Abstract
Intracellular protein degradation, which must be tightly controlled to protect normal proteins, is carried out by ATP-dependent proteases. These multicomponent enzymes have chaperone-like ATPases that recognize and unfold protein substrates and deliver them to the proteinase components for digestion. In ClpAP, hexameric rings of the ClpA ATPase stack axially on either face of the ClpP proteinase, which consists of two apposed heptameric rings. We have used cryoelectron microscopy to characterize interactions of ClpAP with the model substrate, bacteriophage P1 protein, RepA. In complexes stabilized by ATPγS, which bind but do not process substrate, RepA dimers are seen at near-axial sites on the distal surface of ClpA. On ATP addition, RepA is translocated through ≈150 Å into the digestion chamber inside ClpP. Little change is observed in ClpAP, implying that translocation proceeds without major reorganization of the ClpA hexamer. When translocation is observed in complexes containing a ClpP mutant whose digestion chamber is already occupied by unprocessed propeptides, a small increase in density is observed within ClpP, and RepA-associated density is also seen at other axial sites. These sites appear to represent intermediate points on the translocation pathway, at which segments of unfolded RepA subunits transiently accumulate en route to the digestion chamber.
Keywords: ATP-dependent protease, chaperone, protein unfoldase, processivity, cryoelectron microscopy
ATP-dependent proteases have essential functions in controlling the levels of regulatory proteins and in maintaining protein quality control in the cell (1). Structural analyses of two of the four major families of these enzymes suggest common organizational motifs and, by implication, modes of action (2–4). Proteasomes and Clp proteases are all barrel-shaped molecules made up of functionally differentiated tiers, each tier comprising a ring of homologous or identical subunits. Central tiers enclose the proteolytic active sites, whereas outer tiers have various functionalities, including at least one ring of subunits with ATPase activity. Members of two other protease families, Lon/PIM1p and FtsH/YME1p, also have similar domains (5).
The 20 S proteasome (6, 7) and Escherichia coli ClpP (8) have been solved to high resolution by x-ray crystallography. Although their subunit folds and catalytic mechanisms differ, these molecules share a striking architectural feature whereby the active sites are housed in an internal chamber, enclosed between two 7-fold rings. In both cases, the proteolytic chamber is accessed by narrow axial channels that restrict passage to polypeptides in extended conformations. Thus, entry of protein substrates into the digestion chamber should require disruption of their tertiary structure.
Isolated ATPases, such as ClpA and ClpX, are molecular chaperones and have protein remodeling activities that can effect conformational changes and alterations in quaternary interactions of substrates (9, 10). Both proteins have robust protein unfoldase activities that disrupt the tertiary structure of appropriately tagged, stably folded, proteins (11–13). Complexes containing the ATPases of the yeast proteasome also have molecular chaperone activity (14). Thus, in general, unfoldase activity may be used by ATP-dependent proteases to translocate substrates to the proteolytic sites.
Electron microscopy (EM) studies have shown that Clp ATPases form hexameric rings of identical subunits (15, 16), and the 19S component of the yeast proteasome appears to contain hexameric rings of homologous but nonidentical subunits, to which additional subcomplexes are bound (17). In ClpAP complexes, ClpA stacks axially on the faces of ClpP (16), thus controlling access to the channels leading into the proteolytic chamber. This positioning allows for coordination of ClpA's activities, which include specific binding of substrates, unfolding of bound substrates, and translocation of unfolded substrates to ClpP.
Nonhydrolyzable or poorly hydrolyzable ATP analogs such as adenosine 5′-[γ-thio]triphosphate (ATPγS) promote hexamerization of ClpA and its binding to ClpP, sufficing for degradation of polypeptides of up to ≈30 amino acids (18). However, degradation of proteins with significant tertiary structure requires ATP. Folded substrates bind to ClpAP in the presence of ATPγS but are not degraded unless ATP is added (19). These properties make it possible to synchronize degradation in vitro: enzyme–substrate complexes can be formed in the presence of ATPγS and subsequent steps triggered by adding excess ATP. In this study, we exploited these properties to study the binding of RepA by ClpAP and their subsequent interactions during translocation.
Methods
Protein Purification.
Clp proteins, including ClpPSC (12), were purified by standard methods (20); RepA purification was previously described (21). Chemically inactivated ClpPin was prepared by incubating ClpP with excess carbobenzoxy-leucyl-tyrosyl chloromethyl ketone or diisopropyl fluorophosphate (12).
Preparation of ClpA and ClpAP Complexes with RepA.
ClpA was assembled by incubating ClpA (1 mg/ml) in 50 mM Tris/HCl, pH 7.5/0.3 M KCl/25 mM MgCl2/2 mM ATPγS/10% (vol/vol) glycerol. When ClpAP complexes were needed, ClpP or ClpPSC (0.35 mg/ml) was included. RepA was added to give a 2-fold molar excess of RepA dimers over ClpA hexamers and incubated at room temperature for 15 min. For direct analysis, the samples were diluted in the above buffer except that only 1 mM ATPγS was present, and there was no additional glycerol. Complexes were purified by injecting 100 μl of the mixture onto a Superdex200 (Pharmacia Biotech) gel filtration column (3.2 mm × 30 cm) running at room temperature at 0.08 ml/min. Fractions (80 μl) were collected.
EM was performed as described (4), and PIC-III (22) was used for correlation averaging. The resulting images of ClpAP-containing complexes were subjected to further 4-fold averaging (top–bottom and left–right). The number of averaging operations (N) was the number of particles times the order of symmetry. Resolutions (r) are according to the SSNR criterion (23). Contrast transfer function (CTF) effects were corrected by applying ctfmix (24) to the original single-exposure images, with 50% restoration of the low-frequency terms (25) and a Wiener term of 0.3. In control experiments, these parameters were varied (low frequencies, 0–50%, and Wiener term, 0.1–0.3). The averaged images used to calculate difference images were normalized by standardizing the background density and the peak density of ClpP. Each t test compared two sets of images, representing the complexes under comparison pixel by pixel (26).
Results
Binding of RepA to ClpA in the Presence of ATPγS.
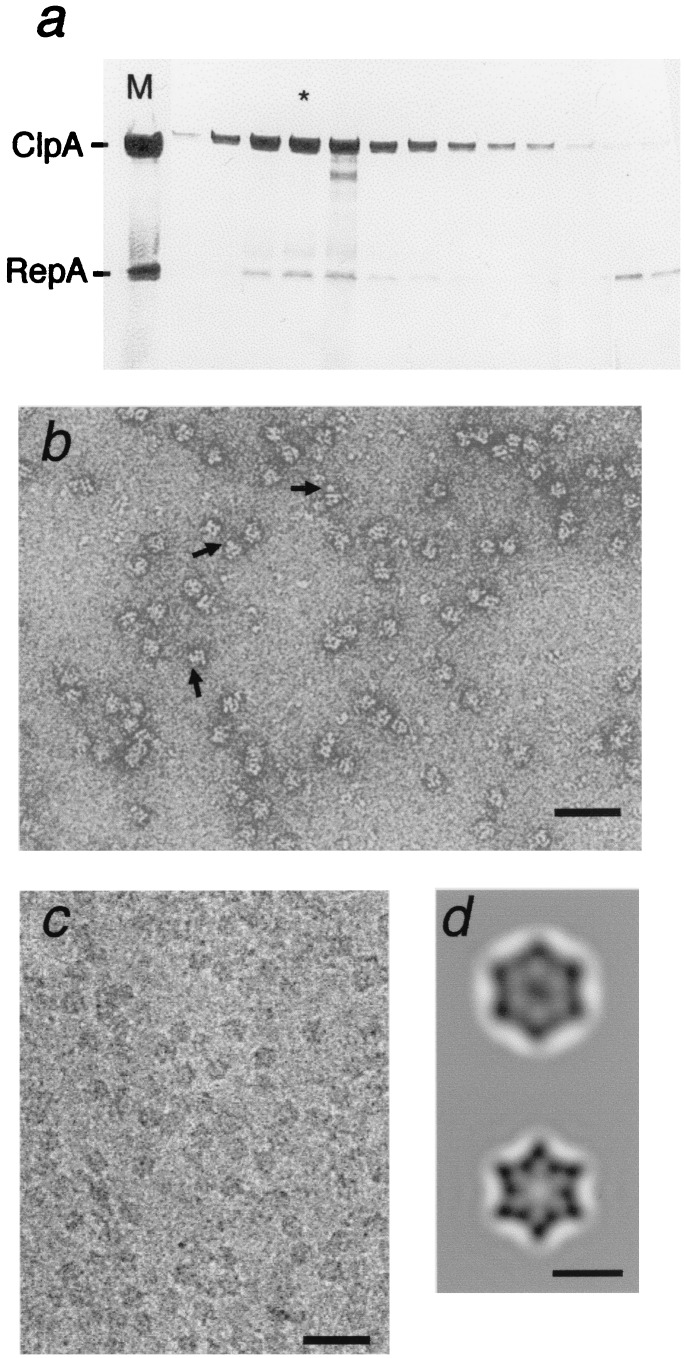
Dimeric RepA (2 × 35 kDa) was added in 2-fold molar excess over hexameric ClpA, and after 15 min the mixture was resolved by gel filtration. RepA coeluted with ClpA at ≈600 kDa, well resolved from unbound RepA (Fig. 1a). When fractions containing ClpA and RepA were examined by negative staining, many side views of ClpA, which distinctively present two parallel striations (16), showed additional density (arrows in Fig. 1b). The complexes were also examined by cryo-EM (Fig. 1c), and “top views” were averaged (Fig. 1d). Compared with ClpA alone, the ClpA–RepA complex has additional density in its central region. This density is not 6-fold symmetric but is elongated (≈45 × 30 Å) and more suggestive of 2-fold symmetry. We conclude that the RepA-binding sites on ClpA are sufficiently close to the axis that when one dimer binds, it blocks access to the other sites. This property accounts for the observed stoichiometry of one RepA dimer per ClpA hexamer (27).
Figure 1.
Isolation and EM of ClpA–RepA complexes. (a) ClpA (200 μg) was added to 90 μl of 50 mM Tris/HCl, pH 7.5/0.2 M KCl/1 mM DTT/25 mM MgCl2/1 mM ATPγS, and incubated for 5 min to allow hexamer formation. RepA (35 μg in 15 μl) was added, and after 15 min at room temperature, 100 μl of the mixture was injected on a Superdex200 column. The eluted fractions were analyzed by SDS/PAGE on a 12% gel (20), with Coomassie blue staining. (b) Negative staining of ClpA/RepA–ATPγS complexes: arrows, side views of ClpA with density attached. (Bar = 500 Å.) (c) Cryomicrograph of ClpA/RepA–ATPγS complexes. (d) Averaged top view (not CTF corrected) of ClpA with RepA bound (Top, n = 182; r = 34 Å) and a ClpA control (Bottom, n = 618; r = 23 Å; from ref. 4).
Binding of RepA to ClpAP in the Presence of ATPγS.
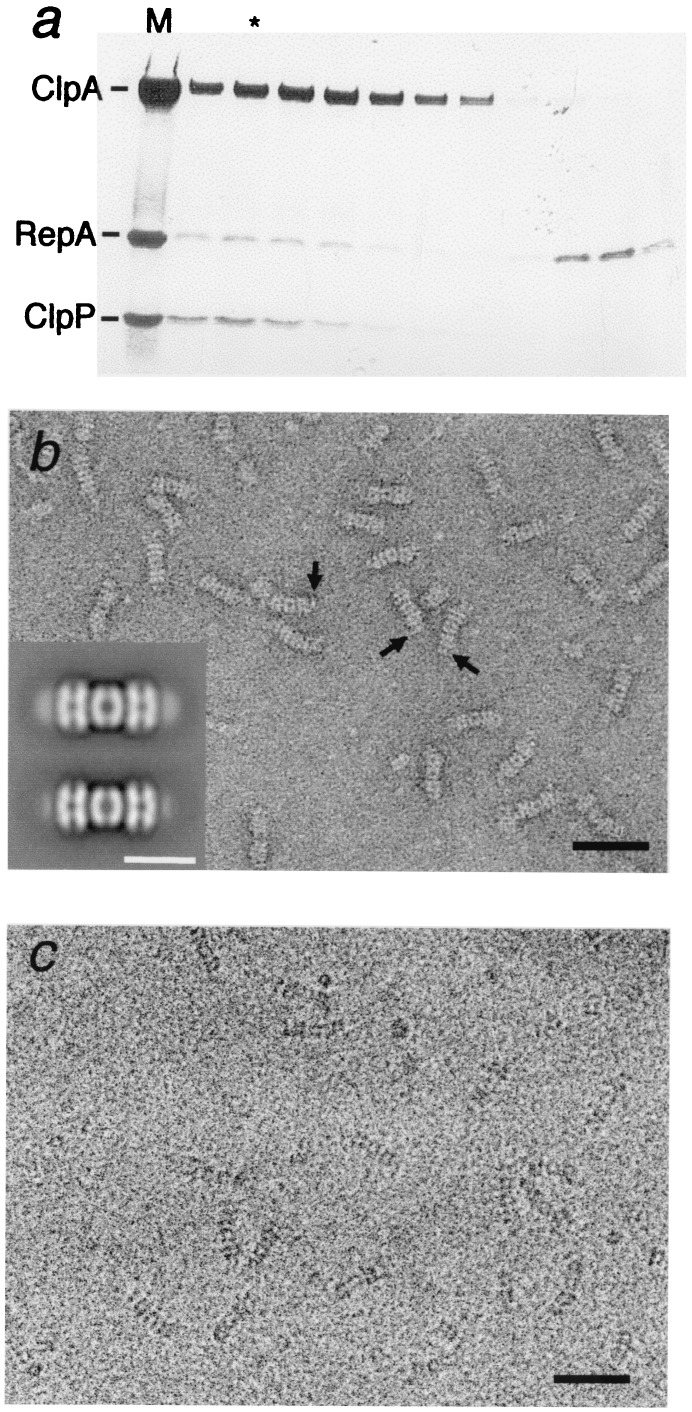
We next examined RepA bound to ClpAP by using proteolytically inactive complexes, ClpAPin (28), with ClpA in 2-fold molar excess to give a majority of complexes with ClpA at both ends of ClpPin. RepA dimers were added in 2-fold excess over ClpA hexamers, and, after 15 min, subjected to gel filtration. SDS/PAGE showed ternary complexes of ClpA, ClpPin, and RepA well separated from unbound RepA (Fig. 2a). The relative intensities of the gel bands indicated one to two dimers of RepA per ClpAPin complex.
Figure 2.
Isolation and microscopy of ClpAP–RepA complexes. (a) ClpA (200 μg) and ClpPin (70 μg) were mixed in the buffer described in Fig. 1 legend and incubated for 15 min to allow assembly of ClpAP complexes. RepA (35 μg) was added, and 15 min later, 0.1 ml was injected on to a Superdex200 column. The fractions were analyzed by SDS/PAGE (20) and fractions containing the ternary complexes were examined by EM. (b) Negatively stained micrograph and (c) cryomicrograph of column-purified ClpAP/RepA–ATPγS complexes. (Bar = 500 Å.) Arrows in b point to some side views showing additional density attached at the ends. These terminal densities, representing bound RepA, are not so evident from direct inspection of cryoelectron micrographs because of their lower contrast and signal-to-noise ratios. (Inset) Averaged side views of negatively stained ClpAP/RepA–ATPγS (Top) and ClpAP-ATPγS (Bottom). (Bar = 200 Å.) The terminal density is much stronger in the top image but nonzero in the control, which we attribute to binding at the same site of some, presumably damaged, ClpA molecules (see Results).
Negative staining showed additional density at one or both ends of most complexes (Fig. 2b), which was confirmed by image averaging (Fig. 2b Inset Top). RepA-free control complexes also showed some density at these sites (Fig. 2b Inset Bottom) but in much lower amounts. Because ClpA is subject to autodigestion by ClpAP (20), this density probably represents dissociated ClpA subunits bound as substrate by some complexes. This density was minimized in freshly prepared ClpAP and increased on storage (data not shown). On this basis, we prefer this explanation to the alternative that this density represents protruding portions of ClpA.
Image Averaging of ClpAP–RepA Complexes.
Many preparations of ClpAP–RepA complexes were examined by cryo-EM (e.g., Fig. 2c). Because of the low signal-to-noise ratio in cryomicrographs, we were not able to ascertain by visual inspection which complexes had bound RepA. Nevertheless, when images were averaged and compared with a RepA-free control, additional density was clearly associated with the distal surface of ClpA (see Fig. 3 b and c). To obtain control images with minimal amounts of terminal density, it was essential to work with freshly isolated complexes.
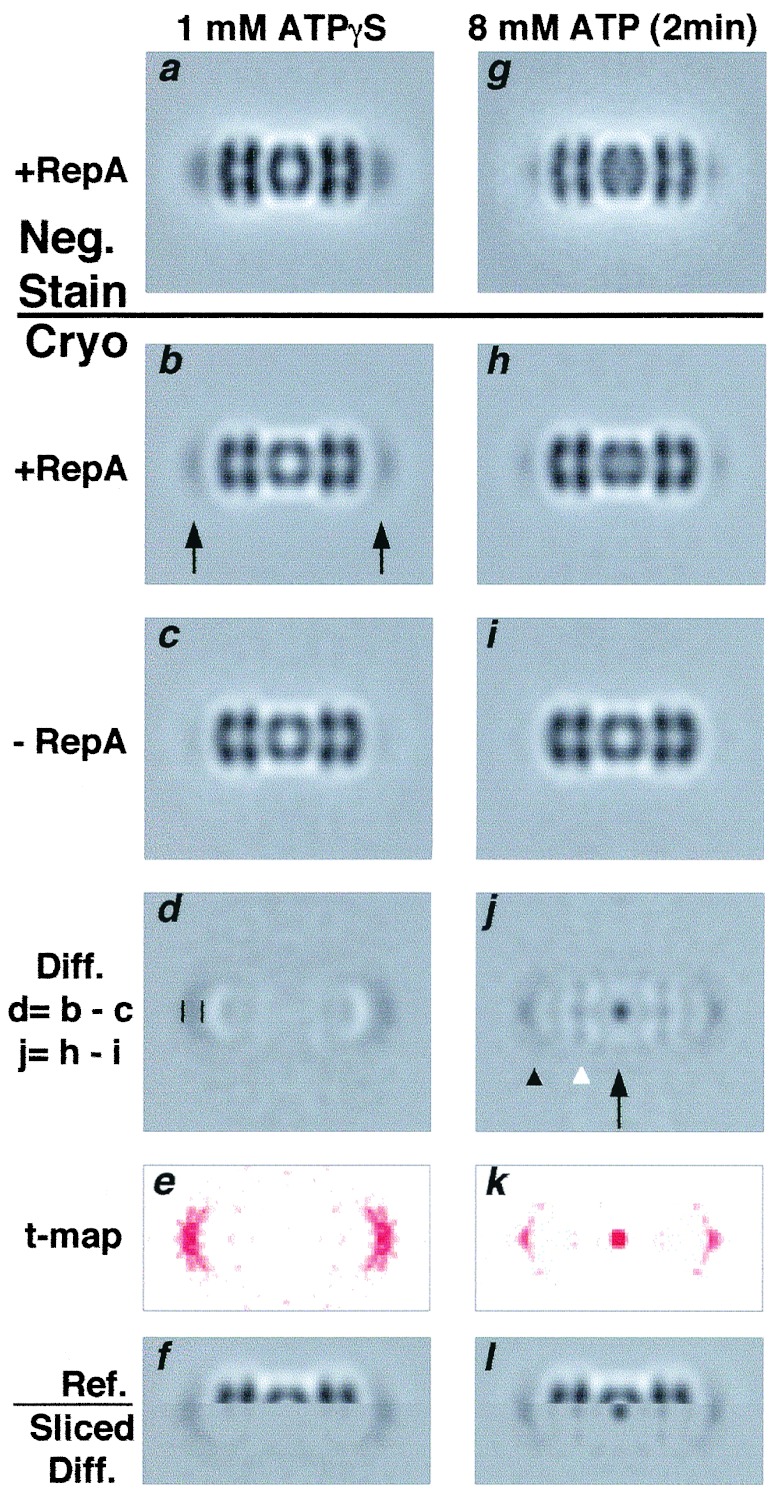
Figure 3.
Substrate translocation in ClpAP on ATP addition. ClpAP/RepA–ATPγS complexes (see Fig. 2 legend) were diluted 4- to 10-fold (in different experiments) into buffer containing 8 mM ATP, a sufficient excess to promote transfer of bound proteins to ClpP (19). Here, chemically inactivated ClpPin was used to avoid degradation. After 2 min, samples were prepared for EM. Control samples (no ATP) were analyzed in parallel. (a, g) Negatively stained ClpAP–RepA complexes before (a; n = 1,800, r = 29 Å) and after ATP addition (g; n = 1,000, r = 29 Å). (b, h) The same complexes analyzed by cryo-EM before (b; n = 6,000, r = 33 Å) and after ATP addition (h; n = 2000, r = 33 Å. (c, i) Cryo-EM of ClpAP without RepA before (c; n = 4,000, r = 33 Å) and after ATP addition (i; n = 1,600, r = 35 Å). (d, j) Difference images of complexes with and without RepA: the densities were amplified 2-fold for greater clarity. (d) Crescents of positive density at the ends of the complex. Vertical bars (Left) mark their axial extent, which we equate with one dimension of the RepA dimer; the transverse dimension is exaggerated by lateral smearing. (j) After ATP addition, strong focal density is visible at the center of the complex (arrow) and smaller densities at axial positions in ClpA and between ClpA and ClpP (arrowheads). (e, k) t-map (26) of d and j: differences coded in red (all positive) are significant at P = 0.99. (f, l) Split images mapping the density differences in ClpAP. (f) Upper half of b with lower half of d. (l) Upper half of h with the lower half of j. The half-difference images were filtered to show only positive densities.
Crescents of RepA-associated density are evident in difference images obtained by subtracting the control ClpAP from ClpAP–RepA complexes (e.g., Fig. 3d). The axial extent of the density is 35–40 Å, but its transverse dimension is greater (≈100 Å), although the density fades toward the edges. To delineate its position in the complex, we assembled a split image with the upper half of ClpAP/RepA juxtaposed on the lower half of the difference image (Fig. 3f). This alignment confirmed that the additional density is associated with the distal surface of ClpA. Because the RepA-binding site is positioned somewhat off-axis, RepA molecules appear in different azimuthal positions on their respective complexes, and this part of the image will be laterally smeared. Off-axis binding of RepA was also seen in some negatively stained images (Fig. 2b).
Translocation of RepA into ClpPin.
ClpAPin/RepA/ATPγS complexes were formed, then diluted into 4 mM ATP and prepared for EM. Averaged side views showed changes from the ATPγS state by both negative staining (see Fig. 3 a and g) and cryo-EM (see Fig. 3 b and h). The terminal densities were much diminished, implying that most RepA had left its initial binding site, and additional density was concentrated at the center of ClpPin, implying that protein had entered the internal cavity. These results are consistent with biochemical data showing that on ATP addition to ClpAP–RepA complexes, a significant fraction of the RepA was degraded without release (19). On comparing cryo-EM images of the ATP and ATPγS state of ClpAP without RepA, little difference was observed (Fig. 3c and i).
Importantly, no increase in density was seen at the center of ClpP. We infer that, on ATP addition, RepA was translocated into the digestion chamber of ClpPin, where it remained intact (refs. 13 and 29, and data not shown). The appearance of RepA-associated density inside ClpP was recorded in at least 10 experiments, including ones in which RepA was added to ClpAPin directly assembled in the presence of ATP (data not shown). In previous studies, the retention of RepA by ClpPin after translocation was shown by separating ClpA from ClpPin, and demonstrating that RepA could be coprecipitated with anti-ClpP antibodies (19). The present results show that the bound RepA entered the central cavity in the assembled ClpAPin complex.
To estimate the efficiency of translocation, we calculated the masses initially bound and eventually observed in the digestion chamber by integrating the positive density in the corresponding regions of the difference images (Fig. 3 d and j). Because cryomicrographs are subject to phase contrast effects that compromise the proportionality between image density and projected mass, the images were corrected for CTF effects (see Methods). The integrals of substrate-attributable density were calibrated against the corresponding integral of additional density at the center of ClpPSC compared with ClpP (Fig. 4c). ClpPSC retains an N-terminal propeptide, and its internal chamber contains an additional mass of 22.5 kDa from the 14 copies (16). Thus estimated, the RepA-attributable mass at the surface averaged 78 kDa per ClpA hexamer, suggesting that the efficiency of binding RepA dimers was close to 100%.
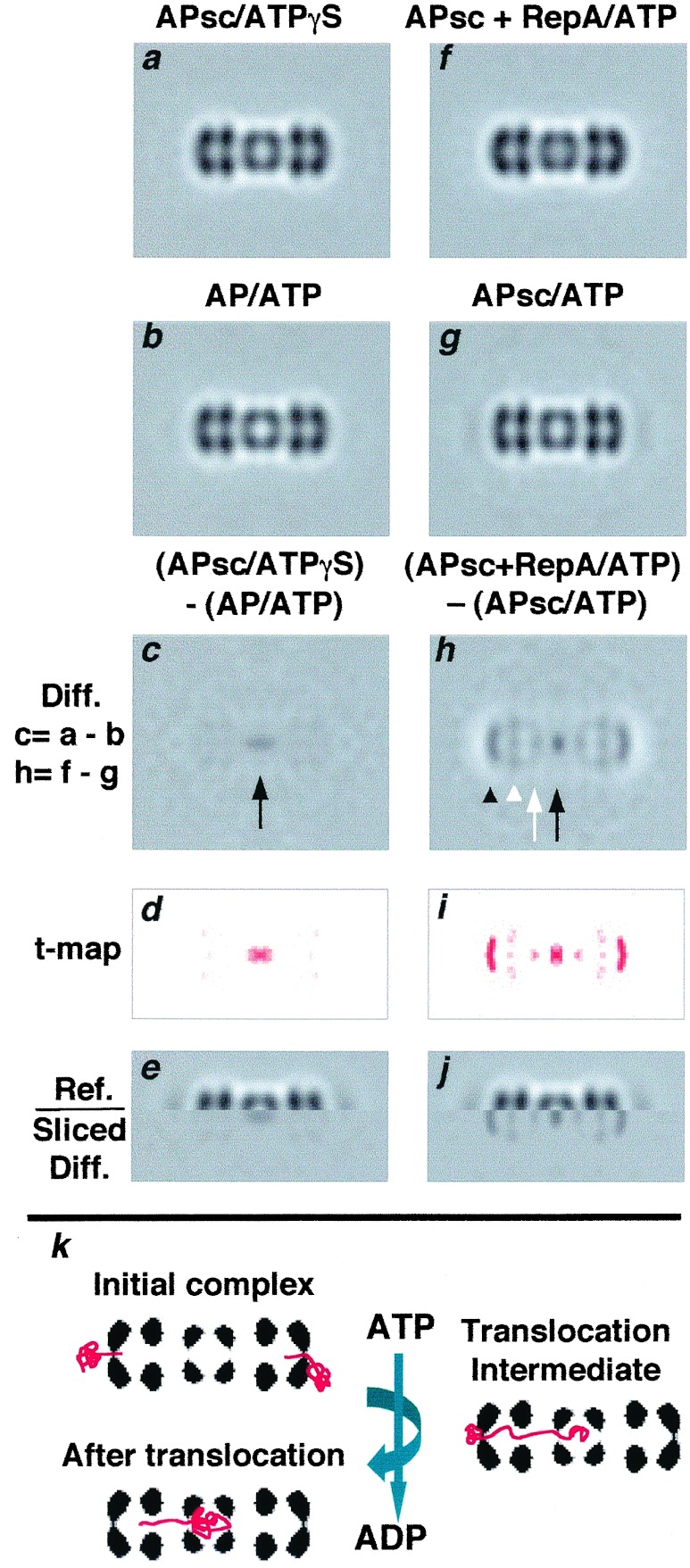
Figure 4.
Intermediate stages of substrate translocation in ClpAP. (a–e) The density difference between ClpP and ClpPSC was measured by cryo-EM. The ClpAPSC complexes were assembled in ATPγS. To obtain ClpP with empty chambers, the ATP state of the wild-type complex was used, because it should translocate and degrade contaminants, eliminating potential sources of extraneous density. (a) ClpAPSC–ATPγS (n = 1,600; r = 35 Å); (b) ClpAP–ATP; (c) difference image of a–b. The only significant feature is positive density coprojecting with the digestion chamber. (d) a t-map of c (see Fig. 3 legend). (e) Split image, juxtaposing the sliced difference image with the complex. (f–j) Changes in ClpAPSC–RepA complexes on ATP addition. (f) RepA was bound to ClpAPSC in the presence of ATPγS and after 20 min, the sample was diluted 10-fold into 4 mM ATP and analyzed by cryo-EM (n = 2,400; r = 34 Å). (g) As a control, ClpAPSC without RepA was treated with ATP and imaged (n = 1,000, r = 35 Å). (h) Difference image between f and g; arrow, additional density at the center of ClpPSC; arrowheads, densities at intermediate axial sites. (i) a t-map of h. (j) Split image marking the positions of these sites on ClpAP. (k) Diagram illustrating initial binding of a dimer at both ends, the translocation end point with one RepA subunit inside ClpAPin, and one possible intermediate.
From Fig. 3j, we calculated that, on average, 28 kDa of protein was translocated into the ClpPin chamber, and 22 kDa remained at each end of the complex. In this experiment, ≈57% of the material initially bound was apparently released on ATP addition, in line with previous observations (19, 30). The margin of error of these calculations was estimated to be <20% (see Discussion).
Translocation of RepA into ClpPSC.
To obtain images of complexes with partially translocated substrates, we used ClpPSC, whose digestion chamber is already partly filled (see above). We reasoned that complexes containing this mutant protein would allow only a portion of the substrate into the chamber, with the remainder blocked at earlier stages of translocation. In Fig. 4 a and b, side views of ClpAPSC/ATPγS and ClpAP/ATP are compared. The difference image (Fig. 4c) shows density in the ClpPSC chamber, which we attribute to the propeptides. We subtracted ClpAP/ATP rather than ClpAP/ATPγS in this experiment, because otherwise extraneous protein scavenged by ClpA might generate negative density on the distal surface (see above); with ATP, any such protein would be translocated and degraded. Moreover, as noted above, we see no significant differences between the ATP and ATPγS states of ClpAP at this resolution.
Although ClpPSC has a lower capacity for incoming substrate, RepA can nevertheless be coimmunoprecipitated with ClpPSC after incubation of RepA with ClpAPSC and ATP, implying that they are stably associated (19). Moreover, ClpX can transfer GFP-SsrA to ClpPSC (S. K. Singh and M.R.M., unpublished work). To investigate translocation into ClpPSC, we diluted ClpAPSC/RepA/ATPγS complexes into ATP and examined the resulting complexes (Fig. 4f). Control images of ClpAPSC/ATP were also obtained (Fig. 4g). The difference map (Fig. 4h) showed additional density (≈19 kDa; black arrow) in the center, indicating some protein was translocated to the chamber. Density also appeared at other axial sites, whose positions are shown in the juxtaposed difference image (Fig. 4j). The site with most density (≈24 kDa; Fig. 4h, black arrowhead) is more closely apposed to the distal surface of ClpA than the initially bound substrate (see Fig. 3 j and l) and may represent the position of RepA after partial unfolding and insertion of an initial segment into ClpA. A second site with ≈15 kDa lies inside ClpA on the inner face of the proximal tier of ATPase subunits (Fig. 4h, white arrowhead). A smaller patch of density (≈6 kDa; Fig. 4h, white arrow) was seen at the “vestibule” between ClpPSC and ClpA. These data suggest that translocation proceeds in several stages along an axial path.
Because the axial patches of density seen in these difference images are relatively small, we assessed their significance by calculating t test maps (26) between the respective data sets (Figs. 3 e and k and 4 d and i). These tests indicated that the features discussed above are significant at the 95% confidence level (data not shown) and all but the 6-kDa density at the 99% level. This conclusion is further supported by the fact that the same set of intermediate sites are visualized in Figs. 3j and 4h, albeit with different occupancies.
Discussion
The images presented here, together with a recent analysis of substrate binding and translocation by ClpXP (29), directly support (and further specify) current concepts of how ATP-dependent proteases effect processive protein degradation (1–3, 31). They point to the following chain of events. Initially, RepA binds to ClpAP at near-axial sites on the distal surface of ClpA. Then RepA is unfolded and fed into the digestion chamber of ClpP. During translocation, RepA migrates ≈150 Å from its initial binding site. Our analysis suggests that translocation is a stepwise process, and portions of substrate accumulate perhaps transiently at two intermediate sites. Given the parallels in structural properties between the Clp proteases [ClpAP, ClpXP, and ClpYQ (HslUV)] and the 26 S proteasome (6–8, 15–17, 32, 33), it is likely that their reaction pathways are also similar.
How Much Protein Can be Accommodated Inside ClpP?
From the dimensions of the ClpP digestion chamber (8), its maximum capacity can be calculated to be ≈51 kDa. However, the practical limit is likely to be lower. Our observation of the propeptides inside ClpPSC (Fig. 4c) shows that at least 22 kDa can be accommodated. The amount of protein internalized by ClpPin (Fig. 3j) was estimated at ≈28 kDa, and the total complement in ClpPSC (propeptide plus translocated protein) at ≈41 kDa (Fig. 4h). These observations imply that about one RepA subunit can be packed into ClpPin, and no more than half a subunit into ClpPSC. However, the amount of substrate protein that accumulates in the digestion chambers of actively degrading complexes is not known.
The protein masses seen in our difference images are not large (6–78 kDa), and the resolution is currently limited to ≈30 Å (largely by variation in the orientations around their common axis of the subcomplexes in different side views). Although the noise level of individual micrographs is such that bound RepA molecules cannot be discerned with confidence (Fig. 2c), they may nevertheless be seen clearly after averaging; for instance, 400-fold averaging boosts the signal-to-noise ratio by a factor of 20. We confirmed their visibility by generating images of proteins of 10–70 kDa adjacent to a complex of comparable size and shape to ClpAP by downloading the corresponding crystal coordinates of ClpYQ (34) and generating side-projections limited to 30 Å resolution from them (data not shown; see ref. 35 and Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org). With or without CTF effects, the 70-kDa protein was strongly visible both directly and in difference images, and the 10-kDa protein was less conspicuous but definitely also visible. The t test data (Figs. 3 e and k, 4 d and i) confirm that masses of this order are visualized in our difference images.
As noted above, quantitation of projected density from such images is complicated by CTF effects: these are suppressed but not entirely eliminated by our corrections. Accordingly, we tested the robustness of our calculations to procedural variations (in normalization and in CTF parameters), and encountered variations of 10–20% in the calculated masses, which we take as the experimental uncertainty. We also synthesized images from blocks whose relative sizes, masses, and positions resembled those of the molecules under study. Phase contrast effects were applied, and the mass associated with each component was recalculated. The resulting “substrate” masses were altered by no more than 10–20%, further supporting the estimated margin of error.
Translocation Proceeds in Several Stages.
Our data indicate that RepA is translocated in stepwise fashion. Its initial binding site is on the distal face of ClpA. Probably, the N-terminal domains of ClpA reside on this face (ref. 16; S. K. Singh and M.R.M., unpublished observations). Evidence for substrate binding to the C-terminal region of ClpA (33, 36) may relate to downstream sites reached after the initial binding event, or there may be more than one substrate-attachment site on ClpA.
In the next step, substrate becomes more closely apposed to the distal surface of ClpA. Presumably this step entails perturbation of RepA, possibly accompanied by insertion of part of RepA into ClpA (akin to “threading the needle”) and/or local structural changes in ClpA. A three-dimensional reconstruction of ClpA–ATPγS at 28-Å resolution found no axial channel through its distal tier (4): however, there may be a small aperture that was not seen because of limited resolution or density contributed by extraneously bound protein. Alternatively, ATP hydrolysis may cause a subtle change in ClpA that creates an opening. In ClpY, this location is characterized by an expandable gateway leading to a concave chamber (34, 37).
Once inside the outer tier, a portion of RepA associates with a site on the inner surface of the distal ATPase domains, the intermediate site seen in Fig. 4 h and j. From that position, the protein is further translocated through ClpA into ClpP. There appears to be a second staging area on the inner surface of the proximal ATPase domains, but our data suggest that only a small portion of RepA accumulates at this site, or that the site had low occupancy in these experiments.
Finally, substrate enters the digestion chamber. Significantly, entry of RepA into the degradation chamber of ClpPin occurred in the absence of peptide bond cleavage. Thus, whereas vectorial transfer of substrates may be aided by their binding to the walls of the degradation chamber, bond breakage or formation of acyl–enzyme intermediates is not required.
Reptation of RepA.
The observation that little more than a single RepA subunit can be accommodated inside ClpPin is consistent with the observed release of some of this initially dimeric protein from ClpAPin–ATPγS on adding ATP (Fig. 3j; refs. 19 and 29). The inference that one subunit at a time is translocated per ClpA hexamer agrees with the requirement that the polypeptide chain must be threaded through the narrowest constrictions on the axial pathway. Recent studies with fusion protein substrates indicate that degradation, and by implication translocation, proceed vectorially from either the N or C terminus of substrates (A. Matouschek, personal communication; S. H. Lee and M.R.M., unpublished work). We interpret the patches of density at axial sites other than the digestion chamber (Figs. 3j and 4h) as portions of subunits at earlier stages of translocation, linked by the extended polypeptide chain to the portion that has entered the digestion chamber. In support of this hypothesis, occupancy of the intermediate sites was higher during translocation with ClpPSC, which cannot accept a complete subunit so that substrates are stranded in partially translocated states. Thus it appears that translocation is a reptation-like process in which the substrate is sometimes extended (to negotiate the constrictions) and sometimes compact, as at the putative staging posts. Some possibilities are shown in Fig. 4k.
Structural Transitions on ATP Hydrolysis.
At 30-Å resolution, we see little change in ClpAP between its ATP and ATPγS states. Thus, any structural changes induced in ClpP by ClpA on changing its nucleotide state are small in scale. It also follows that any changes in ClpA resulting from ATP binding or hydrolysis should be subtle, although our images, which represent cylindrical averages, do not rule out rotational movements between the two tiers of ATPase domains. In any case, they appear smaller than transitions that have been observed between different nucleotide states of chaperones (38–41). Alternatively, the altered states of ClpA are short-lived and thus sparsely represented in the population. In this context, it may be that the internal chamber of ClpA, unlike that of GroEL-like chaperonins, does not represent an unfolding compartment. However, unfolding of substrates is somehow effected during ClpA-mediated translocation. As a corollary, in the absence of ClpP, unfolded proteins should pass through ClpA, to refold on emergence. Efforts to address this question and analysis of other nucleotide states of the ClpAP complex are under way.
Supplementary Material
Acknowledgments
We thank Drs. N. Cheng and D. Winkler for help with EM and Drs. D. Belnap, J. Conway, and B. Trus for help with computing.
Abbreviations
- ATPγS
adenosine 5′-[γ-thio]triphosphate
- EM
electron microscopy
- CTF
contrast transfer function
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wickner S, Maurizi M R, Gottesman S. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt M, Lupas A N, Finley D. Curr Opin Chem Biol. 1999;3:584–591. doi: 10.1016/s1367-5931(99)00013-7. [DOI] [PubMed] [Google Scholar]
- 3.Lupas A, Flanagan J M, Tamura T, Baumeister W. Trends Biochem Sci. 1997;22:399–404. doi: 10.1016/s0968-0004(97)01117-1. [DOI] [PubMed] [Google Scholar]
- 4.Beuron F, Maurizi M R, Belnap D M, Kocsis E, Booy F P, Kessel M, Steven A C. J Struct Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- 5.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 6.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 7.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik H D, Huber R. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 9.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levchenko I, Luo L, Baker T A. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 11.Weber-Ban E U, Reid B G, Miranker A D, Horwich A L. Nature (London) 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 12.Singh S K, Grimaud R, Hoskins J R, Wickner S, Maurizi M R. Proc Natl Acad Sci USA. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoskins J R, Singh S K, Maurizi M R, Wickner S. Proc Natl Acad Sci USA. 2000;97:8892–8897. doi: 10.1073/pnas.97.16.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun B C, Glickman M, Kraft R, Dahlmann B, Kloetzel P M, Finley D, Schmidt M. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- 15.Grimaud R, Kessel M, Beuron F, Steven A C, Maurizi M R. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 16.Kessel M, Maurizi M R, Kim B, Kocsis E, Trus B L, Singh S K, Steven A C. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 17.Glickman M H, Rubin D M, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V A, Finley D. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 18.Thompson M W, Maurizi M R. J Biol Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- 19.Hoskins J R, Pak M, Maurizi M R, Wickner S. Proc Natl Acad Sci USA. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurizi M R, Thompson M W, Singh S K, Kim S H. Methods Enzymol. 1994;244:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- 21.Wickner S, Hoskins J, McKenney K. Proc Natl Acad Sci USA. 1991;88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trus B L, Kocsis E, Conway J F, Steven A C. J Struct Biol. 1996;116:61–67. doi: 10.1006/jsbi.1996.0011. [DOI] [PubMed] [Google Scholar]
- 23.Unser M, Trus B L, Steven A C. Ultramicroscopy. 1987;23:39–51. doi: 10.1016/0304-3991(87)90225-7. [DOI] [PubMed] [Google Scholar]
- 24.Conway J F, Steven A C. J Struct Biol. 1999;128:106–118. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- 25.Belnap D M, Filman D J, Trus B L, Cheng N, Booy F P, Conway J F, Curry S, Hiremath C N, Tsang S K, Steven A C, Hogle J M. J Virol. 2000;74:1342–1354. doi: 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milligan R A, Flicker P F. J Cell Biol. 1987;105:29–39. doi: 10.1083/jcb.105.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pak M, Wickner S. Proc Natl Acad Sci USA. 1997;94:4901–4906. doi: 10.1073/pnas.94.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh S K, Guo F, Maurizi M R. Biochemistry. 1999;38:14906–14915. doi: 10.1021/bi991615f. [DOI] [PubMed] [Google Scholar]
- 29.Ortega J, Singh S K, Maurizi M R, Steven A C. Mol Cell. 2000;6:1515–1521. doi: 10.1016/s1097-2765(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 30.Pak M, Hoskins J R, Singh S K, Maurizi M R, Wickner S. J Biol Chem. 1999;274:19316–19222. doi: 10.1074/jbc.274.27.19316. [DOI] [PubMed] [Google Scholar]
- 31.De Mot R, Nagy I, Walz J, Baumeister W. Trends Microbiol. 1999;7:88–92. doi: 10.1016/s0966-842x(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 32.Gabant P, Newnham P, Taylor D, Couturier M. J Bacteriol. 1993;175:7697–7701. doi: 10.1128/jb.175.23.7697-7701.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith C K, Baker T A, Sauer R T. Proc Natl Acad Sci USA. 1999;96:6678–6682. doi: 10.1073/pnas.96.12.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sousa M C, Trame C B, Tsuruta H, Wilbanks S M, Reddy V S, McKay D. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 35.Ishikawa T, Belnap D, Maurizi M R, Steven A C. Nature (London) 2000;408:667–668. doi: 10.1038/35047165. [DOI] [PubMed] [Google Scholar]
- 36.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 37.Bochtler M, Hartmann C, Song H K, Bourenkov G P, Bartunik H D, Huber R. Nature (London) 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 38.Roseman A M, Chen S, White H, Braig K, Saibil H R. Cell. 1996;18:241–251. doi: 10.1016/s0092-8674(00)81342-2. [DOI] [PubMed] [Google Scholar]
- 39.Xu Z, Horwich A L, Sigler P B. Nature (London) 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 40.Llorca O, Smyth M G, Carrascosa J L, Willison K R, Radermacher M, Steinbacher S, Valpuesta J M. Nat Struct Biol. 1999;6:639–642. doi: 10.1038/10689. [DOI] [PubMed] [Google Scholar]
- 41.Hanson P I, Roth R, Morisaki H, Jahn R, Heuser J E. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.