Abstract
Within organisms, groups of traits with different functions are frequently modular, such that variation among modules is independent and variation within modules is tightly integrated, or correlated. Here, we investigated patterns of trait integration and modularity in Brassica rapa in response to three simulated seasonal temperature/photoperiod conditions. The goals of this research were to use trait correlations to understand patterns of trait integration and modularity within and among floral, vegetative and phenological traits of B. rapa in each of three treatments, to examine the QTL architecture underlying patterns of trait integration and modularity, and to quantify how variation in temperature and photoperiod affects the correlation structure and QTL architecture of traits. All floral organs of B. rapa were strongly correlated, and contrary to expectations, floral and vegetative traits were also correlated. Extensive QTL co-localization suggests that covariation of these traits is likely due to pleiotropy, although physically linked loci that independently affect individual traits cannot be ruled out. Across treatments, the structure of genotypic and QTL correlations was generally conserved. Any observed variation in genetic architecture arose from genotype × environment interactions (GEIs) and attendant QTL × E in response to temperature but not photoperiod.
Keywords: Brassica rapa, flowers, modularity, photoperiod, temperature, trait integration
Introduction
Throughout the natural world, biological systems tend to be modular; organisms are composed of recognizable units that differ in development, structure and function (Wagner et al., 2007; Klingenberg, 2008). Modularity refers to the degree of connectedness; a module is connected internally but independent from other modules (Wagner, 1996). Morphologically, modules are relatively independent groups of phenotypic traits, whereas traits within modules act in an integrated, or coordinated, manner, with coordination arising from interactions among the processes that produce the traits (Cheverud, 1996; Klingenberg, 2008). Evolutionarily, unconnected modules may respond independently to selection and follow different adaptive paths (Wagner, 1996), whereas integration of traits within modules often represents a trade-off; integration reduces the occurrence of maladaptive trait combinations, but may also limit the response of individual traits to selection (Wagner, 1996; Klingenberg, 2008).
The floral organs of plants are commonly viewed as integrated traits. Flowers are composed of multiple organs that frequently act in a coordinated manner to carry out the functions of pollinator attraction, pollen donation and pollen receipt. Given that the singular function of flowers is reproduction, the size and shape of floral organs is frequently coordinated and populations often maintain strong genotypic correlations among floral traits (Conner and Via, 1993; Oneil and Schmitt, 1993; Carr and Fenster, 1994; Conner and Sterling, 1995; Juenger et al., 2000; van Kleunen and Ritland, 2004; Ashman and Majetic, 2006; Brock and Weinig, 2007). One theory for the evolution of floral trait correlations is via natural selection by pollinators on floral phenotypes (Berg, 1960). This theory predicts that plants with specialist pollinators will have highly correlated floral organ sizes, with correlations arising by selection for floral dimensions that fit the dimensions of the pollinator, whereas plants with generalist pollinators are predicted to have less strongly correlated floral traits. However, empirical studies have shown that the relationship between pollinator specificity and the level of covariation among floral traits is not as consistent as predicted by Berg (Conner and Sterling, 1995; Armbruster et al., 1999; Brock and Weinig, 2007), suggesting that other factors may also affect floral trait correlations.
Genetically, several mechanisms may underlie floral trait correlations. If pollinator efficiency has contributed to increased floral trait correlations, selection may have favored specific combinations of alleles at unlinked loci, or ‘adaptive character complexes' (Armbruster and Schwaegerle, 1996), resulting in linkage disequilibrium (LD) among physically unlinked genes. Alternatively, covariation of traits may arise from a common developmental or genetic architecture. For example, correlations among traits may arise because a pleiotropic gene affects variation in several traits (Lande, 1980), as in the ABC model of plant development, where B-class genes affect the development of both petals and stamens (Weigel and Meyerowitz, 1994). From the ABC model for organ identity, one might also predict that correlations for size are stronger between organs residing in adjacent floral whorls than organs in non-adjacent whorls. Empirical research suggests that the genetic mechanisms underlying floral trait correlations may differ among species. All floral organs were equally highly correlated in Mimulus guttatus and Mimulus micranthus (Carr and Fenster, 1994), Phlox drummondii (Waitt and Levin, 1993), Arabidopsis thaliana (Juenger et al., 2000, 2005; Brock and Weinig, 2007) and Brassica nigra (Conner and Sterling, 1995), and QTL mapping and analysis of segregating progenies with extensive generations of recombination suggested that pleiotropic genes (or physically linked loci) were primarily responsible for floral trait correlations in Arabidopsis (Juenger et al., 2000; Conner, 2002). However, in Raphanus raphanistrum and Brassica napus, filament and corolla traits were more strongly correlated than other floral traits, potentially as a consequence of pollinator-mediated selection and LD of physically unlinked genes (Conner and Sterling, 1995). Given these results, patterns of floral trait correlations and their underlying genetic mechanisms may be species-specific.
As floral and leaf traits have a common developmental origin (Ratcliffe et al., 1999), they may also co-vary; however, floral, vegetative and other types of traits have different functions, may have undergone selection for independent development, and may therefore comprise separate modules. Berg's theory of pollinator selection proposes an explanation for the independence of floral and vegetative traits (Berg, 1960); because vegetative traits may respond plastically to environmental variation, plants that buffer the development of floral organs from that of vegetative organs may have more consistent floral organ sizes that better fit the dimensions of specialist pollinators, leading to increased rates of pollination, and ultimately, more independence of floral and vegetative traits (Berg, 1960). Relative to plants with specialist pollinators, this theory predicts that plants with generalist pollinators will experience less selection for consistent floral organ sizes and will have less independent floral and vegetative traits (Berg, 1960). Empirical studies have shown that the degree of independence of floral and vegetative traits is not well predicted by pollination syndrome; modularity of floral and vegetative organs was equally pronounced in species with unspecialized vs specialized pollination syndromes (Armbruster et al., 1999). More generally, several empirical studies have found that floral and vegetative traits are modular, such that traits within floral or vegetative modules were correlated, whereas floral and vegetative traits were generally uncorrelated (Conner and Via, 1993; Conner and Sterling, 1996; Juenger et al., 2005; Ashman and Majetic, 2006; but see Brock and Weinig, 2007), and QTL mapping in a population of recombinant inbred lines (RILs) of Arabidopsis revealed that floral and vegetative traits had largely independent genetic architectures (Juenger et al., 2005). In sum, integration/modularity may depend on genetic, ecological and evolutionary factors that are specific to each species.
It is also noteworthy that the expression of many developmental pathways (such as flowering time, shade avoidance) depends on environmental cues, such that the expression of integration/modularity may vary by environment (Waitt and Levin, 1993; Pigliucci and Marlow, 2001; Conner et al., 2003; Brock and Weinig, 2007). For example, in Phlox drummondii, the correlation structure among traits differed with different nutrient availability (Waitt and Levin, 1993), in Raphanus raphinistrum, the correlation structure among floral traits differed between field and greenhouse environments (Conner et al., 2003), and in A. thaliana, changes in vernalization, season length and light composition affected patterns of trait correlation structure (Pigliucci and Marlow, 2001; Brock and Weinig, 2007). Also of relevance to the current study, photoperiod and temperature may have indirect effects on floral meristem development and floral organ identity genes in A. thaliana (Okamuro et al., 1993, 1996), and such organ-specific effects may be one mechanism by which environment could influence integration/modularity. More generally, these results suggest that patterns of trait integration and modularity may be sensitive to environmental variation, but few studies have used more than a single environment.
In this study, we used a population of 150 RILs to understand the quantitative-genetic and QTL architecture of trait integration and modularity in Brassica rapa, and to asses how this genetic architecture changes across three simulated seasonal settings that the species naturally encounters (see Materials and methods). We were specifically interested in quantifying: (1) patterns of trait integration within flowers, and if floral organs in adjacent whorls show stronger integration than those in non-adjacent whorls as might be expected if genes affecting floral organ size act pleiotropically in adjacent whorls similar to the effects of ABC class genes; (2) the degree of modularity between floral, vegetative and phenological traits; (3) the effect of seasonally variable abiotic factors on the expression of integration/modularity and (4) the genetic mechanisms underlying trait integration/modularity and the response to environmental variation.
Materials and methods
Study system
We investigated the genetic basis of trait integration and modularity in Brassica rapa and the effect of environmental variation on these patterns. B. rapa is an insect-pollinated, oilseed and vegetable crop (Prakash and Hinata, 1980). Like other members of the Brassicaceae, flowers of B. rapa have distinct (unfused) floral organs, including four sepals, four petals, six stamens (four long+two short), and a compound pistil of two carpels. The distinct floral organs make this species well suited to examining the structure of bivariate correlations between floral organs and, specifically, the relative strength of correlations between organs in different whorls.
The original native range of B. rapa is thought to have extended across Eurasia from the Western Mediterranean to Central Asia, and wild individuals of the species still grow throughout this region (Gomez Campo, 1999). Many crops were domesticated from this wild progenitor independently in several locations across Eurasia (Zhao et al., 2005), including varieties cultivated for oil (B. rapa subsp. oleifera), as root vegetables (B. rapa subsp. rapa, or turnip), and as leafy vegetables (B. rapa subsp. chinensis, or pak choi, and B. rapa subsp. pekinensis, or Chinese cabbage). Given that B. rapa was domesticated in many regions and is cultivated in numerous locations worldwide, this species is capable of surviving across a wide range of environmental conditions. Further sources of environmental variation arise from agricultural practices; most leafy vegetables and oilseed varieties are planted as spring or fall annuals, whereas root vegetables such as turnips are planted as fall or winter annuals and biennials. As this species encounters such a wide range of abiotic variation, it is well suited to understanding how integration and modularity of floral, vegetative and phenological traits respond to the environment. Across locations and seasons where B. rapa is cultivated, two main environmental factors that vary are temperature and photoperiod. We studied responses to three different combinations of temperature and photoperiod characteristic of habitats that B. rapa occupies, including: (1) long days, warm temperatures, which are experienced by spring or summer annuals (such as pak choi planted in the spring), (2) long days, cool temperatures, which are experienced by overwintering biennial crops that flower in the spring (such as turnips planted in fall in Europe) and (3) short days, cool temperatures, which are experienced by fall or winter annuals (such as Chinese cabbage or pak choi planted in the fall). We did not examine short photoperiods and warm temperatures, as this combination of conditions is unlikely to exist in any of the locations where B. rapa grows.
The genotypes used in this study were RILs derived from a cross between two highly inbred genotypes of B. rapa. The parents of the RILs were R500, a cultivated seed oil genotype, and IMB211, a rapid cycling genotype that may approximate a weedy member of the species; this cross may therefore be representative of gene flow occurring between cultivated genotypes and their wild neighbors, and can be used to investigate the genetic regions that are responsible for floral, vegetative and phenological trait variation between these two types of populations. Furthermore, the intensity of selection for integration/modularity may have varied in these parents (and between cultivars and wild genotypes in general), and this population of RILs is therefore of interest to investigate integration/modularity in B. rapa. To form the RILs, the two parents were crossed, the F1 generation was selfed, and the progeny were advanced by single seed descent to the S6 generation to form 150 RILs (Iniguez-Luy et al., 2009).
Experimental design and growth conditions
Six growth-chamber compartments (PGC-9/2 with Percival Advanced Intellus Environmental Controller, Percival Scientific, Perry, IN, USA) were used to simulate seasonal settings. As previous research using the same model of growth chamber and similar numbers of B. rapa individuals revealed that plants caused photosynthesis-induced drawdowns of CO2 that affected floral morphology (Edwards et al., 2009), ventilation was increased in each growth-chamber compartment by adding two additional exhaust vents and an intake fan. This modification doubled the number of air exchanges to maintain CO2 at or above 375 p.p.m. For all growth chambers, light intensity during light cycles was maintained at a photosynthetic photon flux density of around 500 μmol m−2 s−1, and the vapor pressure deficit was maintained between 0.45 and 1.65 (45–70% relative humidity).
One replicate of each of the 150 RILs and the two parental genotypes were grown in each of the six growth-chamber compartments. The growth chambers were set to: (1) WL represents warm temperature (24 °C), long photoperiod (14 h/10 h light/dark cycles), (2) CL represents cool temperature (12 °C), long photoperiod (14 h/10 h light/dark cycles) and (3) CS represents cool temperature (12 °C), short photoperiod 10 h/14 h light/dark cycles). Each treatment occupied two of the six growth chambers at one time, and this planting design was temporally replicated three times, totaling six replicates of each genotype in each treatment. The treatments were rotated among growth chambers so that each treatment occupied each growth-chamber compartment once.
For each replicate of each genotype, we planted three seeds that were cold/dark stratified for 3 days at 4 °C and then placed immediately into growth chambers set to the three treatments, with genotypes fully randomized within each chamber. After germination, seedlings were thinned to one plant. Plants were watered regularly to maintain moist soil conditions.
Phenotypic trait measurements
Plants were checked daily for germination, which was scored when the seed coat opened and the radicle became visible. All germinated plants were checked daily for bolting, which was scored when buds differentiated from the apical meristem. At bolting, one young, fully expanded leaf was removed from all bolting plants and scanned. Leaf area was measured from the scanned image using ImageJ (Rasband, 1997–2007). Once a plant had bolted, it was checked every morning for flowering, which was scored when sepals opened and petals became visible or when a flower bud senesced prior to opening. After flowering, each plant was checked for flower maturity, which was recorded every afternoon at 1400 hours for plants in which the anthers on the third and fourth flowers of the main stem dehisced and began to shed pollen. At flower maturity, the third and fourth flowers were removed and placed in 75% ethanol for subsequent dissection, the length of the plants' stem was measured, and the collection date was recorded. As some plants aborted flowers (that is, the flower buds senesced prior to opening), we recorded the number of aborted flowers and collected the first two viable flowers when the third and fourth flowers were not viable (generally not later than the 10th flower). To estimate phenology, we recorded the number of days from germination to flowering (days to flowering) and the number of days from flowering to the collection of the flowers (collection interval), which estimates the length of time necessary for a plant to produce viable flowers. The vegetative traits included in this study were leaf area and stem length.
Preserved flowers were dissected under a dissecting microscope (Nikon SMZ800, Nikon Corporation, Tokyo, Japan) and digitally photographed. As one of our goals was to understand patterns of integration in adjacent and non-adjacent floral whorls, we measured multiple traits from the pistil, stamen and petal whorls. Floral organ sizes were measured from the digital image using ImageJ (Rasband, 1997–2007); measured traits included style length, ovary length, short anther length (that is, the length of anther on a representative short stamen), short filament length (that is, the length of filament on a representative short stamen), long anther length (that is, the length of anther on a long stamen), long filament length (that is, the length of filament on a long stamen), petal blade length, petal claw length, petal width and petal area. Measurements of the two flowers from each individual were averaged for all data analysis, except petal area, which was calculated on only one flower because of the extended time needed for this measurement. From the measurements of individual floral organs, we calculated three composite measurements that estimate floral allometry: (1) herkogamy, or total pistil length minus long filament length, (2) stamen exsertion, or long filament length minus petal claw length and (3) pistil exsertion, or total pistil length minus petal claw length.
Statistical analyses
Within each treatment, we used restricted maximum likelihood (REML in PROC MIXED, SAS ver. 9.2) to estimate the random effects of genotype and temporal block on each phenotypic trait. The variance components estimated from this analysis were used to calculate the ratio VG/VP, where VG is the among-genotype variance component of a trait in each treatment and VP is the total of all estimated variance components of a trait in each treatment.
To test for genotypic differences in the response to treatment and estimate genotypic values, we carried out a mixed-model nested analysis of variance (ANOVA) over all three treatments using restricted maximum likelihood (PROC MIXED, SAS ver. 9.2). We evaluated the fixed effect of treatment and the random effects of genotype, chamber nested within treatment, and genotype × treatment interaction (GEI) on all measured traits. A significant genotype effect was interpreted as genetic differences in mean trait expression, a significant treatment effect was interpreted as phenotypic plasticity (or trait sensitivity) among treatments, and a significant interaction effect was interpreted as genetic variation in plasticity.
Using the full ANOVA model, we estimated the genotypic values for each trait in each treatment using best linear unbiased predictors (Robinson, 1991; Littell et al., 2006). To assess the level of integration and modularity of traits, best linear unbiased predictors of genotypic values in each treatment were used to estimate genotypic correlations among traits within each treatment (SAS PROC CORR, SAS ver. 9.2). The significance values of all correlations were corrected for multiple comparisons by controlling the false discovery rate (Benjamini and Hochberg, 1995). Best linear unbiased predictors of genotypic values in each treatment were also used to estimate rGE, the genotypic correlation of each trait across treatment pairs (for example, across WL/CL, WL/CS, CL/CS; SAS PROC CORR). Estimates of rGE indicate the extent to which the same genetic loci have phenotypic expression and alleles have the same function across treatments; estimates of rGE approaching one suggest that the same genetic loci are expressed and alleles have similar effects across the two treatments, whereas estimates approaching 0 suggest that different genetic loci are expressed or alleles that affect trait variation differ in function across the two treatments (Fry et al., 1996; Gurganus et al., 1998; Vieira et al., 2000). To determine whether the rGE values for each trait across each pair of treatments were significantly different from both 0 and 1, we carried out a mixed-model nested ANOVA using the full model as specified above (evaluating the fixed effect of treatment and the random effects of genotype, chamber nested within treatment, and genotype × treatment interaction), but comparing two treatments at a time (PROC MIXED, SAS ver. 9.2); rGE of a trait across each treatment pair was interpreted as significantly different than 1 when the genotype × treatment interaction was significant, and significantly different from 0 when the among-genotype variance was significant (Gurganus et al., 1998; Vieira et al., 2000).
Prior to analysis, we evaluated the response variables for normality and homoscedasticity. The assumptions of normality for ANOVA were not met for some traits, and these were transformed as follows: style length, petal blade length, and petal width were square-root transformed; ovary length, stem length, petal area and leaf area were log-10 transformed; days to flowering was inverse transformed and short and long anther length, long filament length and petal claw length were transformed to the 1.5 power. We carried out all ANOVA, correlation and QTL-mapping analyses using both non-transformed and transformed data, but we present only the results of non-transformed data because the outcome of significance tests was largely unaffected by transformation (not shown).
QTL mapping
The linkage map used in this study was described previously by Iniguez-Luy et al. (2009) and is composed of 224 RFLP and microsatellite markers spanning 10 linkage groups, with an average marker density of 5.7 cM per marker. QTL mapping of genotypic estimates of each trait in each treatment was carried out using composite interval mapping as implemented in Windows QTL Cartographer ver. 2.5 (Wang et al., 2007). To control for effects of variation segregating elsewhere in the genome, we identified co-factors using a forward–backward regression model and a 10-cM window; a maximum of 10 co-factors was selected for inclusion in the mapping model. We scanned for QTL at 2-cM intervals across the B. rapa genome. The genome-wide significance threshold of the likelihood ratio (LR) test statistic for QTL was determined for each trait using 1000 permutations (Churchill and Doerge, 1994), with a type I error rate of 0.05. We also tested for QTL using a type I error rate of 0.075 to evaluate the presence of marginally significant QTL that might affect the interpretation of environment-specific QTL effects (such as QTL × E interactions). The 2-LOD support limits were calculated as LOD=0.217 × LR (Weber et al., 2001). Closely neighboring QTL, between which LR values dropped below the significance threshold but for which support intervals overlapped, were only considered as separate QTL when additive effects reversed sign; otherwise, only QTL with the largest LR values were retained.
We compared QTL expression of traits within treatments to evaluate the genetic basis of trait integration and modularity and compared QTL expression among treatments to determine the effects of treatment on QTL expression. A given QTL was deemed to affect multiple traits or the same trait across treatments when the 2-LOD support intervals overlapped. To determine whether alleles at QTL were sensitive to treatments, we carried out analyses of QTL × treatment interactions for each significant QTL using single marker, two-way ANOVAs (PROC GLM, SAS ver. 9.2). To differentiate the effects of temperature and photoperiod on QTL expression, these analyses were carried out for all pairwise combinations of treatments in which the QTL was detected (for example, if the QTL was detected in CL, we evaluated QTL × treatment interactions in CL/CS and CL/WL). For these analyses, we evaluated the fixed effects of treatment, the genotype at the marker nearest to each detected QTL, and the marker × treatment interaction. Given the low power of this analysis, we used a 0.1 type I error rate.
Finally, we carried out two-dimensional genome scans to detect pairs of QTL that interact epistatically using R/qtl (Broman et al., 2003). For this test, we scanned at 1-cM steps within each interval. Significance thresholds were determined by performing 1000 permutations and using a 0.05 type I error rate.
Results
Quantitative genetics and correlations within treatments
Within each treatment, all traits demonstrated highly significant effects of genotype (P<0.01; Table 1). Although temporal block was also included as a factor in these ANOVAs, we did not detect a significant effect of this factor (results not shown). The ratios of genotypic to total phenotypic variance in each treatment (VG/VP), or broad-sense heritability, ranged from 0.1 to 0.68. In general, floral, vegetative and composite traits had larger VG/VP (0.28–0.68), phenological traits had smaller VG/VP (0.14–0.54) and the number of aborted flowers had the smallest VG/VP (0.1–0.2; Table 1). The magnitude of VG/VP ratios was generally largest in the CL and smallest in the WL environment.
Table 1. Means and quantitative genetic partitioning of morphological, vegetative and phenological traits in each of three treatments.
| Treatment |
CL |
CS |
WL |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | Mean (s.e.) | VG (s.e.) | VG/VP | Mean (s.e.) | VG (s.e.) | VG/VP | Mean (s.e.) | VG (s.e.) | VG/VP |
| Style length | 2.069 (0.032) | 0.079 (0.011)** | 0.53 | 1.901 (0.032) | 0.056 (0.008)** | 0.58 | 1.896 (0.032) | 0.086 (0.012)** | 0.49 |
| Ovary length | 4.719 (0.057) | 0.280 (0.039)** | 0.54 | 3.993 (0.057) | 0.170 (0.024)** | 0.51 | 3.894 (0.057) | 0.205 (0.029)** | 0.51 |
| Short anther length | 2.161 (0.033) | 0.106 (0.0140)** | 0.68 | 2.147 (0.032) | 0.077 (0.010)** | 0.60 | 2.200 (0.032) | 0.056 (0.008)** | 0.54 |
| Short filament length | 3.464 (0.070) | 0.375 (0.052)** | 0.57 | 3.235 (0.070) | 0.2314 (0.033)** | 0.48 | 2.501 (0.069) | 0.136 (0.020)** | 0.41 |
| Long anther length | 2.246 (0.031) | 0.092 (0.012)** | 0.68 | 2.181 (0.031) | 0.0745 (0.010)** | 0.63 | 2.192 (0.031) | 0.055 (0.007)** | 0.59 |
| Long filament length | 4.572 (0.082) | 0.289 (0.040)** | 0.54 | 4.387 (0.082) | 0.226 (0.031)** | 0.56 | 4.027 (0.082) | 0.183 (0.027)** | 0.44 |
| Petal blade length | 5.697 (0.096) | 0.523 (0.070)** | 0.61 | 5.273 (0.096) | 0.327 (0.045)** | 0.55 | 4.175 (0.095) | 0.195 (0.029)** | 0.40 |
| Petal claw length | 2.872 (0.063) | 0.122 (0.017)** | 0.59 | 2.816 (0.063) | 0.105 (0.014)** | 0.54 | 2.920 (0.063) | 0.063 (0.011)** | 0.28 |
| Petal width | 5.059 (0.106) | 0.6134 (0.081)** | 0.65 | 4.633 (0.105) | 0.478 (0.064)** | 0.61 | 3.132 (0.105) | 0.201 (0.029)** | 0.42 |
| Petal area | 27.818 (0.776) | 49.083 (6.743)** | 0.58 | 23.547 (0.775) | 32.358 (4.483)** | 0.54 | 13.539 (0.771) | 8.208 (1.237)** | 0.36 |
| Flowers aborted | 1.070 (0.251) | 2.537 (0.692)** | 0.18 | 0.649 (0.249) | 0.967 (0.262)** | 0.20 | 0.791 (0.247) | 0.838 (0.276)§ | 0.10 |
| Leaf area | 1.951 (0.835) | 2.954 (0.399)** | 0.53 | 2.698 (0.833) | 7.841 (1.006)** | 0.65 | 12.340 (0.829) | 64.309 (8.262)** | 0.59 |
| Stem length | 27.503 (1.037) | 37.356 (5.071)** | 0.59 | 24.817 (1.036) | 37.356 (5.071)** | 0.65 | 22.815 (1.033) | 44.612 (5.643)** | 0.60 |
| Days to flowering | 63.190 (1.968) | 29.234 (6.076)** | 0.19 | 72.941 (1.965) | 62.813 (9.274)** | 0.36 | 38.853 (1.961) | 39.298 (5.346)** | 0.54 |
| Collection interval | 6.739 (0.577) | 4.734 (1.423)* | 0.14 | 5.626 (0.575) | 9.211 (1.763)** | 0.32 | 1.750 (0.572) | 3.627 (0.545)** | 0.37 |
| Herkogamy | 2.202 (0.081) | 0.198 (0.031)** | 0.40 | 1.498 (0.081) | 0.139 (0.020)** | 0.49 | 1.755 (0.081) | 0.193 (0.029)** | 0.37 |
| Stamen exsertion | 1.707 (0.048) | 0.081 (0.012)** | 0.38 | 1.575 (0.048) | 0.054 (0.008)** | 0.41 | 1.112 (0.047) | 0.076 (0.011)** | 0.43 |
| Pistil exsertion | 3.908 (0.083) | 0.250 (0.038)** | 0.44 | 3.072 (0.082) | 0.170 (0.024)** | 0.51 | 2.866 (0.0820) | 0.217 (0.033)** | 0.36 |
Abbreviations: CL, cool temperature long photoperiod; CS cool temperature short photoperiod; VG, among-genotypic variance; VG/VP, among-genotypic variance divided by total phenotypic variance; WL, warm temperature long photoperiod.
§P<0.01, *P<0.001, **P<0.0001.
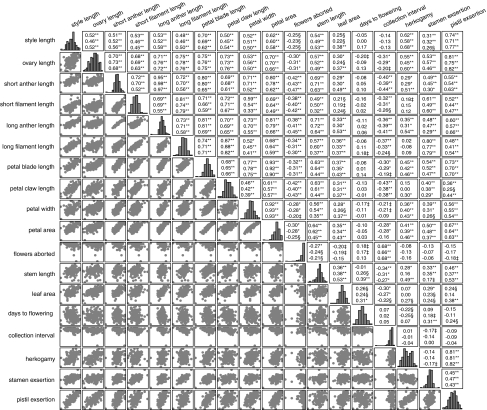
To assess the patterns of integration and modularity within and among floral, vegetative and phenological traits, we estimated genotypic correlations between each pair of traits in each treatment. Consistent with prior studies, floral traits were strongly positively correlated within each treatment (r=0.39–0.97, P<0.0001; Figure 1). Given that LD between physically distant loci is disrupted in RILs, these correlations suggest that either pleiotropy or physical linkage between proximate loci underlies the observed correlations. Adjacent floral whorls were not more strongly correlated than non-adjacent whorls; for example, the average correlation between pistil and stamen traits was 0.61 (range 0.45–0.78), and the average correlation between pistil and petal traits was 0.62 (range 0.45–0.76), and this was consistent across treatments. The number of aborted flowers was negatively correlated with all floral length traits (that is smaller floral organ size was associated with increased floral abortion; r=−0.20 to −0.47, P=0.01 to <0.0001; Figure 1).
Figure 1.
Correlation coefficients and significance of correlations among traits within treatments. Pearson correlation coefficients of correlations among traits within the CL (upper), CS (middle) and WL (bottom) treatments are indicated above the diagonal, with correlations that are significantly different than zero after false discovery rate correction indicated. Histograms of each trait in the CL treatment are shown on the diagonal, and plots of the bivariate correlation among pairs of traits in the CL treatment are shown below the diagonal; histograms and plots are shown only for the CL treatment because correlations are highly similar in all three treatments. ‡P<0.05, §P<0.01, *P<0.001, **P<0.0001.
Contrary to hypothesized modularity, the two vegetative traits were positively correlated with all floral traits (r=0.19–0.72, P=0.02 to <0.0001; Figure 1). Vegetative traits were also marginally negatively correlated with the number of aborted flowers (decreasing plant size was weakly correlated with increased floral abortion; r= −0.15 to −0.27, P=0.07–0.0008; Figure 1). The two phenological traits showed varying degrees of modularity; days to flowering was not correlated with floral or composite traits, marginally positively correlated with number of aborted flowers (that is delayed flowering was marginally associated with increased floral abortion; r=0.13–0.18, P=0.1–0.03; Figure 1), and positively correlated with vegetative traits in all treatments (later-flowering plants were larger; r=0.24–0.31, P=0.004 to <0.0001) except for leaf area in CL (r=−0.01; Figure 1). By contrast, collection interval was marginally to strongly negatively correlated with most floral traits (except style length and petal width), strongly positively correlated with number of aborted flowers (r=0.65–0.68, P<0.0001), negatively correlated with vegetative traits (smaller plants took longer to produce viable flowers once flowering had been initiated, r= −0.09 to −0.44, P=0.27 to <0.0001; Figure 1), and uncorrelated with days to flowering (Figure 1).
Herkogamy (total pistil length minus long filament length) was positively correlated with pistil traits but not filament traits. Stamen exsertion (long filament length minus petal claw length) was positively correlated with both filament and petal claw traits. Pistil exsertion (total pistil length minus petal claw length) was very strongly positively correlated with both pistil traits and petal claw length. In general, for all classes of traits, the correlation structure and magnitude of correlation coefficients were similar across all treatments (Figure 1).
Quantitative genetics and correlations among treatments
Most traits demonstrated highly significant GEI (P<0.0001; Table 2) except collection interval (P=0.026) and the number of aborted flowers (P=0.22). Despite the significant GEI, the effect of genotype remained highly significant for all traits (P<0.0001; Table 2). To measure the relative variance attributable to genotype and GEI, we divided the GEI variance by the among-genotype variance (Gurganus et al., 1998), which ranged from 0.13 to 0.38 (except for leaf area, which was 1.50), indicating that although a significant portion of variation was attributable to GEI for most traits, a larger proportion of variation was attributable to among-genotype differences.
Table 2. Quantitative genetic partitioning of variation in floral, vegetative and reproductive timing traits in B. rapa RILs in three treatments, including variance components of each effect, standard error (in parentheses) and significance of effects.
| Trait | Style length | Ovary length | Short anther length | Short filament length | Long anther length | Long filament length | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Random effects (Z) | ||||||||||||
| Chamber (treatment) | 0.002 (0.001)‡ | 0.009 (0.004)‡ | 0.003 (0.001)§ | 0.017 (0.007)§ | 0.002 (0.001)‡ | 0.029 (0.011)§ | ||||||
| Genotype | 0.053 (0.008)** | 0.173 (0.024)** | 0.067 (0.009)** | 0.186 (0.027)** | 0.071 (0.009)** | 0.203 (0.028)** | ||||||
| Genotype × treatment | 0.021 (0.003)** | 0.046 (0.007)** | 0.015 (0.002)** | 0.071 (0.010)** | 0.010 (0.002)** | 0.048 (0.007)** | ||||||
| Residual | 0.065 (0.002)** | 0.190 (0.007)** | 0.047 (0.002)** | 0.223 (0.008)** | 0.039 (0.001)** | 0.186 (0.006)** | ||||||
| Fixed effects (F) | df | F | df | F | df | F | df | F | df | F | df | F |
| Treatment | F2, 23.7 | 14.87** | F2, 20.4 | 97.29** | F2, 21.3 | 1.17 | F2, 20.1 | 70.59** | F2, 19.7 | 2.56 | F2, 17 | 14.27* |
| Significant genotype × treat contrasts | WL/CL WL/CS | CL/CS WL/CL WL/CS | CL/CS WL/CL WL/CS | CL/CS WL/CL WL/CS | CL/CS WL/CL WL/CS | CL/CS WL/CL WL/CS | ||||||
|
Trait |
Petal blade length |
Petal claw length |
Petal width |
Petal area |
Flowers aborted |
Leaf area |
||||||
| Random effects (Z) | ||||||||||||
| Chamber (treatment) | 0.038 (0.015)§ | 0.019 (0.007)§ | 0.046 (0.018)§ | 2.180 (0.878)** | 0.250 (0.117)‡ | 2.995 (1.135)§ | ||||||
| Genotype | 0.266 (0.038)** | 0.087 (0.012)** | 0.345 (0.047)** | 21.528 (3.083)** | 1.409 (0.245)** | 10.259 (1.999)** | ||||||
| Genotype × treatment | 0.093 (0.012)** | 0.017 (0.003)** | 0.099 (0.013)** | 8.287 (1.102)** | 0.11 (0.145) | 15.345 (1.517)** | ||||||
| Residual | 0.254 (0.009)** | 0.090 (0.003)** | 0.255 (0.009)** | 23.018 (0.786)** | 7.299 (0.248)** | 14.444 (0.480)** | ||||||
| Fixed effects (F) | df | F | df | F | df | F | df | F | df | F | df | F |
| Treatment | F2, 17..8 | 84.34** | F2, 16.1 | 0.71 | F2, 17.5 | 117.8** | F2, 19.3 | 11.43** | F2, 14.5 | 0.87 | F2, 21.7 | 53.95* |
| Significant genotype × treat contrasts | CL/CS WL/CL WL/CS | WL/CL WL/CS | WL/CL WL/CS | WL/CL WL/CS | WL/CL | CL/CS WL/CL WL/CS | ||||||
|
Trait |
Stem length |
Days to flowering |
Collection interval |
Herkogamy |
Stamen exsertion |
Pistil exsertion |
||||||
| Random effects (Z) | ||||||||||||
| Chamber (treatment) | 4.632 (1.759)§ | 20.835 (7.790)§ | 1.599 (0.634)§ | 0.030 (0.012)§ | 0.01 (0.004)§ | 0.03 (0.012)§ | ||||||
| Genotype | 34.074 (4.371)** | 33.542 (4.950)** | 5.3686 (0.821)** | 0.112 (0.018)** | 0.051 (0.008)** | 0.1477 (0.022)** | ||||||
| Genotype × treatment | 6.361 (0.868)** | 10.726 (2.059)** | 0.7516 (0.388)‡ | 0.065 (0.01)** | 0.02 (0.003)** | 0.0638 (0.01)** | ||||||
| Residual | 20.414 (0.68)** | 62.137 (2.097)** | 15.8354 (0.545)** | 0.222 (0.008)** | 0.09 (0.003)** | 0.256 (0.009)** | ||||||
| Fixed effects (F) | df | F | df | F | df | F | df | F | df | F | F | |
| Treatment | F2, 16.6 | 6.55§ | F2, 15.7 | 84.96** | F2, 15.5 | 23.40** | F2, 17.6 | 21.67** | F2, 17.2 | 51.04** | F2, 17.6 | 52.59** |
| Significant genotype × treat contrasts | WL/CL WL/CS | CL/CS WL/CL WL/CS | WL/CL WL/CS | CL/CS WL/CL WL/CS | CL/CS WL/CL WL/CS | CL/CS WL/CL WL/CS | ||||||
Abbreviations: CL, cool temperature long photoperiod; CS cool temperature short photoperiod; RIL, recombinant inbred line; WL, warm temperature long photoperiod.
‡P<0.05, §P<0.01, *P<0.001, **P<0.0001.
To further assess whether the genetic architecture of a given trait changed across environments, we estimated genotypic correlations (rGE) and performed ANOVAs for pairs of treatments. Overall, values of rGE were high (0.77–0.99) for most traits, with the exception of leaf area (0.38–0.47; Table 3). For style length, ovary length, short filament length, long filament length, petal blade length, petal claw length, petal width, petal area, flowers aborted, stem length, collection interval and stamen exsertion across CL/CS, flowers aborted across WL/CS, and flowers aborted and stem length across WL/CS, rGE estimates were significantly different than 0 and not or only moderately (P>0.01) significantly different from 1 (that is, significant genotype effect but non-significant GEI; Table 3), indicating that common loci affected variation and that each allele at a locus had a similar function for these traits across these treatment pairs. For all other comparisons, and more specifically for most comparisons involving WL, estimates of rGE were significantly different than both 0 and 1 (that is, both genotype and GEI effects were highly significant; Table 3), indicating that some different loci affect variation across treatment pairs and/or that some alleles have different functional effects across treatment pairs. These results suggest that for many traits, the expression of loci and the effects of alleles were more conserved across treatments with the same temperatures (yet different photoperiods) and less conserved across treatments with different temperatures.
Table 3. Pearson correlation coefficients of across-treatment genotypic correlations (rGE) for each trait.
| Trait | rGE CL vs CS | rGE WL vs CL | rGE WL vs CS |
|---|---|---|---|
| Style length | 0.93NS | 0.73** | 0.76** |
| Ovary length | 0.93‡ | 0.85** | 0.86** |
| Short anther length | 0.93* | 0.85** | 0.88** |
| Short filament length | 0.91‡ | 0.78** | 0.77** |
| Long anther length | 0.95§ | 0.91** | 0.92** |
| Long filament length | 0.94‡ | 0.83** | 0.86* |
| Petal blade length | 0.93‡ | 0.77** | 0.78** |
| Petal claw length | 0.96NS | 0.89** | 0.89** |
| Petal width | 0.94NS | 0.81** | 0.83** |
| Petal area | 0.93NS | 0.81** | 0.83** |
| Flowers aborted | 0.99NS | 0.99‡ | 0.99NS |
| Leaf area | 0.42** | 0.47** | 0.39** |
| Stem length | 0.95NS | 0.86** | 0.90NS |
| Days to flowering | 0.86§ | 0.86** | 0.88* |
| Collection interval | 0.97NS | 0.97§ | 0.98* |
| Herkogamy | 0.86§ | 0.69** | 0.69** |
| Stamen exsertion | 0.91‡ | 0.74** | 0.83** |
| Pistil exsertion | 0.89§ | 0.76** | 0.78** |
Abbreviations: ANOVA, analysis of variance; CL, cool temperature long photoperiod; CS cool temperature short photoperiod; NS, not significant; WL, warm temperature long photoperiod.
All correlations are significantly different from zero (P<0.0001). P-values indicated below are from tests of significance of the genotype × treatment interaction term from ANOVA analyses of pairs of environments and indicate whether cross-environment correlations are significantly different from 1.
‡P<0.1, ‡P<0.05, §P<0.01, *P<0.001, **P<0.0001.
Patterns of main-effect QTL expression within each treatment
The genome-wide scan detected 181 significant QTL, 61 of which were detected in CL, 61 were detected in CS and 59 were detected in WL (Figure 2; Appendix).
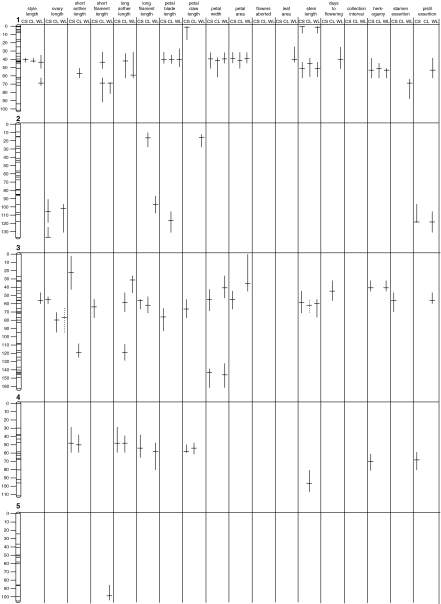
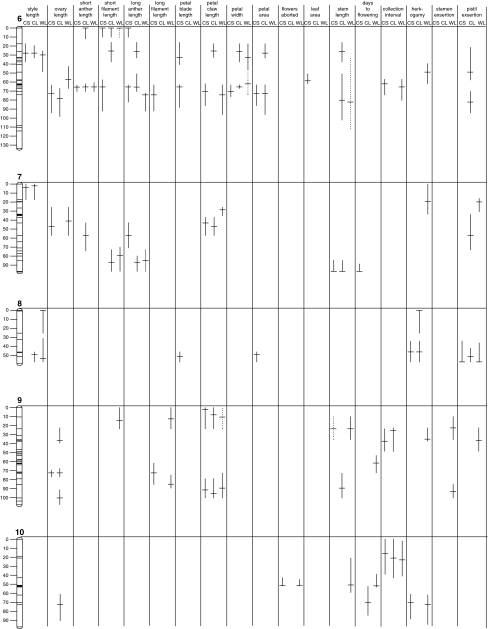
Figure 2.
QTL detected for traits measured in three treatments. The length of bars designate the range of 2-LOD support limits for each QTL, with the peak of the QTL indicated. Solid bars indicate QTL that are significant using a 0.05 type I error rate and dashed bars indicate QTL that are significant using a 0.075 type I error rate.
To evaluate the QTL architecture underlying the strongly positive genotypic correlations among floral traits, we investigated patterns of QTL co-localization among floral organs within each treatment. Consistent with the strongly positive genotypic correlations (Figure 1), almost all floral QTL affected multiple traits, and this pattern was consistent in each of the three treatments. For example, a QTL detected at ∼45 cM on chromosome 1 in CL affected seven of the 10 measured floral traits, or in other words, seven of the eight floral QTL detected on chromosome 1 co-localized, and similarly strong patterns of co-localization were observed on most chromosomes (Figure 2). Overlapping floral QTL always had the same direction of additive effects, and QTL from all combinations of floral whorls overlapped. QTL of organs in adjacent whorls did not overlap with greater frequency than those for organs in non-adjacent whorls; for example, style length and ovary length QTL co-localized with 11 stamen QTL (adjacent whorls) and 12 petal QTL (non-adjacent whorls) in CS, and, coincidentally, with seven stamen QTL (adjacent whorls) and nine petal QTL (non-adjacent whorls) in both CL and WL.
To assess the relationship among floral, vegetative and reproductive timing traits, we investigated patterns of co-localization of QTL. Consistent with strongly positive correlations between floral and vegetative traits (Figure 1), most QTL for vegetative traits co-localized with QTL from one or more floral organs from the same treatment (for example, QTL for floral traits and stem length co-localized at ∼60 cM on chromosome 1 in all three treatments; Figure 2; Appendix). Consistent with a non-significant correlation between days to flowering and floral traits (Figure 1), only a small number of QTL for days to flowering co-localized with floral traits (Figure 2; Appendix). Days to flowering, however, was positively correlated with vegetative traits and had QTL with the same direction of additive effects that co-localized (Figures 1 and 2; Appendix). Consistent with the negative correlation between collection interval and both floral and vegetative traits (Figure 1), QTL for collection interval and both floral and vegetative traits co-localized in all three treatments, and the direction of additive effects of collection interval QTL were opposite those of floral and vegetative QTL (Figure 2; Appendix).
We next compared patterns of QTL expression of composite traits (herkogamy, stamen exsertion and petal exsertion) with those of their underlying floral organs to assess whether their genetic architecture was shared with one or both organs. Herkogamy (total pistil length minus long filament length) was positively correlated with pistil traits but not filament traits (Figure 1), and correspondingly, QTL for herkogamy often co-localized with pistil QTL but not filament QTL (for example, herkogamy QTL co-localized with pistil QTL but not stamen QTL at ∼50 cM on chromosome 1 in CL; Figure 2; Appendix). In contrast, stamen exsertion (long filament length minus petal claw length) was positively correlated with both underlying organs (Figure 2; Appendix), and stamen exsertion QTL co-localized with both petal and stamen QTL in all three treatments (Figure 2; Appendix). Similarly, pistil exsertion (total pistil length minus petal claw length) was positively correlated with both underlying traits (Figure 1) and pistil exsertion QTL co-localized with both petal and pistil QTL in all three treatments (Figure 2; Appendix).
Patterns of main-effect QTL expression across treatments
Consistent with estimates of rGE that were significantly different than 0 for most traits (significant among-genotype effect; Table 3), QTL from all three treatments co-localized for a trait in many cases, and often explained a large percentage of variance (>10%) for a trait in at least one treatment (Figure 2; Appendix).
Despite the large magnitude of most estimates of rGE, we nevertheless detected many across-environment correlations that were significantly different than one, particularly across WL/CS or WL/CL (Table 3), suggesting that somewhat different loci contribute to trait expression across environments with differing temperatures. Using a 0.1 type I error rate, 15 QTL demonstrated significant QTL × E for pistil, stamen, petal, vegetative traits and days to flowering (Appendix). We did not detect significant QTL × E for flowers aborted (Appendix), consistent with expectations given that rGE=1 for this trait. In all but one instance, QTL × E was detected across WL/CL or WL/CS (Appendix), indicating that QTL expression was in fact most sensitive to temperature. We examined the additive effects of these QTL to determine the cause of the significant interaction, and all were due to changes in the magnitude of additive effects (and not sign) across treatments (data not shown).
Epistatically interacting QTL
In the two-dimensional genome scans for QTL × QTL epistasis, only five instances of loci with significant epistatic effects were detected. Four of these (for petal width-WL, petal area-WL, herkogamy-WL and pistil exsertion-CS) were detected between marker BRMS006 on the top of chromosome 8 and marker pX139cH on the top of chromosome 10. The fifth (style length-WL) was detected between marker fito137 in the middle of chromosome 3 and marker pX139cH on the top of chromosome 10.
Discussion
Patterns of integration and modularity
Previous research has shown that floral traits are usually significantly correlated in hermaphroditic plant species, and that plants with zygomorphic flowers have more strongly integrated floral organs than actinomorphic species (Ashman and Majetic, 2006). Despite the fact that Brassicaceae have actinomorphic flowers, previous research in the family revealed that floral traits are frequently highly correlated (Conner and Sterling, 1995; Juenger et al., 2000, 2005; Brock and Weinig, 2007). In the current study, floral traits were highly correlated despite the fact that LD between physically distant loci was reduced by recombination during RIL formation; this suggests that pleiotropic loci (or LD between physically proximate loci) contributed to floral trait integration. The high frequency with which floral QTL co-localized further supports the hypothesis of pleiotropy because it is unlikely that genes that independently affect the size of different floral organs would always be in such close proximity. Previous studies in Arabidopsis also suggested that pleiotropy was likely to be responsible for floral trait covariation (Conner, 2002; Juenger et al., 2005). Interestingly, we did not find stronger correlations between floral organs in adjacent whorls vs non-adjacent whorls; thus, any pleiotropic effects on quantitative variation in floral organ size appear to be uniform throughout the flower, in contrast to the expression pattern of ABC genes, which affect qualitative variation in floral organ identity in adjacent whorls.
Evolutionarily, correlations observed in this study suggest that allometry of floral organs may be genetically or developmentally constrained in B. rapa, and, more specifically, that a response to selection on one floral trait may be accompanied by corresponding responses in other floral traits. This correlation could act as a relevant constraint in this species given that previous research in B. rapa has shown that the allometry of floral organs (herkogamy) strongly affects the rate of reproduction and inbreeding (Brock et al., in review). That said, research in natural populations of close relatives, R. raphanistrum and B. napus, revealed that filament and corolla traits were more strongly correlated than other floral organs, and these enhanced correlations presumably arose from evolutionary responses to pollinator-mediated selection (Conner and Sterling, 1995). It is parsimonious to assume that the initial genetic mechanisms underlying floral development, namely pleiotropy or LD between physically proximate loci, are conserved within this clade, as they appear to determine floral organ size in A. thaliana, B. rapa and B. nigra. Thus, a short-term constraint may exist on floral evolution in B. rapa, but if the behavior of other members of the clade is any guide, then modifiers may arise that enable independent responses of floral organs to selection.
Results of this study also revealed strong genotypic correlations and frequent co-localization of QTL between floral and vegetative traits of B. rapa, suggesting that pleiotropy or LD between physically proximate genes may also be responsible for covariation of the floral and vegetative traits examined in this study. These genotypic and QTL associations again suggest that measured floral and vegetative traits may not evolve completely independently over at least the short-term, assuming that the genetic architecture of the genotypes studied here is representative of the segregating variation of this species. As the parents of the RILs used in this study were a cultivated seed oil genotype (R500) and a rapid cycling (imb211) genotype that may approximate a weedy member of the species, these RILs may be representative of segregating variation for some crosses occurring between cultivated genotypes and their wild neighbors. One interesting avenue for future research would be to assess patterns of modularity of floral and vegetative traits in a more diverse selection of cultivated and wild genotypes of B. rapa to determine the extent to which the results of this study are representative of species-wide patterns of floral integration and modularity.
In contrast to the results of this study, previous research has found that floral and vegetative modules are generally not significantly correlated in plants (Ashman and Majetic, 2006). For example, previous studies in the Brassicaceae found that floral and vegetative organs were uncorrelated (Conner and Sterling, 1996; Juenger et al., 2005), shared few common QTL (Juenger et al., 2005), and therefore represented separate modules (but see Brock and Weinig, 2007). Variation in floral and vegetative traits in B. rapa is thus less modular than has been found previously in other Brassicaceae species. One possible explanation for these differences among studies is that vegetative traits vary in the extent to which they are modular. For instance, in P. drummondii, plant height, biomass and the number of nodes and branches were correlated with floral traits, whereas leaf traits were not (Waitt and Levin, 1993); the choice of vegetative traits could influence the interpretation of modularity. In this study, sampling different vegetative traits may have revealed different patterns of modularity in B. rapa. Another possible reason for experimental differences is that modularity of floral and vegetative organs is not ubiquitous, and may depend more on the evolutionary history and genetic architecture of each species than previously realized.
The patterns of modularity between phenological and floral or vegetative traits varied. Collection interval, or the time elapsed from first flowering until several viable flowers developed, was negatively correlated with floral and vegetative traits and positively correlated with the number of aborted flowers. Thus, the longer the collection interval (or the longer it took to produce a viable flower), the smaller the floral and vegetative organs were, and the more frequently flowers were aborted; potentially, collection interval may therefore be indicative of overall plant vigor. The QTL that co-localized for collection interval, floral traits and vegetative traits may therefore be loci that have a general affect on the overall vigor of the plant. The other phenological trait, days to flowering, was not correlated with floral traits but was significantly positively correlated and had QTL that co-localized with vegetative traits. This relationship between flowering time and vegetative traits is consistent with previous research that found that later-flowering plants are generally larger (Bolmgren and Cowan, 2008), likely because they allocate resources to vegetative growth longer than early-flowering plants. These QTL may directly regulate days to flowering, but only indirectly affect vegetative size by determining the duration of time available to grow prior to reproduction. However, another possibility is that these QTL have direct mechanistic affects in tissues throughout the plant (that is, directly affect flowering time and vegetative organ size). An increase in flowering time, however, did not translate into larger floral organ size. Interestingly, it thus appears that the genetic mechanisms linking flowering time to vegetative traits are independent from those that link vegetative and floral traits.
Composite traits showed differing patterns of QTL architecture in relation to component traits. Herkogamy, which was previously found to be a strong indicator of the rate of self-pollination in B. rapa in the field (Brock et al., in review), was positively correlated and had QTL that co-localized with pistil traits but not filament traits. Thus, in the controlled settings investigated in this experiment, variation in pistil traits more strongly affected herkogamy, and hence, may have a stronger affect on the rate of self-pollination in B. rapa in these conditions. However, these results may be environment-dependent, as stamen traits more strongly affected herkogamy in a field setting (Brock et al., in review). In contrast, stamen exsertion and pistil exertion were correlated and had QTL that co-localized with both underlying traits. Previous studies of pollination efficiency in Raphanus found that the extent to which pistils and stamens were exserted had an effect on the rate of reproduction; genotypes with the most exserted stamens had the most number of pollen grains removed, and genotypes with intermediate pistil exsertion had the highest probability of successful fertilization (Conner et al., 1995). The results of this study suggest that variation in both component traits underlying stamen exsertion or pistil exsertion affect the rate of successful fertilization. These patterns suggest that multiple traits (pistil, stamen or corolla length) could control the degree of organ exsertion and hence the rate of outcrossing, whereas the underlying trait (pistil or stamen length) most strongly affecting herkogamy and hence the rate of inbreeding in B. rapa may vary depending on the environmental setting.
Response of genotypes to differences in temperature and photoperiod
Variance explained by the effect of genotype was typically much greater than that explained by GEI, and this result was supported by QTL that co-localized in all three treatments for most traits. Nonetheless, GEI effects were significant for most traits. rGE was generally greater across treatments with the same temperature (but differing photoperiod) than across treatments with the same photoperiod (but differing temperatures). These results suggest that common genetic loci affected trait variation and that alleles had the same functional effects across treatments with the same temperature (in CL vs CS), whereas differences existed in the causal genetic loci or functional effects of alleles across treatments with different temperatures (in most comparisons involving WL). This significant temperature effect is in agreement with results of research on reproductive traits in other species (Jermstad et al., 2003; Hedhly et al., 2005; Schmuths et al., 2006). Also consistent with the finding that GEI was due to differential sensitivity to temperature, all but one QTL × E interaction was in response to temperature.
One implication of the large effects of temperature detected in this study is that plants growing as fall and winter annuals and as biennials in natural environments may have trait expression similar to each other, but different from spring or summer annuals. The large natural differences in temperature environments that B. rapa experiences across its wide Eurasian distribution, coupled with GEI in response to temperature, may have ecological implications in that different genotypes may be favored in different temperature environments. These results are important because they suggest that future temperature changes associated with climate change may favor specific genotypes. In particular, genotypes may vary in the success of fertilization by pollinators depending on floral responses to temperature; thus, any future temperature changes associated with climate change may therefore exert strong selective pressures that shape patterns of genetic and phenotypic diversity across species ranges.
Conclusions and future research
We found that floral traits were highly integrated in B. rapa and that floral, vegetative and phenological traits were generally not as modular as has been previously found in the Brassicaceae. To assess whether the patterns of trait integration and modularity revealed in this study are representative of those found across the species, future research will focus on understanding the patterns of correlations among traits in a diverse sampling of cultivated and wild genotypes of B. rapa. Results of this study also revealed that large numbers of QTL co-localized for many traits with different functions, suggesting that pleiotropic or closely physically linked genes control variation in many different traits. To further understand the genetic basis of trait variation in B. rapa, future research will focus on fine-mapping QTL identified in this study, starting with genomic regions that contain multiple floral QTL of large effect. Fine-mapping efforts will be facilitated by results of the B. rapa genome project (Kim et al., 2009), which has generated genomic information useful for marker development and selection of candidate genes. Fine mapping of floral QTL may be complementary to research carried out under the floral genome project; the floral genome project identified loci affecting floral development and qualitative variation in floral organ identity across angiosperms, whereas our research aims to identify loci affecting quantitative variation in floral organ size. A further goal of fine-mapping efforts will be to understand whether pleiotropic or tightly linked genes control variation in floral traits and to functionally characterize genetic variants controlling floral quantitative trait variation in B. rapa.
Acknowledgments
We thank M Haselhorst, A Hemenway, O Deninno and A Faulconer for assistance with data collection; M Brock and M Rubin for assistance with data analysis; F Iniguez-Luy for providing seeds and three anonymous reviewers and the evolution laboratory meeting at Wyoming for comments on previous versions of this paper. This research was supported by NSF Grant #DBI 0605736.
Appendix
Results of composite interval QTL mapping in Brassica rapa RILs. The position, 2-LOD support intervals, likelihood ratio (LR) test statistic, additive effect of the imb211 allele, percent variance explained and closest marker are indicated for each significant QTL detected in this study, organized by trait, position and treatment as shown in Figure 2. ### before a trait name indicates marginal QTL that were significant at the P=0.075 level. QTL that demonstrate significant QTL × environment interactions (QTL × E) across pairs of treatments, and the level of significance for these interactions, are also indicated (†P<0.1, ‡P<0.05, §P<0.01).
Table A1. .
| Trait–treatment | Position in cM (2-LOD intervals) | LR | Additive effect | % Variance explained | Closest marker | QTL that demonstrate significant QTL × E across treatment pairs |
|---|---|---|---|---|---|---|
| Chromosome 1 | ||||||
| Style length–CS | 39.66 (38.25–43.22) | 44.148 | −0.097 | 18.248 | fito019d | |
| Style length–CL | 41.83 (38.25–43.22) | 38.319 | −0.101 | 15.373 | fito101a | |
| Style length–WL | 43.22 (34.56–50.6) | 37.516 | −0.109 | 13.803 | pW108aE | |
| Style length–WL | 68.54 (62.24–71.28) | 19.575 | 0.075 | 6.292 | pX128dX | WL/CL† |
| Short anther length–CL | 56.77 (50.6–62.24) | 15.181 | −0.071 | 6.209 | fito222 | |
| Short filament length–CL | 43.22 (30.99–50.77) | 14.019 | −0.160 | 6.851 | pW108aE | WL/CL† |
| Short filament length–CL | 68.54 (62.24–91.28) | 13.822 | 0.159 | 6.434 | pX128dX | |
| Short filament length–WL | 68.54 (67.92–81.28) | 28.216 | 0.150 | 11.600 | pX128dX | |
| Long anther length–CL | 41.66 (32.56–62.24) | 20.036 | −0.073 | 7.068 | fito101a | |
| Long anther length–WL | 58.77 (30.99–62.24) | 18.769 | −0.067 | 7.883 | fito222 | |
| Petal blade–CS | 39.66 (30.99–44.6) | 33.953 | −0.182 | 12.456 | fito019d | |
| Petal blade–CL | 39.66 (34.56–44.6) | 34.553 | −0.217 | 12.441 | fito019d | WL/CL‡ |
| Petal blade–WL | 39.66 (26.99–48.6) | 21.728 | −0.136 | 8.990 | fito019d | See above |
| Petal claw–CS | 0.67 (0.01–15.89) | 14.150 | −0.068 | 4.401 | pW249dX | |
| Petal width–CS | 39.66 (32.56–50.6) | 26.052 | −0.183 | 8.517 | fito019d | |
| Petal width–CL | 41.66 (38.25–60.77) | 23.741 | −0.202 | 8.480 | fito101a | |
| Petal width–WL | 39.66 (32.56–44.6) | 29.924 | −0.172 | 11.601 | fito019d | |
| Petal area–CS | 39.66 (32.56–44.6) | 32.213 | −1.693 | 12.275 | fito019d | |
| Petal area–CL | 41.66 (32.56–50.77) | 31.085 | −1.966 | 12.094 | fito101a | |
| Petal area–WL | 39.66 (32.56–44.6) | 32.843 | −1.285 | 14.047 | fito019d | |
| Leaf area–WL | 39.66 (24.6–43.22) | 12.818 | −1.207 | 7.126 | fito019d | |
| Stem length–CS | 0.01 (0.01–7.86) | 14.996 | −1.514 | 5.734 | pW190aH | |
| Stem length–WL | 0.67 (0.01–7.86) | 15.323 | −1.337 | 4.844 | pW249dX | |
| Stem length–CS | 50.77 (43.22–62.24) | 34.799 | −2.253 | 14.739 | pX136bE | |
| Stem length–CL | 44.6 (38.25–60.77) | 36.122 | −2.131 | 14.115 | pW216aH | |
| Stem length–WL | 50.77 (43.22–60.77) | 47.521 | −2.478 | 17.022 | pX136bE | |
| Days to flowering–WL | 39.66 (24.99–50.77) | 14.473 | −1.335 | 5.162 | fito019d | |
| Herkogamy–CS | 52.77 (38.25–62.24) | 19.946 | −0.103 | 8.534 | pX136bE | |
| Herkogamy–CL | 50.77 (44.6–62.24) | 22.591 | −0.127 | 9.892 | pX136bE | |
| Herkogamy–WL | 52.77 (50.6–62.24) | 38.876 | −0.155 | 15.804 | pX136bE | |
| Stamen exsertion–WL | 68.54 (63.92–87.28) | 20.746 | 0.069 | 7.656 | pX128dX | |
| Pistil exsertion–WL | 52.77 (38.25–62.24) | 24.404 | −0.129 | 10.415 | pX136bE | |
| Chromosome 2 | ||||||
| Ovary length–CS | 104.89 (90.05–117.85) | 22.941 | 0.162 | 10.292 | pX107aX | |
| Ovary length–WL | 100.89 (96.05–129.85) | 13.076 | 0.098 | 4.896 | pW205bH | |
| Ovary length–CS | 135.85 (123.85–135.85) | 12.883 | −0.109 | 5.276 | pX124bX | |
| Long filament length–CL | 15.46 (9.37–26.58) | 12.653 | −0.130 | 5.748 | pX139eH | |
| Long filament length–WL | 96.05 (86.04–106.66) | 21.030 | 0.137 | 7.532 | pW205bH | |
| Petal blade–CL | 115.42 (104.89–129.85) | 13.365 | 0.160 | 6.102 | pW249aX | |
| Petal claw–WL | 14.91 (11.37–26.58) | 14.100 | −0.067 | 5.348 | fito338a | |
| Pistil exsertion–CS | 117.42 (96.05–117.85) | 13.666 | 0.096 | 5.630 | pW249aX | |
| Pistil exsertion–WL | 117.42 (104.89–129.85) | 13.685 | 0.092 | 5.005 | pW249aX | |
| Chromosome 3 | ||||||
| Style length–WL | 55.73 (46.39–59.48) | 17.797 | −0.064 | 6.087 | pW214aX | |
| Ovary length–CS | 54.59 (51.33–59.48) | 25.979 | −0.133 | 10.536 | pW214aX | |
| Ovary length–CL | 79.83 (70.78–94.49) | 16.991 | −0.119 | 6.724 | fito109b | |
| ###Ovary length–WL | 76.61 (65.48–76.61) | 12.107 | −0.085 | 4.017 | fito071d | |
| Short anther length–CS | 22.01 (2.01–42.4) | 14.530 | −0.071 | 7.126 | pX139aH | |
| Short anther length–CL | 118.52 (108.21–124.62) | 23.841 | −0.087 | 8.179 | fito413 | |
| Short filament length–CS | 63.48 (54.59–76.61) | 12.702 | −0.129 | 5.976 | pW147bH | |
| Long anther length–WL | 30.86 (26.01–46.39) | 19.001 | −0.065 | 7.457 | pX144bE | |
| Long anther length–CL | 58.25 (46.39–69.45) | 15.345 | −0.065 | 5.246 | pX141cE | |
| Long anther length–CL | 118.52 (108.87–128.2) | 22.613 | −0.083 | 7.969 | fito413 | |
| Long filament length–CS | 55.73 (54.59–66.06) | 22.798 | −0.201 | 9.753 | pW214aX | |
| Long filament length–CL | 61.48 (51.33–70.78) | 13.908 | −0.166 | 7.401 | pX141cE | |
| Petal blade–CS | 75.72 (65.48–92.49) | 14.105 | −0.118 | 5.172 | fito071d | |
| Petal claw–CS | 66.06 (54.59–76.61) | 14.381 | −0.065 | 4.478 | pW147bH | |
| Petal width–CS | 54.59 (42.4–68.06) | 13.824 | −0.134 | 4.516 | pW214aX | |
| Petal width–WL | 40.4 (26.01–52.59) | 14.191 | −0.108 | 5.193 | pW189aH | |
| Petal width–CS | 142.71 (138.7–161.05) | 18.059 | 0.151 | 5.785 | fito434 | |
| Petal width–WL | 145.76 (132.2–161.05) | 15.013 | 0.117 | 5.770 | pW169aX | |
| Petal area–CS | 54.59 (44.39–66.06) | 17.058 | −1.218 | 6.334 | pW214aX | |
| Petal area–WL | 35.21 (0.01–44.39) | 13.634 | −0.768 | 5.709 | pX144bE | |
| Stem length–CS | 58.25 (44.39–70.78) | 14.227 | −1.361 | 5.102 | pX141cE | |
| ###Stem length–CL | 61.48 (54.59–70.78) | 12.208 | −1.316 | 5.182 | pX141cE | |
| Stem length–WL | 59.48 (54.59–75.72) | 18.529 | −1.538 | 6.035 | pX141cE | |
| Days to flowering–CL | 44.39 (31.83–55.73) | 21.592 | −1.895 | 10.892 | pW174cX | |
| Herkogamy–CS | 40.4 (31.83–44.39) | 16.317 | −0.115 | 6.500 | pW189aH | |
| Herkogamy–WL | 40.4 (31.83–44.39) | 17.725 | −0.097 | 6.224 | pW189aH | |
| Stamen exsertion–CS | 55.73 (46.39–69.45) | 13.586 | −0.056 | 7.297 | pW214aX | |
| Pistil exsertion–WL | 55.73 (46.39–59.48) | 17.005 | −0.104 | 6.509 | pW214aX | |
| Chromosome 4 | ||||||
| Short anther length–CS | 47.35 (28.01–58.53) | 13.772 | −0.057 | 4.945 | pW120dX | |
| Short anther length–CL | 49.35 (37.39–58.53) | 22.018 | −0.081 | 8.140 | pW120dX | |
| Long anther length–CS | 47.35 (28.01–58.53) | 13.111 | −0.055 | 4.677 | pW120dX | |
| Long anther length–CL | 47.35 (38.52–58.53) | 18.930 | −0.069 | 6.525 | pW120dX | |
| Long filament length–CS | 53.35 (37.39–64.62) | 13.728 | −0.110 | 6.124 | pX129dX | |
| Long filament length–WL | 57.35 (47.01–79.62) | 13.455 | −0.105 | 4.948 | pX129dX | |
| Petal claw–CS | 57.35 (49.35–58.53) | 38.401 | −0.118 | 13.804 | pX129dX | |
| Petal claw–CL | 53.35 (47.01–60.63) | 29.217 | −0.115 | 11.974 | pX129dX | |
| Stem length–CL | 96.09 (80.3–106.09) | 24.013 | −1.768 | 9.403 | pW178bE | |
| Herkogamy–CS | 69.62 (60.53–80.3) | 16.314 | 0.093 | 6.741 | pW246fX | WL/CS‡ |
| Pistil exsertion–CS | 67.62 (58.53–79.62) | 20.220 | 0.134 | 8.621 | pW246fX | WL/CS‡ |
| Chromosome 5 | ||||||
| Short filament length–WL | 98.67 (85.84–103.94) | 13.447 | 0.088 | 5.117 | pW143aE | |
| Chromosome 6 | ||||||
| Style length–CS | 27.86 (17.5–37.59) | 23.386 | −0.073 | 9.232 | pW160bH | |
| Style length–CL | 27.86 (19.5–33.27) | 39.796 | −0.105 | 15.227 | pW160bH | |
| Style length–WL | 29.86 (25.5–48.59) | 15.522 | −0.064 | 5.647 | pW160bH | |
| Ovary length–CS | 72.54 (63.4–94.15) | 31.303 | −0.150 | 13.608 | BRMS026b | |
| Ovary length–CL | 78.15 (67.09–98.15) | 24.863 | −0.167 | 13.274 | BRMS026b | |
| Ovary length–WL | 57.03 (42.59–67.09) | 27.071 | −0.134 | 9.947 | fito423 | |
| Short anther length–CL | 0.01 (0.01–12.01) | 13.243 | 0.064 | 4.780 | pW227aE | |
| Short anther length–CS | 65.56 (63.4–70.54) | 32.793 | −0.097 | 12.506 | fito227 | |
| Short anther length–CL | 65.4 (61.95–70.54) | 27.190 | −0.093 | 9.203 | pW219aE | |
| Short anther length–WL | 65.4 (60.63–70.54) | 23.314 | −0.070 | 8.220 | pW219aE | |
| Short filament length–CS | 0.01 (0.01–10.01) | 15.365 | 0.124 | 7.043 | pW227aE | |
| Short filament length–CL | 0.01 (0.01–10.01) | 16.210 | 0.166 | 8.735 | pW227aE | |
| ###Short filament length–WL | 0.01 (0.01–10.01) | 12.124 | 0.088 | 4.835 | pW227aE | |
| Short filament length–cl | 25.5 (16.01–37.59) | 13.702 | −0.160 | 6.692 | pW160bH | |
| Short filament length–CS | 65.4 (57.63–92.15) | 17.622 | −0.135 | 7.575 | pW219aE | WL/CS† |
| Long anther length–CS | 0.01 (0.01–10.01) | 15.183 | 0.063 | 5.771 | pW227aE | |
| Long anther length–CL | 25.86 (16.01–33.27) | 15.074 | −0.073 | 5.136 | pW160bH | |
| Long anther length–CS | 65.4 (63.4–82.15) | 37.633 | −0.102 | 14.547 | pW219aE | |
| Long anther length–CL | 65.56 (51.03–70.54) | 16.076 | −0.070 | 5.395 | pW219aE | |
| Long anther length–WL | 74.15 (72.54–92.15) | 33.840 | −0.088 | 14.071 | BRMS026b | |
| Long filament length–CS | 74.15 (63.09–92.15) | 14.188 | −0.107 | 5.928 | BRMS026b | |
| Petal blade–CS | 32.7 (16.01–40.59) | 13.013 | −0.113 | 4.437 | fito068 | |
| Petal blade–CS | 65.4 (63.09–88.15) | 18.690 | −0.146 | 6.789 | pW219aE | |
| Petal claw–CL | 25.5 (17.5–32.7) | 26.217 | −0.107 | 9.330 | pW160bH | |
| Petal claw–CS | 70.54 (61.95–86.15) | 20.420 | −0.088 | 6.600 | fito227 | |
| Petal claw–WL | 74.15 (63.09–96.15) | 16.827 | −0.071 | 6.470 | BRMS026b | |
| Petal width–CL | 25.86 (17.5–37.59) | 16.366 | −0.186 | 5.650 | pW160bH | |
| Petal width–WL | 32.7 (17.5–46.59) | 14.689 | −0.122 | 5.477 | fito068 | |
| Petal width–CS | 70.54 (63.4–76.15) | 35.125 | −0.241 | 12.709 | fito227 | WL/CS§ |
| Petal width–CL | 65.4 (63.09–67.09) | 30.239 | −0.250 | 11.090 | pW219aE | WL/CL‡ |
| ###Petal width–WL | 61.95 (49.03–74.15) | 12.422 | −0.111 | 4.613 | pX119dX | |
| Petal area–CL | 27.86 (17.5–33.27) | 23.240 | −1.988 | 10.772 | pW160bH | WL/CL‡ |
| Petal area–CS | 72.54 (63.09–96.15) | 28.773 | −1.775 | 12.023 | BRMS026b | WL/CS‡ |
| Petal area–CL | 72.54 (63.09–96.15) | 16.940 | −1.548 | 6.582 | BRMS026b | WL/CL‡ |
| Leaf area–CS | 58.63 (51.03–61.95) | 18.110 | 1.265 | 7.806 | fito378b | WL/CS† |
| Stem length–CL | 25.86 (16.01–37.59) | 13.531 | −1.365 | 5.028 | pW160bH | |
| Stem length–CL | 80.15 (51.03–102.15) | 14.882 | −1.666 | 7.965 | BRMS026b | |
| ###Stem length–WL | 82.15 (32.7–112.44) | 12.633 | −1.479 | 6.416 | BRMS026b | |
| Collection interval–CS | 61.95 (57.03–74.15) | 13.069 | 0.521 | 6.478 | pW200bX | |
| Collection interval–WL | 65.4 (57.03–80.15) | 13.607 | 0.385 | 6.690 | pW219aE | |
| Herkogamy–WL | 49.03 (39.59–61.95) | 24.311 | −0.111 | 8.147 | pW127aE | |
| Pistil exsertion–CL | 49.03 (21.5–57.63) | 19.738 | −0.131 | 9.066 | pW127aE | |
| Pistil exsertion–CL | 82.15 (70.54–94.15) | 12.872 | −0.136 | 9.812 | BRMS026b | |
| Chromosome 7 | ||||||
| Style length–CS | 4.01 (0.01–17.3) | 14.896 | 0.061 | 6.990 | BRMS040 | |
| Style length–CL | 2.01 (0.01–17.3) | 13.517 | 0.058 | 4.928 | BRMS040 | |
| Ovary length–CS | 47.23 (25.82–56.99) | 15.917 | 0.109 | 7.263 | fito348 | |
| Ovary length–WL | 40.96 (25.82–56.99) | 25.484 | 0.133 | 9.551 | fito348 | |
| Short anther length–CL | 56.99 (43.23–74.23) | 13.375 | 0.063 | 3.980 | pW130cE | |
| Short filament length–CL | 86.82 (72.87–96.88) | 13.825 | −0.166 | 7.501 | fito101b | |
| Short filament length–WL | 79.11 (70.08–96.82) | 21.913 | −0.120 | 9.005 | pX101cH | |
| Long anther length–CS | 56.99 (42.96–70.87) | 15.768 | 0.070 | 5.324 | pW130cE | |
| Long anther length–CL | 86.82 (79.91–96.88) | 21.503 | −0.088 | 8.428 | fito101b | |
| ###Long anther length–WL | 84.82 (72.87–84.82) | 12.707 | −0.053 | 4.886 | fito101b | |
| Petal claw–CS | 43.23 (36.96–56.99) | 21.891 | 0.085 | 7.034 | fito348 | |
| Petal claw–CL | 47.23 (36.96–56.99) | 26.090 | 0.109 | 10.685 | fito348 | |
| Petal claw–WL | 28.62 (25.82–35.08) | 16.987 | 0.075 | 7.190 | pW108aH | |
| Stem length–CS | 96.88 (84.82–96.9) | 24.563 | −1.985 | 9.994 | pW150cH | |
| Stem length–CL | 96.88 (84.82–96.9) | 15.932 | −1.475 | 6.010 | pW150cH | |
| Days to flowering–CS | 96.88 (88.82–96.9) | 19.008 | −2.052 | 9.320 | pW150cH | CL/CS‡ WL/CS† |
| Herkogamy–WL | 19.43 (0.01–33.4) | 12.736 | 0.081 | 4.134 | pW200cX | |
| Pistil exsertion–WL | 19.82 (16.01–30.62) | 16.842 | 0.106 | 6.175 | fito105 | |
| Pistil exsertion–CL | 56.99 (33.79–72.87) | 14.566 | 0.115 | 6.810 | pW130cE | |
| Chromosome 8 | ||||||
| Style length–WL | 0.01 (0.01–25.18) | 19.053 | −0.066 | 6.264 | BRMS006 | |
| Style length–CL | 49.04 (46.23–57.19) | 14.197 | −0.055 | 4.580 | pW146aH | |
| Style length–WL | 53.19 (31.18–57.19) | 16.734 | −0.065 | 6.005 | pW245bX | |
| Petal blade–CS | 51.04 (46.07–57.19) | 22.502 | −0.171 | 8.002 | pW245bX | |
| Petal area–CS | 49.04 (46.07–57.19) | 17.594 | −1.524 | 6.760 | pW146aH | |
| Herkogamy–CL | 0.01 (0.01–25.18) | 13.657 | 0.100 | 5.687 | BRMS006 | |
| Herkogamy–CS | 46.07 (34.07–57.19) | 17.133 | −0.092 | 6.728 | fito429 | |
| Herkogamy–CL | 46.07 (34.07–57.19) | 17.916 | −0.114 | 7.770 | fito429 | |
| Pistil exsertion–CS | 57.19 (34.07–57.19) | 15.446 | −0.103 | 6.884 | pW120cX | |
| Pistil exsertion–CL | 51.19 (42.07–57.19) | 23.714 | −0.155 | 12.526 | pW245bX | |
| Pistil exsertion–WL | 57.19 (36.07–57.19) | 15.872 | −0.099 | 6.015 | pW120cX | |
| Chromosome 9 | ||||||
| Ovary length–CL | 36.64 (22.46–39.31) | 13.365 | −0.136 | 5.223 | pW188cE | |
| Ovary length–CS | 72.45 (69.65–76.89) | 21.745 | −0.175 | 8.779 | pX119bX | |
| Ovary length–CL | 72.45 (67.65–76.89) | 15.395 | −0.154 | 6.251 | pX119bX | |
| Ovary length–CL | 100.29 (91.38–107.79) | 14.652 | −0.138 | 5.789 | pW246cX | |
| Short filament length–WL | 14.46 (0.01–23.46) | 19.844 | −0.118 | 8.727 | pX123dH | |
| Long filament length–WL | 12.46 (0.01–23.46) | 13.254 | −0.097 | 4.874 | pX123dH | |
| Long filament length–CS | 72.45 (61.71–85.38) | 13.388 | −0.113 | 5.510 | pX119bX | |
| Long filament length–WL | 85.13 (74.89–89.38) | 37.831 | −0.165 | 14.533 | fito151a | |
| Petal claw–CS | 2.01 (0.01–23.46) | 15.139 | −0.070 | 5.018 | fito097b | |
| Petal claw–CL | 8.01 (0.01–23.46) | 13.419 | −0.075 | 5.146 | pX123dH | |
| ###Petal claw–WL | 10.46 (0.01–23.46) | 12.774 | −0.062 | 4.879 | pX123dH | |
| Petal claw–CS | 91.38 (78.89–100.29) | 23.582 | −0.092 | 8.970 | fito151a | |
| Petal claw–CL | 95.38 (78.89–100.29) | 15.779 | −0.082 | 6.592 | pW246cX | |
| Petal claw–WL | 89.38 (72.89–100.29) | 18.244 | −0.081 | 8.260 | fito151a | |
| ###Stem length–CS | 23.46 (10.01–35.52) | 12.195 | −1.491 | 4.791 | pX123dH | |
| Stem length–WL | 23.46 (10.01–35.52) | 13.966 | −1.253 | 4.370 | pX123dH | |
| Stem length–CL | 89.38 (72.89–100.29) | 13.080 | −1.332 | 5.504 | fito151a | |
| Days to flowering–WL | 61.57 (53.24–72.45) | 13.934 | −1.438 | 5.047 | fito100b | |
| Collection interval–CS | 37.31 (23.46–48.43) | 15.106 | 0.490 | 7.626 | pW130cX | |
| Collection interval–CL | 25.46 (22.46–48.74) | 14.275 | 0.489 | 8.919 | pX123dH | |
| Herkogamy–WL | 34.96 (22.46–37.31) | 13.421 | −0.109 | 4.674 | BRMS016 | |
| Stamen exsertion–WL | 22.46 (10.01–35.52) | 14.400 | −0.060 | 5.565 | pX123dH | |
| Stamen exsertion–WL | 93.38 (85.13–100.29) | 21.824 | −0.082 | 11.276 | pW246cX | |
| Pistil exsertion–WL | 36.64 (22.46–48.43) | 17.156 | −0.106 | 6.684 | pW188cE | |
| Chromosome 10 | ||||||
| Ovary length–CL | 72.54 (61.1–90.55) | 16.635 | 0.119 | 6.517 | pW120aX | |
| Flowers aborted–CS | 51.25 (42.79–51.83) | 18.311 | 0.491 | 9.354 | pW256aE | |
| Flowers aborted–WL | 51.25 (44.79–51.83) | 19.488 | 0.514 | 10.862 | pW256aE | |
| Stem length–WL | 50.79 (20.94–59.1) | 14.568 | −1.350 | 4.743 | pW255aE | WL/CL† |
| Days to flowering–CL | 70.54 (52.39–84.55) | 15.355 | −1.552 | 7.304 | fito520a | |
| Days to flowering–WL | 51.83 (38.94–53.1) | 32.071 | −2.127 | 12.489 | pW174aX | |
| Collection interval–CS | 16.01 (0.01–38.94) | 17.460 | 0.586 | 10.471 | pX139cH | |
| Collection interval–CL | 20.94 (0.01–42.79) | 12.981 | 0.449 | 7.347 | FLC1aE | |
| Collection interval–WL | 22.94 (2.01–40.94) | 28.733 | 0.539 | 16.509 | FLC1aE | |
| Herkogamy–CS | 70.54 (61.1–88.55) | 14.427 | 0.085 | 5.521 | fito520a | |
| Herkogamy–WL | 72.54 (62.04–94.55) | 14.833 | 0.087 | 4.835 | pW120aX | |
Abbreviations: CL, cool temperature long photoperiod; CS cool temperature short photoperiod; WL, warm temperature long photoperiod.
The authors declare no conflict of interest.
References
- Armbruster WS, Di Stilio VS, Tuxill JD, Flores TC, Runk JLV. Covariance and decoupling of floral and vegetative traits in nine neotropical plants: a re-evaluation of Berg's correlation-pleiades concept. Am J Bot. 1999;86:39–55. [PubMed] [Google Scholar]
- Armbruster WS, Schwaegerle KE. Causes of covariation of phenotypic traits among populations. J Evol Biol. 1996;9:261–276. [Google Scholar]
- Ashman TL, Majetic CJ. Genetic constraints on floral evolution: a review and evaluation of patterns. Heredity. 2006;96:343–352. doi: 10.1038/sj.hdy.6800815. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B-Methodol. 1995;57:289–300. [Google Scholar]
- Berg RL. The ecological significance of correlation pleiades. Evolution. 1960;14:171–180. [Google Scholar]
- Bolmgren K, Cowan PD. Time and size tradeoffs: a phylogenetic comparative study of flowering time, plant height and seed mass in a north-temperate flora. Oikos. 2008;117:424–429. [Google Scholar]
- Brock MT, Maloof J, Weinig C.Floral genetic architecture: an examination of QTL architecture underlying floral (co)variation across environments (in review). [DOI] [PMC free article] [PubMed]
- Brock MT, Weinig C. Plasticity and environment-specific covariances: an investigation of floral-vegetative and within flower correlations. Evolution. 2007;61:2913–2924. doi: 10.1111/j.1558-5646.2007.00240.x. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Carr DE, Fenster CB. Levels of genetic variation and covariation for Mimulus (Scrophulariaceae) floral traits. Heredity. 1994;72:606–618. [Google Scholar]
- Cheverud JM. Developmental integration and the evolution of pleiotropy. Am Zool. 1996;36:44–50. [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J, Via S. Patterns of phenotypic and genetic correlations among morphological and life-history traits in wild radish, Raphanus raphanistrum. Evolution. 1993;47:704–711. doi: 10.1111/j.1558-5646.1993.tb02128.x. [DOI] [PubMed] [Google Scholar]
- Conner JK. Genetic mechanisms of floral trait correlations in a natural population. Nature. 2002;420:407–410. doi: 10.1038/nature01105. [DOI] [PubMed] [Google Scholar]
- Conner JK, Davis R, Rush S. The effect of wild radish floral morphology on pollination efficiency by 4 taxa of pollinators. Oecologia. 1995;104:234–245. doi: 10.1007/BF00328588. [DOI] [PubMed] [Google Scholar]
- Conner JK, Franks R, Stewart C. Expression of additive genetic variances and covariances for wild radish floral traits: comparison between field and greenhouse environments. Evolution. 2003;57:487–495. doi: 10.1111/j.0014-3820.2003.tb01540.x. [DOI] [PubMed] [Google Scholar]
- Conner JK, Sterling A. Testing hypotheses of functional relationships--a comparative survey of correlation patterns among floral traits in 5 insect-pollinated plants. Am J Bot. 1995;82:1399–1406. [Google Scholar]
- Conner JK, Sterling A. Selection for independence of floral and vegetative traits: evidence from correlation patterns in five species. Can J Bot. 1996;74:642–644. [Google Scholar]
- Edwards CE, Haselhorst MSH, McKnite AM, Ewers BE, Williams DG, Weinig C. Genotypes of Brassica rapa respond differently to plant-induced variation in air CO2 concentration in growth chambers with standard and enhanced venting. Theor Appl Genet. 2009;119:991–1004. doi: 10.1007/s00122-009-1103-5. [DOI] [PubMed] [Google Scholar]
- Fry JD, Heinsohn SL, Mackay TFC. The contribution of new mutations to genotype-environment interaction for fitness in Drosophila melanogaster. Evolution. 1996;50:2316–2327. doi: 10.1111/j.1558-5646.1996.tb03619.x. [DOI] [PubMed] [Google Scholar]
- Gomez Campo C. Biology of Brassica coenospecies. Elsevier: Amsterdam; 1999. [Google Scholar]
- Gurganus MC, Fry JD, Nuzhdin SV, Pasyukova EG, Lyman RF, Mackay TFC. Genotype-environment interaction at quantitative trait loci affecting sensory bristle number in Drosophila melanogaster. Genetics. 1998;149:1883–1898. doi: 10.1093/genetics/149.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedhly A, Hormaza JI, Herrero M. Influence of genotype-temperature interaction on pollen performance. J Evol Biol. 2005;18:1494–1502. doi: 10.1111/j.1420-9101.2005.00939.x. [DOI] [PubMed] [Google Scholar]
- Iniguez-Luy F, Lukens L, Franham MW, Amasino RM, Osborn TC. Development of public immortal mapping populations, molecular markers and linkage maps for rapid cycling Brassica rapa and B. oleracea. Theor Appl Genet. 2009;120:31–43. doi: 10.1007/s00122-009-1157-4. [DOI] [PubMed] [Google Scholar]
- Jermstad KD, Bassoni DL, Jech KS, Ritchie GA, Wheeler NC, Neale DB. Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas fir. III. Quantitative trait loci-by-environment interactions. Genetics. 2003;165:1489–1506. doi: 10.1093/genetics/165.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger T, Perez-Perez JM, Bernal S, Micol JL. Quantitative trait loci mapping of floral and leaf morphology traits in Arabidopsis thaliana: evidence for modular genetic architecture. Evol Dev. 2005;7:259–271. doi: 10.1111/j.1525-142X.2005.05028.x. [DOI] [PubMed] [Google Scholar]
- Juenger T, Purugganan M, Mackay TFC. Quantitative trait loci for floral morphology in Arabidopsis thaliana. Genetics. 2000;156:1379–1392. doi: 10.1093/genetics/156.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Choi SR, Bae J, Hong CP, Lee SY, Hossain MJ, et al. Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics. 2009;10:432. doi: 10.1186/1471-2164-10-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP. Morphological integration and developmental modularity. Annu Rev Ecol Evol Syst. 2008;39:115–132. [Google Scholar]
- Lande R. The genetic covariance between characters maintained by pleiotropic mutations. Genetics. 1980;94:203–215. doi: 10.1093/genetics/94.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littell RC, Milliken GA, Stroup WW, Wolfinger RD, Oliver S. SAS for Mixed Models, Second Edition. SAS Institute, Inc.: Cary, NC; 2006. [Google Scholar]
- Okamuro JK, Denboer BGW, Jofuku KD. Regulation of Arabidopsis flower development. Plant Cell. 1993;5:1183–1193. doi: 10.1105/tpc.5.10.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, denBoer BGW, LotysPrass C, Szeto W, Jofuku KD. Flowers into shoots: photo and hormonal control of a meristem identity switch in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:13831–13836. doi: 10.1073/pnas.93.24.13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oneil P, Schmitt J. Genetic constraints on the independent evolution of male and female reproductive characters in the tristylous plant Lythrum salicaria. Evolution. 1993;47:1457–1471. doi: 10.1111/j.1558-5646.1993.tb02168.x. [DOI] [PubMed] [Google Scholar]
- Pigliucci M, Marlow ET. Differentiation for flowering time and phenotypic integration in Arabidopsis thaliana in response to season length and vernalization. Oecologia. 2001;127:501–508. doi: 10.1007/s004420000613. [DOI] [PubMed] [Google Scholar]
- Prakash S, Hinata K. Taxonomy, cytogenetics and origin of crop Brassicas, a review. Opera Botanica. 1980;55:1–57. [Google Scholar]
- Rasband WS. ImageJ 1.38x. National Institutes of Health: Bethesda, MD, USA; 1997–2007. [Google Scholar]
- Ratcliffe O, Bradley D, Coen E. Separation of shoot and floral identity in Arabidopsis. Development. 1999;126:1109–1120. doi: 10.1242/dev.126.6.1109. [DOI] [PubMed] [Google Scholar]
- Robinson G. That BLUP is a good thing: the estimation of random effects. Stat Sci. 1991;6:15–51. [Google Scholar]
- Schmuths H, Bachmann K, Weber WE, Horres R, Hoffmann MH. Effects of preconditioning and temperature during germination of 73 natural accessions of Arabidopsis thaliana. Ann Bot. 2006;97:623–634. doi: 10.1093/aob/mcl012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kleunen M, Ritland K. Predicting evolution of floral traits associated with mating system in a natural plant population. J Evol Biol. 2004;17:1389–1399. doi: 10.1111/j.1420-9101.2004.00787.x. [DOI] [PubMed] [Google Scholar]
- Vieira C, Pasyukova EG, Zeng ZB, Hackett JB, Lyman RF, Mackay TFC. Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics. 2000;154:213–227. doi: 10.1093/genetics/154.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP. Homologues, natural kinds and the evolution of modularity. Am Zool. 1996;36:36–43. [Google Scholar]
- Wagner GP, Pavlicev M, Cheverud JM. The road to modularity. Nat Rev Genet. 2007;8:921–931. doi: 10.1038/nrg2267. [DOI] [PubMed] [Google Scholar]
- Waitt DE, Levin DA. Phenotypic integration and plastic correlations in Phlox drummondii (Polemoniaceae) Am J Bot. 1993;80:1224–1233. [Google Scholar]
- Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer. 2.5. Department of Statistics, North Carolina State University: Raleigh, NC; 2007. [Google Scholar]
- Weber K, Eisman R, Higgins S, Morey L, Patty A, Tausek M, et al. An analysis of polygenes affecting wing shape on chromosome 2 in Drosophila melanogaster. Genetics. 2001;159:1045–1057. doi: 10.1093/genetics/159.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Wang XW, Deng B, Lou P, Wu J, Sun RF, et al. Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor Appl Genet. 2005;110:1301–1314. doi: 10.1007/s00122-005-1967-y. [DOI] [PubMed] [Google Scholar]