appears to impair early control of infection, resulting in more extensive immune cell infiltration; this corresponds to data from murine models of Mtb infection (24).
Factors that substantially contribute to control of infection within the granuloma
To identify which parameters significantly contributed to control of Mtb infection within the granuloma, we performed a global uncertainty and sensitivity analysis, varying parameters in Tables I and II. Using the outcomes of this, we performed a sensitivity analysis to statistically determine which factors control bacterial numbers within the granuloma (Fig 4A). Approximately 2/3 of the simulations led to clearance of the infection. Some combinations of parameter values promote elimination of bacteria prior to granuloma formation due to apoptosis-induced killing and innate clearance by Mr (data not shown). The parameters that emerged as especially important were bacterial growth rates, T cell movement, and certain TNF-related effects. We then assessed the effects of these identified parameters on variables such as T cell functions, granuloma size, macrophage numbers and level of caseation in the model (Table IV).
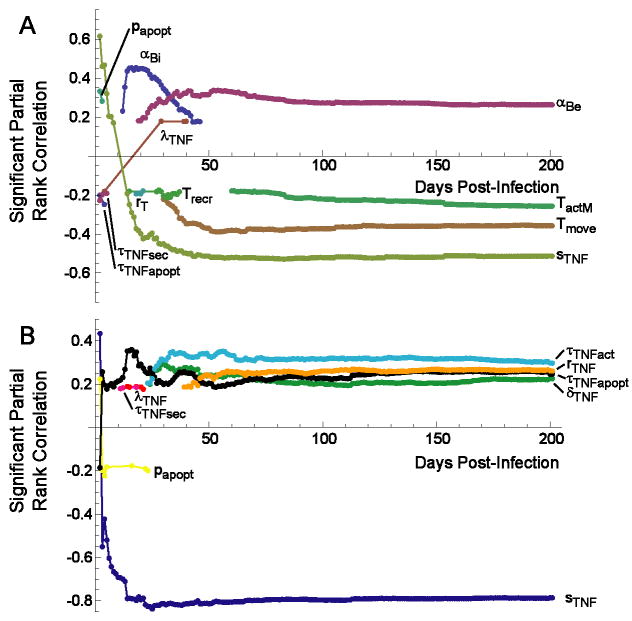
Figure 4.
Correlation of mechanisms in the model with extracelluar bacterial load over the course of infection. Graphs depict significant partial rank correlations (p < 0.01). A. Global sensitivity analysis reveals four dominant parameters. B. TNF-focused sensitivity analysis predicts the contribution of individual TNF-related mechanisms over time. Non-TNF parameters are set equal to the baseline control scenario (Tables 1-3) in panel B. αBe: extracellular Mtb growth rate; αBi: intracellular Mtb growth rate; papopt: probability of TNF-induced apoptosis in one ten-minute interval; Tmove: probability of T cell movement onto an occupied location; sTNF: rate of TNF secretion by macrophages; τTNFact: threshold for TNF-induced activation by macrophages; rMTNF: effect of TNF on trans-endothelial migration; δTNF: rate of TNF degradation; τTNFapopt: threshold for TNF-induced apoptosis by macrophages.
Table IV.
Significant partial rank correlations between parameters identified in the Sensitivity Analysis in Figure 4 and various granuloma outcome measures 20 and 200 days post-infection forseveral different outcome variables (column 1).1
| Parameter | αBi | αBe | sTNF | Tmove | ||||
|---|---|---|---|---|---|---|---|---|
| Outcome Day Measure | 20 | 200 | 20 | 200 | 20 | 200 | 20 | 200 |
| Be | +++ | + | +++ | --- | --- | --- | ||
| Bi | +++ | --- | --- | --- | ||||
| Total T cells | --- | --- | ||||||
| Tγ | --- | --- | ||||||
| “Secretor” Tγ2 | +++ | -- | -- | |||||
| Tc | --- | --- | ||||||
| Treg | --- | --- | ||||||
| Total Macs | --- | --- | --- | |||||
| Mr | --- | --- | --- | |||||
| Mi | --- | --- | --- | |||||
| Mci | +++ | + | --- | --- | --- | |||
| Ma | ++ | --- | --- | |||||
| TNF | + | --- | --- | --- | ||||
| Chemokines | ++ | --- | --- | --- | ||||
| Caseation | +++ | --- | --- | |||||
| Be growth rate | +++ | +++ | --- | --- | --- | |||
| Granuloma Size | ND3 | ND3 | ND3 | --- | ND3 | --- | ||
Parameter definitions are given in Tables 1-3. Significant positive correlations: +++ (p < 0.0001); ++ (p < 0.001); + (p < 0.01). Significant negative correlations: --- (p < 0.0001); -- (p < 0.001); - (p < 0.01).
Number of Tγ cells actively secreting IFN-γ
ND: Not done due to relative lack of granuloma formation by day 20.
The analysis indicated that growth rates of intracellular and extracellular Mtb (αBi and αBe, respectively) are positively correlated with increased bacterial numbers in the granuloma4 (Figure 4A). This result suggests that the growth rate of mycobacteria is a virulence factor, consistent with the published data that more virulent clinical strains of Mtb grow more quickly in macrophages (53). A faster intracellular growth rate leads to increased secretion of TNF and chemokines early in infection (due to increased antigenic stimulation), and to increased T cell activity by 200 days post-infection (Table IV). However, the heightened immune response is apparently incapable of controlling the infection in the face of the increased bacterial growth rate. This is recapitulated in animal models, where increased bacterial numbers results in increased inflammation and more T cell activation, but not necessarily better control of infection (54).
Not surprisingly, the probability (Tmove) of a T cell moving to a location occupied by another cell (either a T cell or a macrophage) is significantly negatively correlated with bacterial levels (Fig 4A). A micro-compartment can hold either two T cells or a T cell and a macrophage, but not two T cells and a macrophage or more than one macrophage. At 200 days, an increase in the rate of Tmove negatively impacts every measure in our model. In other words, a more tightly packed granuloma is more effective in controlling bacterial numbers, since this increases cell-cell interactions leading to activation of infected macrophages by T cells.
As expected from the human and animal model data and our previous studies (17, 20), TNF is crucial to the outcome of infection at the level of the granuloma (Fig 3). In this analysis, the overall rate of TNF production (sTNF) was strongly negatively correlated with bacterial numbers (Fig 4A), confirming the necessity for TNF in controlling infection. Increased TNF production from the beginning of infection resulted in clearance of infection in more than half of the simulations. The rate of TNF secretion strongly and negatively influences most variables in the model – particularly in the long term (see Table IV) - suggesting that the rate of TNF secretion from macrophages and Tγ cells (sTNF) has a global regulatory role in the system, with a major impact on immune cell and bacterial populations.
Contribution of individual TNF-dependent mechanisms to infection outcome
The original analysis confirmed a major role for TNF in control of the infection within the granuloma. In this model and in vivo, TNF has numerous functions, including inducing secretion of TNF and chemokines, activation of macrophages to a bactericidal state (in concert with IFN-γ), recruitment of cells, and apoptosis of macrophages. To assess the correlation of individual TNF-related functions with control of bacterial infection, we performed a focused sensitivity analysis, varying only the 7 TNF parameters, with all other parameters at the values shown in Tables II-III.
This analysis (Figure 4B) reveals positive correlations between extracellular Mtb numbers and four TNF-related parameters: τTNFact (threshold for TNF-induced activation by macrophages); τTNFapopt (threshold for TNF-induced apoptosis by macrophages); δTNF (rate of TNF degradation); and rTNF (effect of TNF on cell recruitment via trans-endothelial migration). Not surprisingly, the rate of TNF secretion from macrophages and Tγ cells (sTNF) is still the most significantly correlated mechanism (Figure 4B). These results suggest that multiple TNF-dependent mechanisms contribute to the observed effect of TNF on bacterial control.
The role of individual TNF-dependent mechanisms in control of infection
To identify the contribution of each TNF-dependent mechanism individually to the outcome of infection at the granuloma level, we simulated deletion and depletion of each mechanism (both single and in combination).
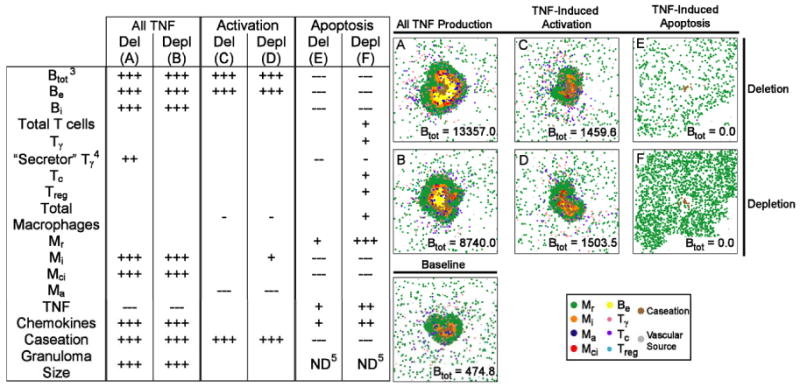
Virtual deletion and depletion of TNF-related macrophage activation shows significantly higher levels of extracellular bacteria and caseation, with lower numbers of activated macrophage and unchanged granuloma size (Table V; c.f. Supplement 3 Figure S2 for all simulated granulomas). The granuloma structures resemble an intermediate between the tightly packed form of the baseline scenario and the irregular core observed with complete TNF deletion or depletion (see Figure 3). Caseation is most likely increased because bacterial numbers increase, leading to more macrophage cell death (which contributes to caseation).
Table V.
Significant changes in granuloma variables at the final time point1 for deletion and depletion of all TNF activity (“All TNF”), TNF-induced activation activity (“Activation”), and TNF-induced apoptosis activity (“Apoptosis”) versus the baseline control scenario2. Representative granuloma structures for each deletion and depletion are shown. A full list of all simulated granulomas for ach deletion/depletion is given in Supplement 3.
|
200 days post-infection (Activation), or the day when complete bacterial elimination occurs (Apoptosis).
+ denotes a higher variable value for the deletion or depletion than the control scenario; - denotes a lower value. +++ (p < 0.0001); ++ (p < 0.001); + (p < 0.01). Significant negative correlations: --- (p < 0.0001); -- (p < 0.001); - (p < 0.01).
Btot = Be + Bi
Number of Tγ cells actively secreting IFN-γ.
Nearly complete bacterial elimination prevented testing.
Deletion and depletion of TNF-induced secretion activity (Table VI) suggests different roles for this activity before and after T cell infiltration begins. Loss of secretion activity (that is, TNF inducing TNF and chemokine production from macrophages) leads to higher levels of caseation and extracellular bacteria during a simulated deletion. No significant effects on these variables are observed during depletion, which has elevated intracellular bacteria and TNF/chemokines (compare Table VI E and F). In contrast, during both deletion and depletion (Table VI C and D), loss of TNF recruitment activity leads to a reduction of immune cells indicating that this function of TNF is not infection stage specific.
Table VI.
Significant changes in granuloma variables at 200 days post-infection for deletion and depletion of all TNF activity (“All TNF”), TNF effects on cellular transendothelial migration activity (“Recruitment”), and TNF-induced secretion of chemokines/TNF activity (“Secretion”) versus the baseline control scenario1. Represenative granuloma structures for each deletion and depletion are shown. A full list of all simulated granulomas for ach deletion/depletion is given in Supplement 3.
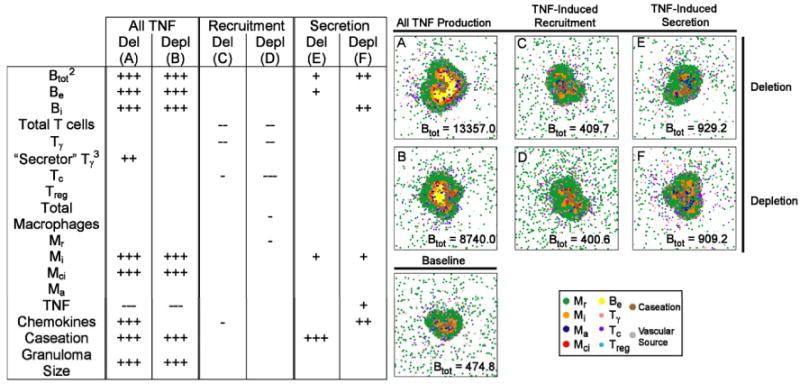
|
+ denotes a higher variable value for the deletion or depletion than the control scenario; - denotes a lower value. +++ (p < 0.0001); ++ (p < 0.001); + (p < 0.01). Significant negative correlations: --- (p < 0.0001); -- (p < 0.001); - (p < 0.01).
Btot = Be + Bi
Number of Tγ cells actively secreting IFN-γ.
When comparing infection initiated with one bacterium (above) with that of 15 bacteria as the inoculum, there are differences in the TNF analysis. While examining the role of each TNF activity in the higher inoculum scenario, we found that the effects of depleting TNF-induced secretion is not significant in the 15 bacteria case, as it was in the single bacteria scenario. This implies that aspects of TNF activities may be sensitive to the initial bacterial inoculum. We are exploring this dose-response topic further in a manuscript in preparation.
Is there a synergy or trade-off between TNF activities?
To determine the effects of interactions between specific TNF activities on granuloma structure, we performed virtual deletions and depletions of pairs and triplets of individual activities. In this section we present results for TNF-dependent secretion, recruitment and activation, deferring exploration of apoptosis activity to the section below.
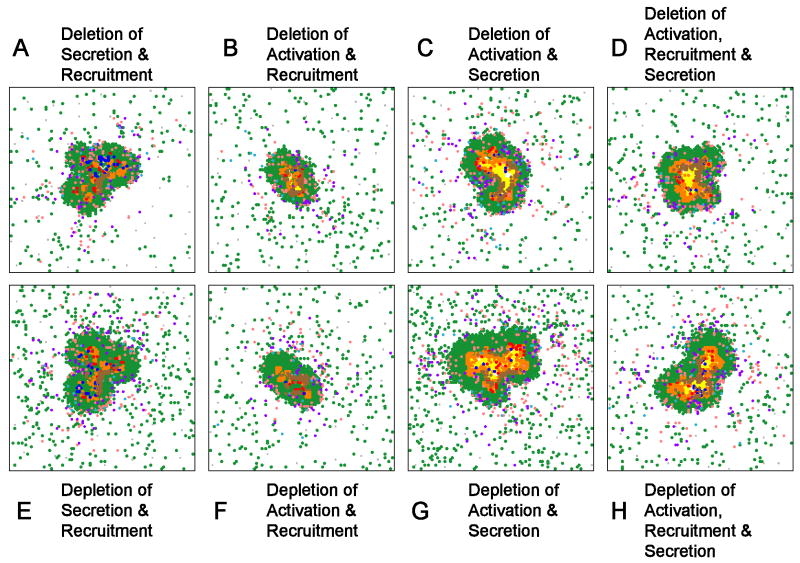
Performing virtual double deletion/depletion of TNF-dependent recruitment and either secretion or activation activity results in the same outcome as either single depletion for secretion and activation (compare Figure 5A-B to Tables V and VI). This suggests that TNF-dependent recruitment is not playing a major role in the control of infection at the level of the granuloma. In contrast, virtual double deletion/depletion with TNF-dependent activation and secretion yields results similar to deletion/depletion of all TNF activities where the granuloma is larger in size, bacterial levels are increased and there is increased caseation (compare Figure 5C with Tables V and VI). A triple deletion/depletion of these three activities also results in poorly formed granulomas with high bacterial loads and caseation (Figure 5D,H). Together these results suggest that both activation and secretion activities of TNF contribute important and distinct roles to granuloma function, formation and/or maintenance, and that TNF-dependent recruitment is not a significant factor. Supplement 3 Figure S3 shows all simulated granulomas at day 200 for all combinations of deletions and depletions.
Figure 5.
Representative double and triple deletions/depletions of TNF activities. A-D. Deletion of TNF-induced secretion and recruitment (A), activation and recruitment (B), activation and secretion (G) and activation, recruitment and secretion. E-H depict analogous combinations of targeted TNF depletions. Structures for multiple replicates of each simulation, and for depletions of each combination at 100 days post-infection, are given in Supplement 3.
Granuloma Structure
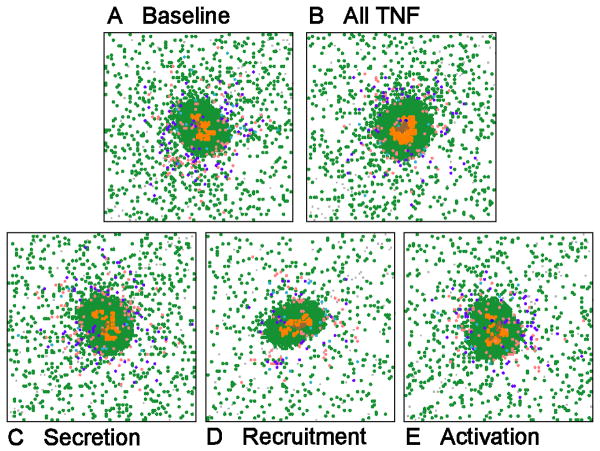
For many of the results obtained thus far it is difficult to distinguish the role of bacterial numbers versus TNF activity in driving the structure of the granuloma. To address this, we perform a simulation that is impossible to do in vivo, namely, to allow the granuloma to form and then to fix the total number of bacteria present (see Methods for more details). This allows us to isolate the effects of TNF without the contribution of changing bacterial numbers. Figure 6 shows results of fixing the bacterial levels and performing virtual depletions performed at day 100 for: all TNF, secretion, recruitment and activation activities, respectively. Depletion of TNF recruitment in these simulations did have an impact on the number of T cells at the site, and the granulomas were on average smaller (but not significantly so). However, the granuloma structures were not affected. In all other cases, depletion of the single TNF activities did not affect the shape or size of the granuloma once bacterial numbers were fixed. This suggests that bacterial load is the primary factor responsible determining granuloma structure and shape.
Figure 6.
Characteristic granulomas resulting from holding total bacterial numbers fixed after day 100. Shown are: baseline (A), virtual depletion of all TNF activities (B), and virtual depletion of TNF secretion activity (C), TNF recruitment activity (D) and TNF activation activity (E).
Role of TNF-induced Apoptosis
An unanticipated finding was that deletion and depletion of TNF-induced apoptosis results in effective clearance of bacteria (“Apoptosis” in Table V). At the time after all bacteria were cleared, there is disrupted granuloma structure with a robust increase in cell infiltration and TNF/chemokine production (Table V). Separate depletion of TNF-induced apoptosis activity from infected and uninfected macrophages shows that this phenomenon is primarily driven by uninfected macrophages, which constitute the largest macrophage sub-population (data not shown). These data suggest that TNF-induced apoptosis of uninfected macrophages within the granuloma prevents excessive inflammation, but this leads to persistent Mtb infection.
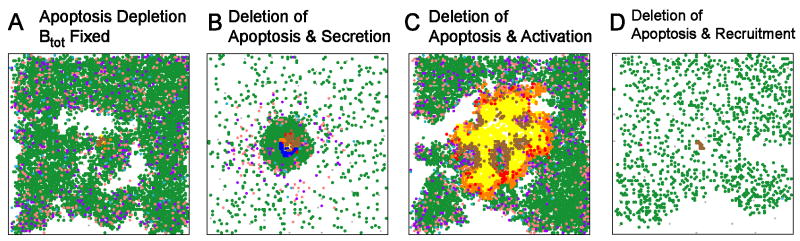
To directly assess the contribution to granuloma structure, we fixed bacterial numbers at 100 days as described above and eliminated TNF-dependent apoptosis. We again observed the hyper-inflammatory state described above (Figure 7A). This supports a major role for uninfected macrophages driving granuloma structure. One possible explanation is that in the absence of apoptotic cell death of macrophages, induction of TNF at the site of infection continues to induce TNF in surrounding macrophages, forming a positive feedback loop- a process normally limited by TNF-induced apoptosis in this model.
Figure 7.
Effects of losing TNF-induced apoptosis activity on granuloma structures 200 days post-infection. (A) Two characteristic structures that are attainable under apoptosis depletion with total bacterial numbers held constant after day 100. (B) Deletion of apoptosis and TNF-induced secretion. (C) Deletion of both TNF-induced apoptosis and activation. (D) Deletion of both TNF-induced apoptosis and recruitment. In the latter structures, continued bacterial growth after day 100 was permitted. A full list of apoptosis-related deletions are presented in Supplement 3.
To test this possibility, we performed virtual deletion or depletion of both apoptosis and TNF-induced secretion of both TNF and chemokines (Figure 7B). Without secretion of TNF and chemokines, loss of apoptosis did not result in enhanced inflammation and resolution of infection, clearly demonstrating that the robust inflammatory response in the single deletion and depletion of apoptosis was dependent on TNF-mediated induction of TNF and chemokines from uninfected macrophages. This may be a feedback loop where bacteria released by dying macrophages induce secretion of TNF, and higher subsequent levels of TNF and chemokines. Apoptosis of macrophages likely prevents this TNF-induced cytokine/chemokine response by both reducing the bacteria released from macrophages and eliminating excess macrophages from the granuloma; these macrophages are the source of the cytokines and chemokines that influence the inflammatory response. Combining a loss of TNF-mediated apoptosis with loss of TNF-induced macrophage activation (Figure 7C) results in substantially worse inflammation, but no clearance of bacteria, because macrophage activation is essential for Mtb killing. Deleting or depleting both apoptosis and TNF-mediated cell recruitment (Figure 7D) is not different than the individually simulated apoptosis deletion and depletion, indicating that the observed excess inflammation is not due to changes in total recruitment of cells to the lungs.
Discussion
TNF is clearly a central factor in control of M. tuberculosis infection. It has been repeatedly demonstrated in murine models that granuloma function, formation and maintenance are dependent on TNF (24). However, TNF is a pleiotropic cytokine and the relative contributions of its primary activities to the functional and structural characteristics of the granuloma have not been elucidated. The complex nature of TNF in the immune response to M. tuberculosis likely accounts for the dramatic effects observed when neutralizing this cytokine in humans with latent M. tuberculosis infection (21) and in animal models (16, 19).The agent based model of a tuberculous granuloma developed in this study is unique in its ability to address the roles of individual functions of TNF in granuloma formation and control of infection. The major findings from this study are: 1. Both TNF-induced macrophage activation and TNF-dependent secretion of chemokines and cytokines are crucial factors in control of infection within the granuloma; 2. TNF-induced apoptosis functions to reduce inflammation at the expense of impairing bacterial clearance; 3. Structural alterations in granuloma size and shape resulting from perturbation of TNF activity is directly driven by bacterial growth, and indirectly by TNF activities.
Our agent-based model of granuloma formation reproduces major features of infection by representing interactions of individual cell agents and molecular effectors with a representation of a growing mycobacterial population. This work was based on a previous model (38) with major extensions that include representations of TNF, a simple chemokine network, and distinct T cell sub-populations, including effector and regulatory cells. A baseline parameter set demonstrates bacterial control and variable granuloma structures, both of which are disrupted by virtual deletion or depletion of TNF. This sophisticated model allows for a spatial and temporal representation of events occurring during granuloma formation and function allowing for exploration of this complex biological system in ways that are currently not tractable with experimental methods. A next logical step is to consider a three-dimensional spatial representation, and this may be important for making further predictions about elements within the system. On the other hand, a three-dimensional granuloma model (55) gave similar results to our previous two-dimensional model (38), suggesting that the two-dimensional approach is sufficient to study many aspects of granuloma formation.
One further important simplification in our model is a reduction in the number of cell types to only those with well-characterized roles in Mtb granulomas. Multinucleate giant cells may modulate chemokine production (56) without taking up extracellular Mtb (57); dendritic cells are necessary for optimal antigenic stimulation of T cells (58, 59); foamy cells are dendritic-like cells (60) that may be a nutrient source for Mtb (61); B cells form ectopic germinal centers in Mtb-infected mouse lungs, with some role in containment (62). Each of these cell types may have important quantitative or qualitative roles, but are not sufficiently characterized to include in this model. Future work can easily incorporate these into the model when mechanistic information becomes available.
Our model provides the opportunity to study various combinations of effector functions controlled by TNF. Virtual deletion or depletion of each individual TNF-related function demonstrated that TNF-dependent macrophage activation was the primary factor involved in control of bacterial numbers, but that TNF-induced chemokine and TNF secretion also played a role in this process, probably by bringing macrophages and T cells close together in the granuloma, as well as producing more TNF in the granuloma. In contrast, loss of TNF-mediated recruitment, where TNF acts on the vasculature to bring more cells into the lungs, had little effect on bacterial numbers, but resulted in recruitment of fewer immune cells. This is consistent with reduced recruitment in TNF-blockaded M. bovis BCG infection in mice (63); lower virulence of BCG compared to Mtb may explain why complete loss of TNF activity did not cause a severe phenotype in that study.
Virtual elimination of both TNF-dependent macrophage activation and cytokine/chemokine secretion resulted in large granulomas with extensive caseation and lack of bacterial control that was essentially identical to the scenario of total loss of TNF, and was worse than loss of either function independently. In contrast, eliminating TNF-mediated recruitment had no additional detrimental effects when coupled with the loss of TNF-dependent macrophage activation or secretion. Thus, both macrophage activation and cytokine/chemokine secretion induced by TNF are important and distinct functions in control of bacterial numbers in the granuloma.
Based on data in the murine system, including our own data, it has been widely believed that TNF is the major factor that controls the actual formation, structure and maintenance of a granuloma (17, 64). Humans treated with TNF-neutralizing drugs for inflammatory diseases have an increased incidence of reactivation tuberculosis. In an initial report (21), lung biopsies from a patient with disseminated tuberculosis due to TNF neutralization did not reveal granulomas, supporting the murine data. However, another human study (65) has indicated that granulomas are present in tuberculous lungs from TNF-neutralized patients, calling into question whether TNF is required for maintenance of granulomas. Indeed, data from our non-human primate model of tuberculosis also indicate that granulomas can form and be maintained even when TNF is neutralized (Lin, Flynn, manuscript submitted).
The current virtual model provides an opportunity to test the functions of TNF that are related to granuloma function, formation and structure. The flexibility of this model, compared to in vivo systems, allows us to hold the bacterial numbers steady while varying other parameters. This provides an opportunity to distinguish between effects of granuloma structure that are due to bacterial numbers or to TNF-dependent factors. In fact, when bacterial numbers cannot increase as a result of TNF depletion, granuloma structure is essentially unaffected by loss of TNF-dependent factors. This suggests that bacterial numbers are a driving force in the aberrations in granuloma formation and maintenance due to loss of TNF. This does not negate a role for TNF in the formation and maintenance of granulomas, but instead suggests that bacterial numbers play a greater role in this than previously believed.
One surprising finding was that deletion and depletion of TNF-induced apoptosis results in effective clearance of bacteria. Simulations predicted disrupted granuloma structure, increased cell infiltration and TNF/chemokine production in this case. Virtual depletion of TNF-induced apoptosis activity from infected or uninfected macrophages demonstrated that this phenomenon is primarily driven by uninfected macrophages. Comparing dual and single depletions, as well as the “fixed bacterial numbers” simulations, it became clear that the increased inflammation observed in the absence of apoptosis was due to TNF-induced secretion of chemokines and TNF from uninfected macrophages. This function of TNF enhances bacterial clearance, but can lead to excessive pathology. Thus a role for TNF-induced apoptosis seems to be control of pathology at the expense of bacterial clearance. This provides evidence that the outcome of infection in any one granuloma is a balance of different factors, including TNF-induced functions. This result may differ if certain anti-inflammatory cytokines such as IL-10 were included in the model, which could modulate the effects of TNF; this possibility has been proposed previously (66).
The size of the initial bacterial inoculum may have some effects on progression and containment of Mtb infection, particularly in cases with immunological deficiencies (e.g. 67). We explored the effects of increasing the bacterial load in the first infected macrophage from one bacterium to 15. Cell and bacterial kinetics were different preceding T cell infiltration at 20 days post-infection, but became very similar after (not shown). The primary difference in dose as it relates to TNF activities is for TNF-induced secretion of chemokines and TNF. For the low inoculum, depleting this activity significantly affected aspects of the granuloma (Table VI), where for the higher inoculum, no significant effects for the depletion were observed (not shown). A very high inoculum, above the small range we tested here, would likely have dramatic effects on the outcome of infection, as demonstrated in our previous studies (6, 20), although the effect may not manifest at the level of a single granuloma. We have a manuscript in preparation exploring the role of initial dose in a more detailed fashion.
In this work we have used an advanced mathematical modeling approach to identify the relative contribution of different TNF-dependent activities in granuloma formation and function. Model results predicted mechanisms involved in control of both bacterial numbers within the granuloma as well as inflammation. These mechanisms can now serve as putative targets for vaccine and treatment strategies. The approach applied here contributes groundwork for the use of computational approaches in identifying biological pathways that can be targeted for modulation of tuberculosis.
Supplementary Material
Acknowledgments
We thank members of JCJR's PhD thesis committee (Gary Huffnagle, Jennifer Linderman, Alex Ninfa, and Joel Swanson); Shlomo Ta'asan and other members of the Center for Modeling Pulmonary Immunity; P. Ling Lin; Steven Kunkel and Simeone Marino for helpful comments.
Footnotes
This work was supported by NIH grants HL68526 (DEK, JLF), NO1 AI50018 (JLF, DEK), HL72682 (DEK) and LM00902701 (DEK, JLF), the F. G. Novy Fellowship (JCJR), and the Ellison Foundation (JLF).
This result differs slightly from our previous results in Segovia-Juarez et al (38), which predicted that intracellular growth rates are transiently negatively correlated with extracellular Mtb numbers between days 30 and 150 post-infection. This discrepancy is due to a peak in chronically infected macrophage bursting in that model that is not reproduced here since we hold the initial number of macrophages constant. This allows uncertainty analysis to have identical initial conditions between different parameter sets.
Contributor Information
J. Christian J. Ray, Email: jjray@rice.edu.
JoAnne L. Flynn, Email: joanne@pitt.edu.
Denise E. Kirschner, Email: kirschne@umich.edu.
References
- 1.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 2.Ridley M, Heather C, Brown I, Willoughby D. Experimental epithelioid cell granulomas tubercle formation and immunological competence: An ultrastructural analysis. The Journal of Pathology. 1983;141:97–112. doi: 10.1002/path.1711410202. [DOI] [PubMed] [Google Scholar]
- 3.Emile JF, Patey N, Altare F, Lamhamedi S, Jouanguy E, Boman F, Quillard J, Lecomte-Houcke M, Verola O, Mousnier JF, Dijoud F, Blanche S, Fischer A, Brousse N, Casanova JL. Correlation of granuloma structure with clinical outcome defines two types of idiopathic disseminated BCG infection. Journal of Pathology. 1997;181:25–30. doi: 10.1002/(SICI)1096-9896(199701)181:1<25::AID-PATH747>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 4.Lin PL, Pawar S, Myers A, Pegu A, Fuhrman C, Reinhart TA, Capuano SV, Klein E, Flynn JL. Early events in Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun. 2006;74:3790–3803. doi: 10.1128/IAI.00064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoebe K, Janssen E, Beutler B. The interface between innate and adaptive immunity. Nat Immunol. 2004;5:971–974. doi: 10.1038/ni1004-971. [DOI] [PubMed] [Google Scholar]
- 6.Sud D, Bigbee C, Flynn JL, Kirschner DE. Contribution of CD8+ T cells to control of Mycobacterium tuberculosis infection. J Immunol. 2006;176:4296–4314. doi: 10.4049/jimmunol.176.7.4296. [DOI] [PubMed] [Google Scholar]
- 7.Vignali D, Collison L, Workman C. How regulatory T cells work. Nature Reviews Immunology. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason CM, Porretta E, Zhang P, Nelson S. CD4+CD25+transforming growth factor--producing T cells are present in the lung in murine tuberculosis and may regulate the host inflammatory response. Clinical & Experimental Immunology. 2007;148:537–545. doi: 10.1111/j.1365-2249.2007.03371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guyot-Revol V, Innes JA, Hackforth S, Hinks T, Lalvani A. Regulatory T cells are expanded in blood and disease sites in patients with tuberculosis. Am J Respir Crit Care Med. 2006;173:803–810. doi: 10.1164/rccm.200508-1294OC. [DOI] [PubMed] [Google Scholar]
- 10.Ordway D, Henao-Tamayo M, Harton M, Palanisamy G, Troudt J, Shanley C, Basaraba R, Orme I. The Hypervirulent Mycobacterium tuberculosis Strain HN878 Induces a Potent TH1 Response followed by Rapid Down-Regulation. J Immunol. 2007;179:522–531. doi: 10.4049/jimmunol.179.1.522. [DOI] [PubMed] [Google Scholar]
- 11.Roberts T, Beyers N, Aguirre A, Walzl G. Immunosuppression during active tuberculosis is characterized by decreased interferon- gamma production and CD25 expression with elevated forkhead box P3, transforming growth factor- beta, and interleukin-4 mRNA levels. J Infect Dis. 2007;195:870–878. doi: 10.1086/511277. [DOI] [PubMed] [Google Scholar]
- 12.Asano M, Toda M, Sakaguchi N, Sakaguchi S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J Exp Med. 1996;184:387–396. doi: 10.1084/jem.184.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 14.Thornton AM, Shevach EM. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J Exp Med. 1998;188:287–296. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baatar D, Olkhanud P, Sumitomo K, Taub D, Gress R, Biragyn A. Human Peripheral Blood T Regulatory Cells (Tregs), Functionally Primed CCR4+ Tregs and Unprimed CCR4- Tregs, Regulate Effector T Cells Using FasL. J Immunol. 2007;178:4891–4900. doi: 10.4049/jimmunol.178.8.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, Schreiber R, Mak TW, Bloom BR. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 17.Bean A, Roach D, Briscoe H, France M, Korner H, Sedgwick J, Britton W. Structural Deficiencies in Granuloma Formation in TNF Gene-Targeted Mice Underlie the Heightened Susceptibility to Aerosol Mycobacterium tuberculosis Infection, Which Is Not Compensated for by Lymphotoxin. J Immunol. 1999;162:3504–3511. [PubMed] [Google Scholar]
- 18.Bekker LG, Freeman S, Murray PJ, Ryffel B, Kaplan G. TNF-alpha controls intracellular mycobacterial growth by both inducible nitric oxide synthase-dependent and inducible nitric oxide synthase-independent pathways. J Immunol. 2001;166:6728–6734. doi: 10.4049/jimmunol.166.11.6728. [DOI] [PubMed] [Google Scholar]
- 19.Mohan VP, Scanga CA, Yu K, Scott HM, Tanaka KE, Tsang E, Tsai MM, Flynn JL, Chan J. Effects of tumor necrosis factor alpha on host immune response in chronic persistent tuberculosis: possible role for limiting pathology. Infect Immun. 2001;69:1847–1855. doi: 10.1128/IAI.69.3.1847-1855.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marino S, Sud D, Plessner H, Lin PL, Chan J, Flynn JL, Kirschner DE. Differences in reactivation of tuberculosis induced from anti-TNF treatments are based on bioavailability in granulomatous tissue. PLoS Comput Biol. 2007;3:1909–1924. doi: 10.1371/journal.pcbi.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keane J, Gershon S, Wise RP, Mirabile-Levens E, Kasznica J, Schwieterman WD, Siegel JN, Braun MM. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N Engl J Med. 2001;345:1098–1104. doi: 10.1056/NEJMoa011110. [DOI] [PubMed] [Google Scholar]
- 22.Winthrop KL. Risk and prevention of tuberculosis and other serious opportunistic infections associated with the inhibition of tumor necrosis factor. Nat Clin Pract Rheumatol. 2006;2:602–610. doi: 10.1038/ncprheum0336. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z, Connell MC, Macewan DJ. TNFR1-induced NF-kappaB, but not ERK, p38MAPK or JNK activation, mediates TNF-induced ICAM-1 and VCAM-1 expression on endothelial cells. Cell Signal. 2007;19:1238–1248. doi: 10.1016/j.cellsig.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Algood HM, Lin PL, Flynn JL. Tumor necrosis factor and chemokine interactions in the formation and maintenance of granulomas in tuberculosis. Clin Infect Dis. 2005;41 3 doi: 10.1086/429994. [DOI] [PubMed] [Google Scholar]
- 25.Roach D, Bean A, Demangel C, France M, Briscoe H, Britton W. TNF Regulates Chemokine Induction Essential for Cell Recruitment, Granuloma Formation, and Clearance of Mycobacterial Infection. J Immunol. 2002;168:4620–4627. doi: 10.4049/jimmunol.168.9.4620. [DOI] [PubMed] [Google Scholar]
- 26.van Buul J, Hordijk P. Signaling in Leukocyte Transendothelial Migration. Arterioscler Thromb Vasc Biol. 2004;24:824–833. doi: 10.1161/01.ATV.0000122854.76267.5c. [DOI] [PubMed] [Google Scholar]
- 27.Stein J, Nombela-Arrieta C. Chemokine control of lymphocyte trafficking: a general overview. Immunology. 2005;116:1–12. doi: 10.1111/j.1365-2567.2005.02183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flesch I, Kaufmann S. Mycobacterial growth inhibition by interferon-gamma-activated bone marrow macrophages and differential susceptibility among strains of Mycobacterium tuberculosis. J Immunol. 1987;138:4408–4413. [PubMed] [Google Scholar]
- 29.Flesch IE, Kaufmann SH. Activation of tuberculostatic macrophage functions by gamma interferon, interleukin-4, and tumor necrosis factor. Infect Immun. 1990;58:2675–2677. doi: 10.1128/iai.58.8.2675-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rook GA, Steele J, Ainsworth M, Champion BR. Activation of macrophages to inhibit proliferation of Mycobacterium tuberculosis: comparison of the effects of recombinant gamma-interferon on human monocytes and murine peritoneal macrophages. Immunology. 1986;59:333–338. [PMC free article] [PubMed] [Google Scholar]
- 31.Laster SM, Wood JG, Gooding LR. Tumor necrosis factor can induce both apoptic and necrotic forms of cell lysis. J Immunol. 1988;141:2629–2634. [PubMed] [Google Scholar]
- 32.Keane J, Balcewicz-Sablinska MK, Remold HG, Chupp GL, Meek BB, Fenton MJ, Kornfeld H. Infection by Mycobacterium tuberculosis promotes human alveolar macrophage apoptosis. Infect Immun. 1997;65:298–304. doi: 10.1128/iai.65.1.298-304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohan K, Ding Z, Hanly J, Issekutz TB. IFN-gamma-inducible T cell alpha chemoattractant is a potent stimulator of normal human blood T lymphocyte transendothelial migration: differential regulation by IFN-gamma and TNF-alpha. J Immunol. 2002;168:6420–6428. doi: 10.4049/jimmunol.168.12.6420. [DOI] [PubMed] [Google Scholar]
- 34.Kristensen NN, Gad M, Thomsen AR, Lu B, Gerard C, Claesson MH. CXC chemokine receptor 3 expression increases the disease-inducing potential of CD4+ CD25- T cells in adoptive transfer colitis. Inflamm Bowel Dis. 2006;12:374–381. doi: 10.1097/01.MIB.0000217337.15442.e1. [DOI] [PubMed] [Google Scholar]
- 35.Valente AJ, Graves DT, Vialle-Valentin CE, Delgado R, Schwartz CJ. Purification of a monocyte chemotactic factor secreted by nonhuman primate vascular cells in culture. Biochemistry. 1988;27:4162–4168. doi: 10.1021/bi00411a039. [DOI] [PubMed] [Google Scholar]
- 36.Peters W, Cyster JG, Mack M, Schlöndorff D, Wolf AJ, Ernst JD, Charo IF. CCR2-dependent trafficking of F4/80dim macrophages and CD11cdim/intermediate dendritic cells is crucial for T cell recruitment to lungs infected with Mycobacterium tuberculosis. J Immunol. 2004;172:7647–7653. doi: 10.4049/jimmunol.172.12.7647. [DOI] [PubMed] [Google Scholar]
- 37.Yurchenko E, Tritt M, Hay V, Shevach E, Belkaid Y, Piccirillo C. CCR5-dependent homing of naturally occurring CD4+ regulatory T cells to sites of Leishmania major infection favors pathogen persistence. J Exp Med. 2006;203:2451–2460. doi: 10.1084/jem.20060956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segovia-Juarez JL, Ganguli S, Kirschner D. Identifying control mechanisms of granuloma formation during M. tuberculosis infection using an agent-based model. J Theor Biol. 2004;231:357–376. doi: 10.1016/j.jtbi.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 39.Krombach F, Munzing S, Allmeling AM, Gerlach JT, Behr J, Dorger M. Cell size of alveolar macrophages: an interspecies comparison. Environ Health Perspect. 1997;105 5:1261–1263. doi: 10.1289/ehp.97105s51261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadek MI, Sada E, Toossi Z, Schwander SK, Rich EA. Chemokines induced by infection of mononuclear phagocytes with mycobacteria and present in lung alveoli during active pulmonary tuberculosis. Am J Respir Cell Mol Biol. 1998;19:513–521. doi: 10.1165/ajrcmb.19.3.2815. [DOI] [PubMed] [Google Scholar]
- 41.Brice GT, Graber NL, Hoffman SL, Doolan DL. Expression of the chemokine MIG is a sensitive and predictive marker for antigen-specific, genetically restricted IFN-gamma production and IFN-gamma-secreting cells. J Immunol Methods. 2001;257:55–69. doi: 10.1016/s0022-1759(01)00446-x. [DOI] [PubMed] [Google Scholar]
- 42.Kang SG, Piniecki RJ, Hogenesch H, Lim HW, Wiebke E, Braun SE, Matsumoto S, Kim CH. Identification of a chemokine network that recruits FoxP3(+) regulatory T cells into chronically inflamed intestine. Gastroenterology. 2007;132:966–981. doi: 10.1053/j.gastro.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Iijima W, Ohtani H, Nakayama T, Sugawara Y, Sato E, Nagura H, Yoshie O, Sasano T. Infiltrating CD8+ T Cells in Oral Lichen Planus Predominantly Express CCR5 and CXCR3 and Carry Respective Chemokine Ligands RANTES/CCL5 and IP-10/CXCL10 in Their Cytolytic Granules: A Potential Self-Recruiting Mechanism. Am J Pathol. 2003;163:261–268. doi: 10.1016/S0002-9440(10)63649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staruch MJ, Camacho R, Dumont FJ. Distinctive calcineurin-dependent (FK506-sensitive) mechanisms regulate the production of the CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES vs IL-2 and TNF-alpha by activated human T cells. Cell Immunol. 1998;190:121–131. doi: 10.1006/cimm.1998.1411. [DOI] [PubMed] [Google Scholar]
- 45.O'Brien AD, Standiford TJ, Christensen PJ, Wilcoxen SE, Paine R. Chemotaxis of alveolar macrophages in response to signals derived from alveolar epithelial cells. J Lab Clin Med. 1998;131:417–424. doi: 10.1016/s0022-2143(98)90142-1. [DOI] [PubMed] [Google Scholar]
- 46.Tan J, Deleuran B, Gesser B, Maare H, Deleuran M, Larsen CG, Thestrup-Pedersen K. Regulation of human T lymphocyte chemotaxis in vitro by T cell-derived cytokines IL-2, IFN-gamma, IL-4, IL-10, and IL-13. J Immunol. 1995;154:3742–3752. [PubMed] [Google Scholar]
- 47.Van Furth R, Diesselhoff-den Dulk MC, Mattie H. Quantitative study on the production and kinetics of mononuclear phagocytes during an acute inflammatory reaction. J Exp Med. 1973;138:1314–1330. doi: 10.1084/jem.138.6.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sprent J. Lifespans of naive, memory and effector lymphocytes. Current opinion in immunology. 1993;5:433–438. doi: 10.1016/0952-7915(93)90065-z. [DOI] [PubMed] [Google Scholar]
- 49.Lazarevic V, Nolt D, Flynn JL. Long-term control of Mycobacterium tuberculosis infection is mediated by dynamic immune responses. J Immunol. 2005;175:1107–1117. doi: 10.4049/jimmunol.175.2.1107. [DOI] [PubMed] [Google Scholar]
- 50.Francis K, Palsson BO. Effective intracellular communication distances are determined by the relative time constants for cyto/chemokine secretion and diffusion. Proceedings of the National Academy of Sciences. 1997;94:12258–12262. doi: 10.1073/pnas.94.23.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marino S, Hogue I, Ray C, Kirschner D. A Methodology For Performing Global Uncertainty And Sensitivity Analysis In Systems Biology. J Theor Biol. 2008;254:178–196. doi: 10.1016/j.jtbi.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulrichs T, Kosmiadi GA, Trusov V, Jörg S, Pradl L, Titukhina M, Mishenko V, Gushina N, Kaufmann SH. Human tuberculous granulomas induce peripheral lymphoid follicle-like structures to orchestrate local host defence in the lung. The Journal of pathology. 2004;204:217–228. doi: 10.1002/path.1628. [DOI] [PubMed] [Google Scholar]
- 53.Theus S, Eisenach K, Fomukong N, Silver RF, Cave MD. Beijing family Mycobacterium tuberculosis strains differ in their intracellular growth in THP-1 macrophages. Int J Tuberc Lung Dis. 2007;11:1087–1093. [PubMed] [Google Scholar]
- 54.Flynn JL. Lessons from experimental Mycobacterium tuberculosis infections. Microbes Infect. 2006;8:1179–1188. doi: 10.1016/j.micinf.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 55.Warrender C, Forrest S, Koster F. Modeling intercellular interactions in early Mycobacterium infection. Bull Math Biol. 2006;68:2233–2261. doi: 10.1007/s11538-006-9103-y. [DOI] [PubMed] [Google Scholar]
- 56.Zhu XW, Friedland JS. Multinucleate giant cells and the control of chemokine secretion in response to Mycobacterium tuberculosis. Clinical immunology (Orlando, Fla) 2006;120:10–20. doi: 10.1016/j.clim.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 57.Lay G, Poquet Y, Salek-Peyron P, Puissegur MP, Botanch C, Bon H, Levillain F, Duteyrat JL, Emile JF, Altare F. Langhans giant cells from M. tuberculosis-induced human granulomas cannot mediate mycobacterial uptake. The Journal of pathology. 2007;211:76–85. doi: 10.1002/path.2092. [DOI] [PubMed] [Google Scholar]
- 58.Uehira K, Amakawa R, Ito T, Tajima K, Naitoh S, Ozaki Y, Shimizu T, Yamaguchi K, Uemura Y, Kitajima H, Yonezu S, Fukuhara S. Dendritic Cells Are Decreased in Blood and Accumulated in Granuloma in Tuberculosis. Clinical Immunology. 2002;105:296–303. doi: 10.1006/clim.2002.5287. [DOI] [PubMed] [Google Scholar]
- 59.Giri P, Schorey J. Exosomes Derived from M. Bovis BCG Infected Macrophages Activate Antigen-Specific CD4+ and CD8+ T Cells In Vitro and In Vivo. PLoS ONE. 2008;3:e2461. doi: 10.1371/journal.pone.0002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ordway D, Henao-Tamayo M, Orme IM, Gonzalez-Juarrero M. Foamy macrophages within lung granulomas of mice infected with Mycobacterium tuberculosis express molecules characteristic of dendritic cells and antiapoptotic markers of the TNF receptor-associated factor family. Journal of immunology (Baltimore, Md : 1950) 2005;175:3873–3881. doi: 10.4049/jimmunol.175.6.3873. [DOI] [PubMed] [Google Scholar]
- 61.Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffé M, Emile JF, Marchou B, Cardona PJ, de Chastellier C, Altare F. Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS pathogens. 2008;4 doi: 10.1371/journal.ppat.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maglione PJ, Xu J, Chan J. B cells moderate inflammatory progression and enhance bacterial containment upon pulmonary challenge with Mycobacterium tuberculosis. Journal of immunology (Baltimore, Md : 1950) 2007;178:7222–7234. doi: 10.4049/jimmunol.178.11.7222. [DOI] [PubMed] [Google Scholar]
- 63.Egen J, Rothfuchs A, Feng C, Winter N, Sher A, Germain R. Macrophage and T Cell Dynamics during the Development and Disintegration of Mycobacterial Granulomas. Immunity. 2008;28:271–284. doi: 10.1016/j.immuni.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chakravarty SD, Zhu G, Tsai MC, Mohan VP, Marino S, Kirschner DE, Huang L, Flynn J, Chan J. Tumor necrosis factor blockade in chronic murine tuberculosis enhances granulomatous inflammation and disorganizes granulomas in the lungs. Infection and immunity. 2008;76:916–926. doi: 10.1128/IAI.01011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iliopoulos A, Psathakis K, Aslanidis S, Skagias L, Sfikakis P. Tuberculosis and granuloma formation in patients receiving anti-TNF therapy. Int J Tuberc Lung Dis. 2006;10:588–590. [PubMed] [Google Scholar]
- 66.Garcia-Ramallo E, Marques T, Prats N, Beleta J, Kunkel SL, Godessart N. Resident cell chemokine expression serves as the major mechanism for leukocyte recruitment during local inflammation. J Immunol. 2002;169:6467–6473. doi: 10.4049/jimmunol.169.11.6467. [DOI] [PubMed] [Google Scholar]
- 67.Scott HM, Flynn JL. Mycobacterium tuberculosis in chemokine receptor 2-deficient mice: influence of dose on disease progression. Infection & Immunity. 2002;70:5946–5954. doi: 10.1128/IAI.70.11.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.