Abstract
The evolution of subterranean animals following multiple colonisation events from the surface has been well documented, but few studies have investigated the potential for species diversification within cavernicolous habitats. Isolated calcrete (carbonate) aquifers in central Western Australia have been shown to contain diverse assemblages of aquatic subterranean invertebrate species (stygofauna) and to offer a unique model system for exploring the mechanisms of speciation in subterranean ecosystems. In this paper, we investigated the hypothesis that microallopatric speciation processes (fragmentation and isolation by distance (IBD)) occur within calcretes using a comparative phylogeographic study of three stygobiontic diving beetle species, one amphipod species and a lineage of isopods. Specimens were sequenced for the mitochondrial cytochrome c oxidase 1 gene from three main sites: Quandong Well, Shady Well (SW) and Mt. Windarra (MW), spanning a 15 km region of the Laverton Downs Calcrete. Phylogenetic and haplotype network analyses revealed that each species possessed a single divergent clade of haplotypes that were present only at the southern MW site, despite the existence of other haplotypes at MW that were shared with SW. IBD between MW and SW was evident, but the common phylogeographic pattern most likely resulted from fragmentation, possibly by a salt lake adjacent to MW. These findings suggest that microallopatric speciation within calcretes may be a significant diversifying force, although the proportion of stygofauna species that may have resulted from in situ speciation in this system remains to be determined.
Keywords: stygofauna, in situ fragmentation, calcrete aquifer, mitochondrial DNA
Introduction
The shift from an epigean existence to cave life and subsequent isolation can typically lead to dramatic speciation events and morphological adaptation (Niemiller et al., 2008). This process of speciation by transition from surface to subterranean environments is suggested as the main mechanism for the evolution of subterranean species, although there is some debate about whether it requires extinction of the surface ancestral species (climate relict hypothesis; for example Peck and Finston (1993)) or can proceed in the presence of gene flow between the surface and subterranean populations (adaptive shift hypothesis; for example Desutter-Grandcolas and Grandcolas (1996)). However, given the extraordinary number of species observed in cave systems (Poulson and Culver, 1969; Humphreys, 2000, 2008; Buhay et al., 2007), this transition, from surface to subterranean environments, is unlikely to be the sole source of species diversification in cave animals and it is possible that some species evolved from troglomorphic ancestors fully adapted to life underground (Juan and Emerson, 2010; Juan et al., 2010; Ribera et al., 2010). The difficult nature of gaining access to cave habitats makes this latter hypothesis difficult to investigate; however, a number of key studies have examined these questions (Caccone, 1985; Buhay and Crandall, 2005; Guzik et al., 2009). They observe that, internally, cavernicolous habitats maintain complex genetic systems that, undisturbed, are largely stable over time and can maintain significant levels of genetic diversity (Culver, 1970a, 1970b; Culver et al., 1973). This, despite the substantial climatic oscillations during the Pleistocene, suggests that cave systems may share similarities with other well-known closed ecosystems in which sympatric, parapatric or microallopatric (allopatry in situ) speciation has been documented (for example, sticklebacks in Nicaraguan crater lakes (Taylor and McPhail, 2000), palms on Lord Howe Island (Savolainen et al., 2006), spiders on Hawaiian Islands (Gillespie, 2004)). A cave system in the desert of outback Australia provides a further opportunity to investigate the possibility of in situ diversification of subterranean animals.
Calcrete (carbonate) aquifers in the Yilgarn region of Western Australia comprise some of the greatest diversity of short-range endemic (sensu; Harvey (2002)) cave species in the world. Each aquifer comprises similar stygobiontic organisms: crustaceans (for example, amphipods, copepods, isopods, bathynellids), oligochaetes (segmented worms) and water beetles (Dytiscidae; Humphreys, 2000), but each maintains a distinct set of species. The subterranean Dytiscidae (Watts and Humphreys, 2006) exemplify this diversity and are thought to represent the world's largest groundwater beetle fauna by a factor of 10 (Humphreys, 2006). The origin of this fauna is thought to lie in the mid-late Miocene when Central and Western Australia was a stable mesic habitat (Leys et al., 2003; Byrne et al., 2008). Subsequent aridification led to relictualisation of the fauna to habitats that retained water (for example, calcrete aquifers, mound springs and groundwater-fed lakes). Very likely, significant speciation probably occurred through vicariance at the time of colonisation and calcrete enclosure.
Evidence suggests that de novo speciation may also have occurred in situ well after these vicariance events (Cooper et al., 2002; Leys et al., 2003). A repeated pattern of two to three species of stygobiontic diving beetles is consistently observed within isolated aquifers, some of which are clear sympatric sister species (Leys et al., 2003; Watts and Humphreys, 2006; Guzik et al., 2009). To date, this pattern has been observed in 29 calcretes in which each species occupies non-overlapping size classes, ranging from large (∼5 mm) to very small (∼1 mm) beetles (Watts and Humphreys, 1999, 2000, 2001, 2003, 2004, 2006, 2009). Phylogenetic analyses using mitochondrial DNA (mtDNA) data have shown that 13 of these calcretes contain sympatric sister species (two with a sister triplet) and have further suggested that species were isolated within single calcretes ∼3–10 million years ago (mya; Cooper et al., 2002; Leys et al., 2003; Leys and Watts, 2008). Microallopatric or sympatric speciation is a plausible scenario for the origin of these species, in addition to other geographic modes of speciation between these two extremes (Butlin et al., 2008). Alternatively, repeated colonisation from the same ancestral source may also lead to such patterns. Currently, it is difficult to test the scenario of sympatric speciation, but identifying whether microallopatric speciation processes, such as population fragmentation, have operated within calcrete aquifers is a distinct possibility.
Guzik et al. (2009) investigated the population genetic and phylogeographic structure of three sympatric sister species of dytiscid diving beetle from a single aquifer. Their results revealed evidence for isolation by distance (IBD) effects in two of the three beetle species over a very small spatial scale. However, the study area of 3.5 km2 at Sturt Meadows Calcrete (SMC), although large in size relative to the inhabiting organisms, was not large enough to detect possible evidence of population fragmentation in the three species. In the present study, we gained access to a substantially larger sampling region (up to 15 km between samples compared with ∼3.5 km) in a calcrete aquifer that lies ∼135 km northeast of SMC and on a different palaeodrainage channel, Carey drainage (east), known as the Laverton Downs Calcrete (LDC). As in the previous study, we used a comparative phylogeographic approach using three dytiscid beetle species, Limbodessus lapostae Watts and Humphreys (1999), L. windarraensis Watts and Humphreys (1999) and L. palmulaoides Watts and Humphreys (2006), which vary in size from the smallest to the largest (1.3, 2.2 and 4.2 mm, respectively). The three species are congeneric and sympatric but are not sister species (Leys et al., 2003; Leys, unpublished). It is most likely that these three species evolved from three distinct ancestral species that were already reproductively isolated before their colonisation of and enclosure within the aquifer. An undescribed chiltoniid amphipod species and a lineage of Haloniscus isopods were also analysed to further examine whether similar geographic patterns of population subdivision applied to other taxa in the aquifer. The Haloniscus isopods were examined previously using mtDNA sequence analyses (Cooper et al., 2008), in which they were shown to form a reciprocally monophyletic clade, and a number of lineages were observed from LDC specimens with some evidence for phylogeographic structure. We included all of them in the current analyses and use the term ‘isopods' to broadly identify them.
The specific aim of this study was to investigate broad-scale (distances up to 15 km in an aquifer ∼90 km2 in size) population genetic structuring within a single calcrete aquifer for three dytiscid beetles, an amphipod and a lineage of isopods. We have used a mitochondrial gene marker and a multispecies phylogeographic approach to determine whether there were concordant patterns of genetic structure among taxa. It was predicted that if fragmentation of the range of each species had occurred in the past, the signal of this event should (a) be present and shared in the genes of each species and (b) reflect robust monophyletic groups of haplotypes with long branches connecting each group (Avise, 1994). In the absence of or with reduced recombination, as in mitochondrial markers, this fragmentation signal may still be present within extant populations even if gene flow subsequently recommenced. In the event of evidence for fragmentation, we aimed to test a hypothesis of IBD or vicariance (that is, potential microallopatry) within the subterranean aquatic habitat.
Materials and methods
Sampling
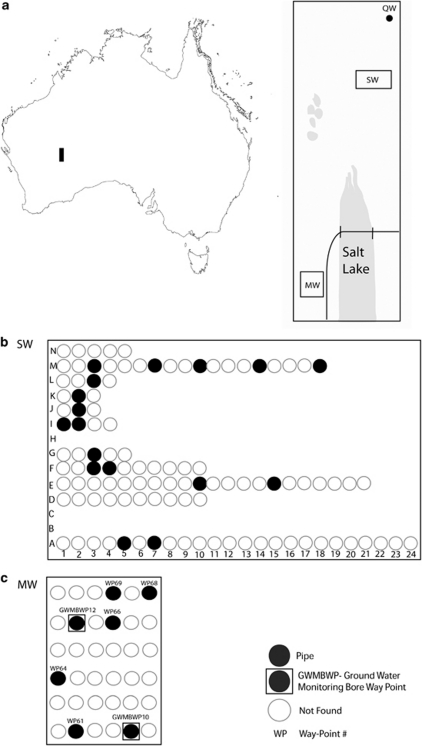
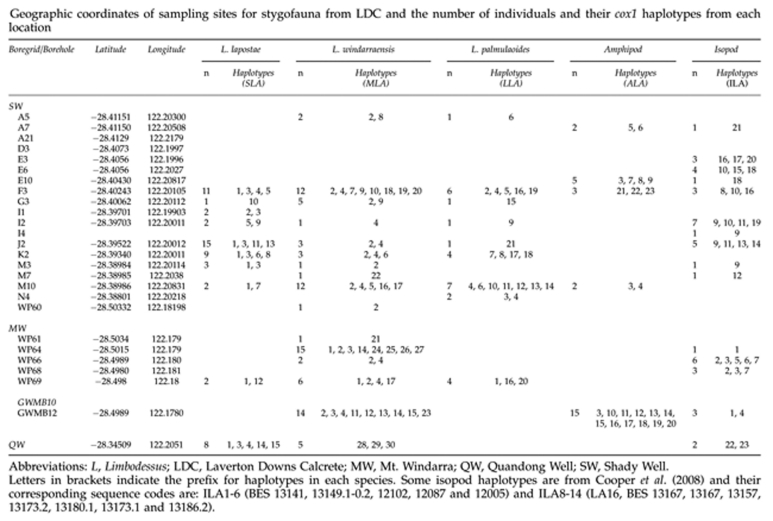
The three species of Dytiscidae, the amphipod and isopods were collected (years 2005–2007) from three key locations in the LDC of the Yilgarn region (Figure 1 and Appendix 1). Two of these locations comprise borefields, similar to that of SMC (Guzik et al., 2009), that are ∼9.5 km apart. These borefields are named Mt. Windarra (MW) and Shady Well (SW) consecutively from south to north. Each borefield contains a grid of up to 20 mineral exploration bores, but MW has substantially fewer bores than SW. At the time these bores were established, their primary purpose was not for biological study, which was also the case at the SMC (Guzik et al., 2009). Another site ∼6.5 km further north of the SW borefield, Quandong Well (QW), an active pastoral well, provided additional access to the calcrete. Specimens were collected using plankton nets as per Allford et al. (2008). The use of this method captured many different stygobiontic invertebrates and yielded large numbers of L. windarraensis but fewer specimens of L. lapostae and L. palmulaoides. The amphipod was collected relatively frequently, whereas the isopods were rare and considered potentially benthic. Specimens were preserved in the field using 100% ethanol and stored at −20 °C until DNA extraction. Voucher specimens of each of the three beetle species have been lodged in the South Australian Museum. Frozen DNA vouchers have also been lodged at the South Australian Museum's Evolutionary Biology Unit, located at the University of Adelaide.
Figure 1.
(a) Schematic map of Australia with location of LDC and a diagram of borefield locations at LDC, MW, SW and QW. Schematic diagram of borefields at (b) SW and (c) MW.
Sequencing and alignment
PCR was used to amplify a 689-bp fragment of the mtDNA cytochrome c oxidase subunit 1 (cox1) gene. The primers used for beetles were C1-J-2183 (5′-CAACATTTATTTTGATTTTTTGG-3′ Simon et al., 1994) and UEA10 (5′-TCCATTGCACTAATCTGCCATATTA-3′ Lunt et al., 1996); for the amphipods: LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) (Folmer et al., 1994); and the same primers as used in Cooper et al. (2008) for the isopods. PCR was carried out on a PalmCycler v.2.1.7 (Corbett Research, Sydney, Australia) thermocycler using 25 μl reaction volumes consisting of 17.4 μl of nuclease-free H2O, 2.5 μl of 2.5 × buffer, 0.5 m of dNTP's, 1.0 μl of 5 μ of each primer and 0.1 μl of HotmasterTaq (Eppendorf AG, Hamburg, Germany). The cycling conditions for cox1 consisted of one cycle at 94 °C for 2 min and 40–45 cycles (94 °C, 30 s; 48–54 °C, 30 s; and 72 °C, 60 s), followed by a final incubation step at 24 °C for 3 min. Amplified PCR products were then identified using gel electrophoresis. PCR products were purified using the AMPure (Agencourt Bioscience Corporation, Danvers, MA, USA) system according to the manufacturer's protocol. Purified products were sequenced using the ABI Prism Big Dye Terminator Cycle sequencing kit (PE Applied Biosystems, Foster City, CA, USA) in 10 μl reaction volumes, according to the manufacturer's protocol.
The sequence alignment program BioEdit v7.0.5.3 (Hall, 1999) was used to edit and align sequences. Unique haplotypes were identified using the program FaBox (Villesen, 2007). PAUP* 4.0b10 (Swofford, 2002) was used to compute corrected (HKY) pairwise genetic distances among haplotypes.
Phylogenetic analysis
To examine relationships within and among species and haplotypes, mtDNA cox1 sequences were analysed using a phylogenetic approach in the beetles and isopods. Phylogenetic relationships of amphipod populations were not presented here because of the high similarity between haplotypes among sampled individuals. Instead, haplotype networks were a better representation of these data (see below). Two out-group beetle taxa were chosen, Bidessodes limestonensis (Watts and Humphreys, 2003) and Bidessodes gutteridgei (Watts and Humphreys, 2003), which are distant relatives from the same tribe of diving beetles as the Limbodessus sampled from LDC (Bidessini) (Leys et al., 2003), and their sequences were obtained from Genbank (Genbank accession: AY350880 and AY350894, respectively). For the LDC isopod out-groups, we chose their nearest relatives on the basis of the phylogeny of Cooper et al. (2008). In that study, the taxa (BES10582: EU364601 and BES 11811: EU64602) from the Mt. Morgan calcrete on the Carey palaeodrainage channel formed a monophyletic relationship with those from LDC. In addition to new sequence data for the isopods, a number of sequences were obtained from Genbank (BES12005, 12087, 12102, 13141, 13149.1: EU364595-600 and BES10291.1 (13167), 10291.2 (13157), 12021.1 (13173.2), BES13173.1, 13180.1, 13186.2: EU364589-4). Phylogenetic reconstruction using a Bayesian approach was implemented with MrBayes 3.1.2 (Huelsenbeck and Ronquist, 2001). The model that best fit the higher level sequence data for both beetles and isopods was estimated with MODELTEST 3.7 (Posada and Crandall, 1998) for nucleotide data under an Akaike Information Criterion framework. Models were tested for all three codon positions: the HKY+I+G model was favoured for the first and second, and GTR+I+G for the third position. The nucleotide sequence data were partitioned by codon position and each partition was started independently with a different model (listed above). All parameters were unlinked and the rates were allowed to vary over the partitions. This approach circumvents part of the problem of saturation at the third codon position because each position is treated independently and not assumed to be evolving at the same rate. Four chains were run simultaneously for 5 000 000 generations in two independent runs, sampling trees every 100 generations. To evaluate convergence to the stationary distribution, the program Tracer 1.4 (Drummond and Rambaut, 2007) was used. The likelihood values converged to relative stationarity after about 75 000 generations. A burn-in of 1 250 000 was chosen and a 50% consensus tree was constructed from the remaining trees.
Population analyses
A hierarchical approach was taken to examine the partitioning of genetic variation within and between populations. The highest population level consisted of the three main sites from south to north that were separated by more than 6.5 km: MW, SW and QW, described here as ‘borefield'. Although QW is a single bore, we also use the term ‘borefield' as a general term for the three broad populations. The next hierarchical level was that of individual ‘bore holes', which permitted a fine-scale examination of the genetic variance. ARLEQUIN v.3.1 (Excoffier et al., 2005) was used to carry out analysis of molecular variance, determine nucleotide compositions, as well as carry out population comparisons. ARLEQUIN was also used to test for population differentiation using 100 000 Markov chain steps and 10 000 dememorisation steps, at P=0.05. TCS v.1.21 (Clement et al., 2000) was used to generate and arrange haplotype networks for each of the three beetles and the amphipod at a 95% connection limit. Networks were not presented for the isopods as this parsimony-based analysis does not adequately reflect the distinctive relationships among some of the haplotypes.
It must be noted here that, distinct from our a priori population groupings discussed above, our results also revealed genetically distinct clades among all taxa. Individuals bearing haplotypes for one of these clades were localised in their distribution to the MW borefield (MW-only clade) and the other haplotypes were found to be widespread in their distribution and were generally found in all borefields (widespread-LAV clade). This pattern was found particularly for the three beetle species and to a lesser extent for the isopods (see results section for details). The amphipods showed some evidence of genetic differentiation relative to geographic location, but haplotypes were too closely related to warrant discussion of an MW-only clade. Therefore, these were also included in some of the population analyses.
The demographic history of individual populations was assessed with ARLEQUIN v3.1 by computing Fu's (Fu, 1997) Fs (Fs), Tajima's (Tajima, 1989) D (D), parameters for the model of population expansion (time since expansion (τ), and relative population sizes before (θ1) and after (θ2) expansion) and for the continent-island model of demographic expansion (τ=2Tμ, θ=2Nμ and M=2 Nm, where T=number of generations before spatial expansion, μ=mutation rate, N=size of deme (assumed constant) and m=fraction of individuals from a deme exchanging with other demes). The generalised least-squares approach (Schneider and Excoffier, 1999) in ARLEQUIN was used to test the empirical mismatch distribution against a model of demographic expansion. We used DnaSP v4.20.2 (Rozas et al., 2003) to calculate R2 (Ramos-Onsins and Rozas, 2002) for detecting population growth in the same genetic populations on the basis of the number of segregating sites: an assumption of no recombination and 10 000 replicates. The model of IBD was tested using a Mantel test following the approach of Slatkin (1993) using the Alleles In Space program (Miller, 2005) with 10 000 randomisations. The tests were conducted with pairwise comparisons between each of the three borefields rather than all three at once. The rationale for this was that an artificial IBD signature was observed when all three locations were tested because of the presence of genetically distinct haplotypes at the MW borefield only (MW-only clade see below). This IBD signature did not reflect the presence of widespread and shared haplotypes at all borefields (widespread-LAV clade haplotypes) because of the MW-only clade haplotypes being found only in the southern borefield. The pairwise Mantel test allowed us to test for IBD between borefields while minimising the impact of the MW-only clade.
Estimating coalescent time of mtDNA sequences
BEAST v1.4.7 (Drummond and Rambaut, 2007) was used to estimate the coalescence time (that is, time to most recent common ancestor of mtDNA sequences for each species). The subprogram BEAUti v1.4.7 (Drummond and Rambaut, 2007) was used to create input .xml files, and Tracer v1.4 (Drummond and Rambaut, 2007) was used to analyse the parameter distributions estimated from BEAST. An UPGMA starting tree was estimated under the HKY+I+G model in which (a) base frequencies were estimated, (b) codon positions were partitioned (positions 1+2, 3) and (c) the parameters, substitution model across codon positions and rate heterogeneity model were unlinked. The substitution rate was fixed at 0.0115 (standard arthropod mtDNA molecular clock of 2.3% divergence per million years (Brower, 1994)), and a relaxed clock (uncorrelated lognormal) was used. A number of different models built earlier were implemented separately on each of the LDC species using BEAST (for example, Yule speciation, and coalescent: expansion, exponential, constant and Bayesian Skyline). We found that each of the previous models made little difference to the final time estimate and that these estimates were very similar (within 0.2 mya); therefore, we present results using the Yule model only. Each analysis was run for 10 000 000 generations, with sampling every 100 generations, and the burn-in was 25% of the total sampled trees (that is, 25 000). Each analysis was run multiple times and all estimated dates were consistent with those presented here.
Results
Mitochondrial cox1 sequences of 689-bp length were obtained from L. lapostae (n=55, h=15), L. windarraensis (n=84, h=30) and L. palmulaoides (n=27, h=21), of 618 bp for the amphipod species (n=80, h=43) and of 605 bp (n=42, h=23) for the isopods (see Appendix 1 for details of haplotypes and location data). All sequences were free of insertions and deletions, and comprised an open reading frame, as expected of a functional gene, suggesting that the sequences were unlikely to represent mitochondrial pseudogenes. Sequences were deposited at Genbank: amphipods: ALA1-43, HQ535666-708; beetles: L. palmulaoides: LLA1-21, HQ535709-29; L. lapostae: SLA1-15, HQ535730-44; L. windarraensis: MLA1-30, HQ535745-74; and isopods: ILA7-8 (HQ535781-82), ILA15-23 (HQ535789-97).
Phylogenetic analysis
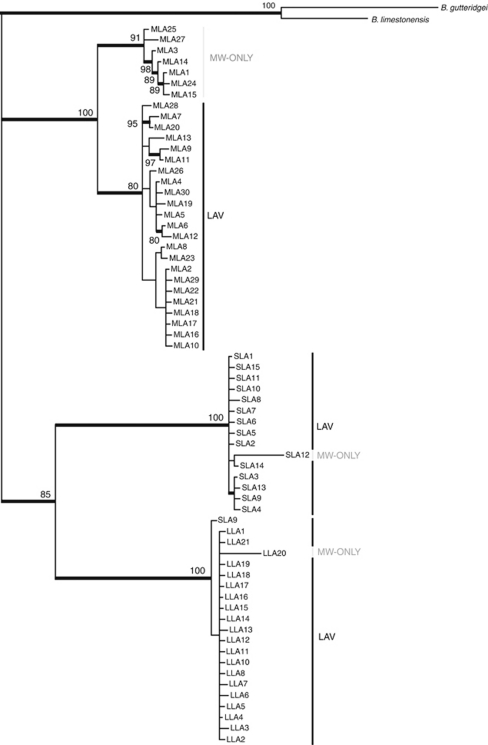
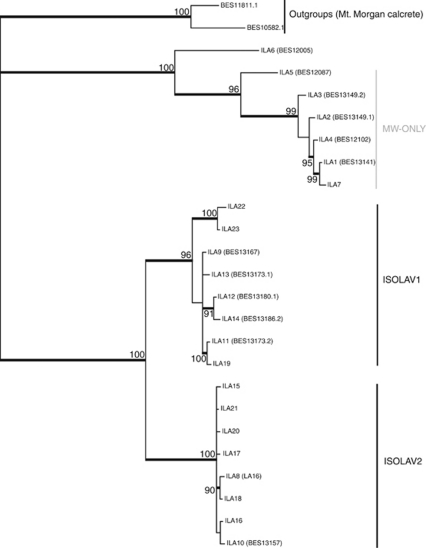
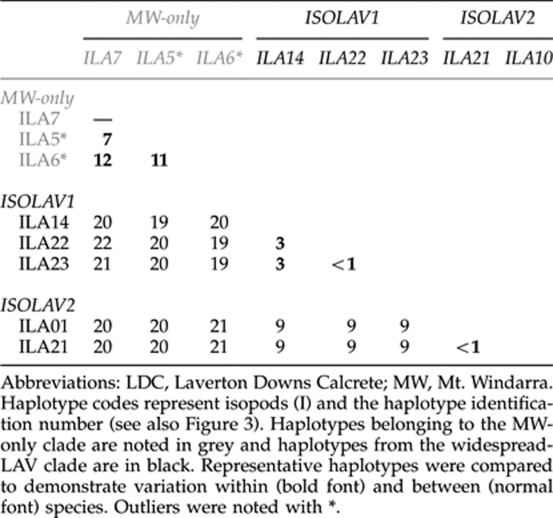
Each species showed significant phylogenetic structuring, with multiple divergent monophyletic groups of haplotypes. The Bayesian tree of haplotypes for all three beetle species (Figure 2) showed that each species possessed a small clade of individuals found only from the southern MW location (herein coded MW-only clades). All remaining individuals were widespread and represented throughout the LAVerton region by what we refer to here as the widespread-LAV clade for which haplotypes were generally found at all three borefields (that is, MW, SW and QW). The isopods (Figure 3) showed a similar pattern, with the presence of one divergent clade almost entirely restricted to MW and a second major clade restricted to SW+QW, the latter containing two subclades, referred to here as ISOLAV1 (SW+QW haplotypes) and ISOLAV2 (SW haplotypes). The only discrepancies in the geographic patterns between isopods and all other taxa were that the MW clade contained a single divergent haplotype (BES 12005: ILA6) from the SW region and there were no shared haplotypes between the MW and SW+QW regions.
Figure 2.
Consensus phylogeny estimated using a Bayesian approach that employed the HKY+I+G model of nucleotide evolution for haplotypes of L. lapostae, L. windarraensis and L. palmulaoides, with out-group taxa Bidessodes gutteridgei and B. limestonensis. Lists of specimens and haplotypes are given in the Appendix 1. Haplotype acronyms reflect small (S), medium (M) and large (L) size of the beetle species from LDC: MLA, L. windarraensis; SLA, L. lapostae; and LLA, L. palmulaoides. Both the MW-only clades and the widespread-LAV (LAV) clades are noted. Thickened black lines and numbers on branches indicate Bayesian posterior probabilities >80%.
Figure 3.
Consensus phylogeny estimated using a Bayesian approach that employed the GTR+I+G model of nucleotide evolution for haplotypes of the Haloniscus isopod. Numbers in brackets indicate sequences taken from Cooper et al. (2008) from LDC, with unknown out-group taxa (BES 11811.1 and BES 10582.1) from Mt. Morgan calcrete. Both the MW-only clades and the two widespread-LAV clades (ISOLAV1 and ISOLAV2) are noted. Thickened black lines indicate Bayesian posterior probabilities >80%.
Genetic distances
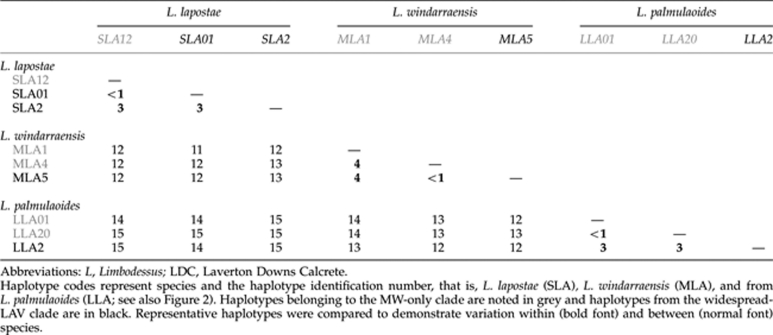
Corrected genetic distances (HKY model of base substitution) among and within the three beetle species showed that, for most individuals of the same species, genetic distance was rarely more than 2% (data not shown). Tables 1 and 2 show the percentage genetic distance between clades from locations throughout LDC. Consistent with the phylogenetic trees, the MW-only clade individuals in each species were substantially more divergent than the rest (widespread-LAV clade individuals; Table 1). In L. lapostae and L. palmulaoides, divergences were 0–3% among all haplotypes of each species. In L. windarraensis, seven haplotypes from southern MW sites (WP12, WP64 and WP69) showed high divergence (1–4%) compared with all others. Further, several haplotypes from these bores were also shared with individuals from the SW bores (widespread-LAV clade). Genetic distances were the highest between ‘known' species at 11–15% (Table 1), which is consistent with levels of inter-specific divergence for stygobiontic (Guzik et al., 2009) and epigean dytiscid congeners for cox1 (Ribera et al., 2003). The amphipod showed low levels of divergence among individuals, ranging between 0 and 2% (for instance, 2% divergence between ALA24 and ALA46). The most divergent haplotypes were from MW but these differences were not as great as those within beetle species. In contrast to the other species, the two major clades of isopods were highly divergent from each other (19–21% among haplotypes from each clade) and genetic divergence among haplotypes was also high within each clade (0–12% Table 2). Within the ISOLAV1 and ISOLAV2 subclades, divergences were typically 0–3%. Two private haplotypes that grouped closely with the MW-only clade were found only once in our samples (BES 12005 (ILA5) and 12087 (ILA6)) and were very divergent (9–12%) from all others from MW.
Table 1. Percentage of genetic distance (HKY model of substitution) between major clades in each of three stygobiontic beetle species from LDC.
Table 2. Percentage of genetic distance (HKY model of substitution) between major clades of stygobiontic Haloniscus isopods from LDC.
Molecular diversity
Haplotype diversity (h) was generally high for all regions in all species (Table 3) with the lowest estimates observed at SW and QW for L. windarraensis and L. lapostae (h=0.66–0.86). In contrast, nucleotide diversity (polymorphism) represented by the mean number of pairwise differences (π) was consistently low at SW and QW for each of the beetle species and in the amphipod at all locations, whereas the MW borefield showed very high nucleotide diversity for the beetles and isopods (π>8 average differences) because of the presence of highly divergent haplotypes in the MW borefield. The isopods also showed very high nucleotide diversity (π=23.9) at SW.
Table 3. Molecular diversity indices and population demographic parameters for three beetle species, the amphipod and isopods from three borefields on LDC: MW, SW and QW.
| Statistics | n | h | S | h | π | Fs | D | Demographic expansion |
R2 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| τ | θ0 | θ1 | SSD | |||||||||
| L. lapostae | ||||||||||||
| MW | 2 | 2 | 17 | 1.0±0.1 | 17.0 | 2.8 | 0 | — | — | — | — | 0.1 |
| SW | 45 | 12 | 13 | 0.7±0.1 | 0.9 | −9.1* | −2.1* | 1.0 | 0 | Inf. | 0.004 | 0.1* |
| QW | 8 | 5 | 4 | 0.9±0.1 | 1.4 | −2.0* | −0.5 | 1.5 | 0 | Inf. | 0.02 | 0.2 |
| L. windarraensis | ||||||||||||
| MW | 39 | 16 | 46 | 0.8±0.1 | 14.2 | 2.2 | 1.1 | 26.1 | 0.002 | 24.0 | 0.06* | 0.1 |
| SW | 40 | 14 | 20 | 0.8±0.1 | 4.3 | −1.9 | −0.3 | 7.7 | 0.002 | 7.0 | 0.06 | 0.1 |
| QW | 5 | 3 | 11 | 0.7±0.2 | 5.4 | 2.5 | 0.2 | 9.5 | 0.002 | 11.8 | 0.3 | 0.2 |
| L. palmulaoides | ||||||||||||
| MW | 4 | 3 | 17 | 0.8±0.2 | 8.7 | 2.4 | −0.7 | 17.1 | 0 | 3.4 | 0.2 | 0.3 |
| SW | 23 | 19 | 23 | 1.0±0.03 | 2.6 | −19.8* | −2.2* | 2.6 | 0 | Inf. | 0.01 | 0.1* |
| Amphipod | ||||||||||||
| MW | 35 | 23 | 27 | 1.0±0.03 | 4.9 | −12.6 | −0.9 | 6.6 | 0 | 14.3 | 0.006 | 0.1 |
| SW | 25 | 18 | 15 | 1.0±0.02 | 1.6 | −21.3 | −2.1* | 1.7 | 0.01 | Inf. | 0.003 | 0.1* |
| QW | 20 | 12 | 19 | 0.9±0.1 | 2.5 | −6.3 | −2.0* | 1.3 | 1.0 | Inf. | 0.003 | 0.1* |
| Isopods | ||||||||||||
| MW | 13 | 7 | 85 | 0.9±0.1 | 19.5 | 4.8 | −1.3 | 8.1 | 0.004 | 12.1 | 0.04 | 0.2 |
| SW | 27 | 17 | 55 | 0.9±0.04 | 23.9 | 1.3 | 2.6 | 45.6 | 0 | 61.87 | 0.07* | 0.1 |
| QW | 2 | 2 | 3 | 1.00±0.5 | 3.0 | 1.1 | 0 | 0 | 0 | 0 | 0 | 0.3 |
Abbreviations: D, Tajima's (1989) D; Fs, Fu's (1997) Fs; h, haplotype diversity; inf, infinite estimate; L, Limbodessus; LDC, Laverton Downs Calcrete; MW, Mt. Windarra; n, total number of individuals; QW, Quandong Well; R2, Ramos-Onsins and Rozas's (2002) R2; S, number of polymorphic sites; SSD, sum of squatter deviations between the observed and the expected mismatch as a test statistic; SW, Shady Well; π, nucleotide diversity as mean number of pairwise differences in the population; −, not estimated.
Model of demographic expansion parameters, where τ is an index of time since the expansion expressed in units of mutational time; θ0 and θ1 are pre- and post-expansion values for the mutation parameter (that is, 2Nμ, where N is the effective female population size and μ is the mutation rate per gene per generation); *, significance tests where P<0.05.
Haplotype networks
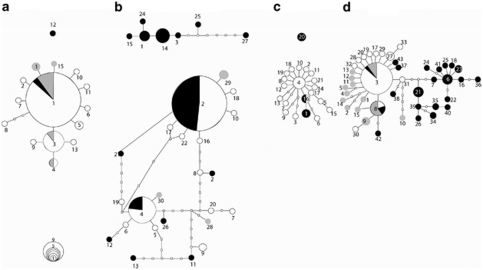
All species showed typical patterns of expansion (or selection) in haplotype networks in which a single dominant haplotype has experienced multiple single-point mutations, leading to many novel and rare haplotypes (Figure 4). In addition, as mentioned above, the beetle species all showed strikingly divergent haplotypes that were solely from MW populations and did not join the networks at 95% confidence in statistical parsimony analyses (Figure 4). The amphipod (Figure 4d) also showed some geographic divergence among individuals from this site; however, they differed less than that observed in the beetles. Amphipod haplotypes were mostly shared between the major populations of MW, SW and QW, excluding individuals from the MW-only clades. We have not shown isopod networks because of the lack of connectivity between numerous clades/haplotypes at the 95% confidence interval. We consider that the relationships between haplotypes were better represented by the phylogenetic analyses.
Figure 4.
Haplotype network of (a) L. lapostae, (b) L. windarraensis, (c) L. palmulaoides and (d) the amphipod. Each haplotype is represented by a circle, with the size of the circle corresponding to the number of individuals sharing the haplotype (key bottom left corner) and the proportion of haplotypes shaded indicating the fraction of individuals from a given location possessing that haplotype, with black representing MW; white, SW; and grey, QW. Each line represents a single base difference between two haplotypes, and missing haplotypes are the smallest circles.
Population structure
In an analysis of molecular variance, we examined three borefields that provided access to LDC (MW, SW and QW), with borefields comprising numerous individual bore holes at the former two locations and one at QW. Overall, substantial genetic differentiation was observed in the MW borefield (Table 4). In beetles, the majority of genetic variation was observed within individual bores and was significant in L. windarraensis, the amphipod and the isopods (60.53–80.27%, P⩽0.003). We attribute this diversity within bores to the two major haplogroups existing within sites at MW. However, some of the genetic variation was also attributed between borefields for the beetles (24.34–41.07%), but only significantly so for the isopod and the amphipod (35.11–74.16%, P⩽0.01). A confounding factor in beetle data was the presence of both types of highly divergent haplotypes at the southern MW borefield, but not so in the northern borefields (SW and QW). The clearest association between genetic data and spatial location was observed in isopods and the amphipod.
Table 4. AMOVA results estimated with ARLEQUIN (Excoffier et al., 2005) using three borefields on LDC (MW, SW and QW) and individual bores within each borefield to examine patterns of spatial genetic structure in three stygobiontic beetles L. lapostae, L. windarraensis and L. palmulaoides, the amphipod and isopods.
| Species | Sum of squares | Variance | Variation (%) | Fixation index |
|---|---|---|---|---|
| L. lapostae | ||||
| Among borefields | 8.86 | 0.46 Va | 41.07 | ΦSC: −0.03 |
| Among bores within borefields | 4.73 | −0.02 Vb | −1.60 | ΦST: 0.39 |
| Within bores | 29.50 | 0.67 Vc | 60.53 | ΦCT: 0.41 |
| L. windarraensis | ||||
| Among borefields | 71.76 | 1.43 Va | 24.34 | ΦSC: −0.06 |
| Among bores within borefields | 45.39 | −0.27 Vb | −4.61 | ΦST: 0.20a |
| Within bores | 320.46 | 4.71 Vc | 80.27 | ΦCT: 0.24a |
| L. palmulaoides | ||||
| Among borefields | 6.93 | 0.80 Va | 32.92 | ΦSC: 0.03 |
| Among bores within borefields | 10.87 | −0.05 Vb | −1.92 | ΦST: 0.31 |
| Within bores | 30.13 | 1.67 Vc | 69.00 | ΦCT: 0.33 |
| Amphipod | ||||
| Among borefields | 55.90 | 0.93 Va | 35.11 | ΦSC: 0.11a |
| Among bores within borefields | 30.82 | 0.20 Vb | 7.35 | ΦST: 0.42a |
| Within bores | 96.50 | 1.53 Vc | 57.54 | ΦCT: 0.35a |
| Isopod | ||||
| Among borefields | 768.29 | 34.94 Va | 74.16 | ΦSC: 0.52a |
| Among bores within borefields | 276.02 | 6.30 Vb | 13.35 | ΦST: 0.88a |
| Within bores | 152.98 | 5.88 Vc | 12.49 | ΦCT: 0.74a |
Abbreviations: AMOVA; analysis of molecular variance; L, Limbodessus; LDC, Laverton Downs Calcrete; MW, Mt. Windarra; QW, Quandong Well; SW, Shady Well.
Significance tests where P<0.05.
All Mantel test results (Table 5) for the isopods, by borefield, showed a moderate to strong correlation between genetic and geographic distance (r=0.28–0.93, P<0.01). A confounding factor was the presence of three clear genetic lineages (MW-only; ISOLAV1; and ISOLAV2) that were each restricted in their distribution to distinct geographic locations in the LDC. We deemed it inappropriate to continue with these tests because of the confounding influence of the deep genetic differences among individuals that, in some cases, may represent species level differences. The remaining taxa showed some evidence of IBD between sites. Between SW and MW (∼9.5 km), L. windarraensis showed IBD that was significant (r=0.14, P<0.01) and the amphipod showed similar correlations between genetic and geographic distance (r=0.30, P<0.01). Between QW and MW, the amphipod showed significant IBD (r=0.23, P<0.01) and for other taxa, the r values were high but not significant over this ∼15 km distance. For L. windarraensis, these results were unsurprising, again because of the presence of highly divergent lineages, such as those at MW. Interestingly, the comparison between SW and QW revealed no evidence of IBD over a substantial distance (∼6.5 km) in any of the species (excluding Haloniscus discussed above). It should also be noted that L. palmulaoides was not sampled at QW.
Table 5. Results of Mantel test for IBD where a pairwise comparison between three borefields on LDC: MW, SW and QW were conducted for three stygobiontic beetles L. lapostae, L. windarraensis and L. palmulaoides, an amphipod and the isopods.
| MW | SW | |
|---|---|---|
| SW | ||
| L. windarraensis | 0.14 (P<0.001)a | — |
| L. palmulaoides | 0.40 (P=0.04) | — |
| L. lapostae | 0.63 (P=0.03) | — |
| Amphipod | 0.30 (P<0.001)a | — |
| Isopods | 0.93 (P<0.001)a | — |
| QW | ||
| L. windarraensis | 0.04 (P=0.7) | 0.08 (P=0.13) |
| L. palmulaoides | NA | NA |
| L. lapostae | 0.51 (P=0.11) | 0.08 (P=0.20) |
| Amphipod | 0.23 (P<0.001)a | 0.03 (P=0.18) |
| Isopods | 0.88 (P=0.009) | 0.28 (P<0.001) |
Abbreviations: IBD, isolation by distance; L, Limbodessus; LDC, Laverton Downs Calcrete; MW, Mt. Windarra; NA, not available; QW, Quandong Well; SW, Shady Well.
Indicates significant difference from the null hypothesis of no IBD with Bonferroni correction.
Historical demography
Some evidence for a departure from neutrality and population expansion in cox1 was observed in the LDC system. Negative and significant estimates of Fs, D and significant R2 estimates were observed in SW and QW populations of L. lapostae and L. palmulaoides, and the amphipod (Table 3). Selection is typically indicated by an excess of identical haplotypes, which is seen in only a few of these populations, although these tests are not able to distinguish between selection and demographic processes (Ballard and Whitlock, 2004). It has been demonstrated that R2 and Fs are powerful tests of population expansion and R2 is particularly useful when sample size is low, whereas Fs is useful when sample size is high (Ramos-Onsins and Rozas, 2002). Evidence of demographic expansion was also found using the mismatch analyses. There was no significant difference between the observed and expected mismatch distributions under the demographic expansion model in all species, excluding L. windarraensis at MW and the isopod at SW (Table 3).
Estimates of the parameter τ (time since expansion) under the model of demographic expansion (Table 3) indicated most bore populations of L. lapostae and L. palmulaoides, and the amphipod showed relatively recent (low) demographic expansion events in all northern bore holes (τ<3), whereas the QW population of L. windarraensis and the MW borefield of the amphipod showed medium ages of τ (6.6–9.5). Particularly interesting was the ancient demographic expansion found at populations MW in the two species that had high sample sizes at these sites (L. windarraensis and L. palmulaoides; 17.1–26.1). The estimates of τ, θ0 and θ1 under the demographic expansion model indicated that, in most cases, the relative population size is likely to have undergone substantial demographic expansion in all three beetle species at some point in time.
Coalescent timing
The BEAST analyses showed a total coalescent time of approximately 1–2 mya for L. lapostae and L. palmulaoides (Table 6), whereas an older total time to coalescence of 2.6 mya was observed for L. windarraensis. Further, dating of time to coalescence for the divergent clades revealed dates of 3–400 000 years for L. windarraensis (MW-only), L. lapostae (widespread-LAV) and L. palmulaoides (MW-only), whereas L. windarraensis (widespread-LAV) had a date of 800 000 years. Estimates of time since the last common ancestor for the LDC amphipods was a mean of ∼7.5 mya, suggestive of a divergence time much earlier in history than for the L. windarraensis (MW-only), L. lapostae (widespread-LAV) and L. palmulaoides (MW-only) clades. In contrast, the BEAST analyses on the isopod lineages failed to reach convergence, with effective sample size values below 100, despite running chains of >10 000 000 generations. Given the very large margins of error and our low confidence in the estimates, we excluded these results from the study and considered the results of the isopods qualitatively.
Table 6. Estimates of time since most recent common ancestor (time per million years) for three stygobiontic beetle and the amphipod based on Yule (speciation) coalescent model using a Bayesian coalescent approach with BEAST (Drummond and Rambaut, 2007).
| Species | Clade | Mean | s.d. of mean | Median | 95% HPD lower | 95% HPD upper | ACT | ESS |
|---|---|---|---|---|---|---|---|---|
| L. lapostae | Widespread-LAV | 0.3 | 3.3E-03 | 0.2 | 0.1 | 0.4 | 1.6E+04 | 623.8 |
| Total | 1.4 | 9.5E-03 | 1.3 | 0.7 | 2.2 | 5870.2 | 1702.0 | |
| L. windarraensis | MW-only | 0.4 | 4.0E-03 | 0.4 | 0.2 | 0.7 | 6711.9 | 1488.6 |
| Widespread-LAV | 0.8 | 6.2E-03 | 0.8 | 0.5 | 1.2 | 1.1E+04 | 918.5 | |
| Total | 2.6 | 1.7E-02 | 2.5 | 1.6 | 3.8 | 8398.1 | 1189.7 | |
| L. palmulaoides | Widespread-LAV | 0.3 | 4.4E-03 | 0.3 | 0.1 | 0.4 | 2.6E+04 | 387.3 |
| Total | 1.3 | 9.1E-03 | 1.2 | 0.7 | 2.0 | 6276.4 | 1591.9 | |
| Amphipod | Total | 7.5 | 4.0E-02 | 7.4 | 4.7 | 10.7 | 6355.4 | 1572.0 |
Abbreviations: ACT, auto-correlation time; ESS, effective sample size; HPD, highest posterior density; L, Limbodessus; MW, Mt. Windarra.
Each species was examined as a whole group (total) and within each of the two major intra-specific clades observed in this study, that is, the MW-only clade and the widespread-LAV clade.
Discussion
The inherently parapatric and allopatric nature of the epigean-subterranean nexus (Niemiller et al., 2008) is indisputably a key speciation mechanism in cave environments; however, in situ speciation (that is, speciation within cavernicolous habitats) is also predicted to have had an impact (Juan et al., 2010). Speciation from an obligate subterranean-adapted ancestor within the cave environment has rarely and only recently been explored (Caccone, 1985; Buhay and Crandall, 2005; Guzik et al., 2009). There is some evidence for intra-calcrete speciation by the presence of consistent size class variation among beetles species within calcretes of the Yilgarn region of Western Australia, some of which are sister species (Cooper et al., 2002; Leys et al., 2003). In this paper, we have investigated whether there is evidence for population fragmentation with a possibility for microallopatric population processes within a single calcrete for several cave-adapted species that may potentially result in de novo speciation following adaptation to obligate subterranean life.
Our previous study demonstrated that stygobiontic dytiscid diving beetles maintain thriving populations with high genetic (haplotype) diversity despite their likely isolation from the surface environment since the last ∼3–10 million years (Guzik et al., 2009). The levels of intra-calcrete diversification were high within a very small sample region with evidence of IBD detected in two of the three species of beetles. However, evidence for population fragmentation within species was not observed. Here, we examined genetic diversity and population genetic structure of stygobionts on a much broader spatial scale (LDC sample region of ∼15 km transects through a calcrete ∼25 km long and ∼90 km2 in area). We used a comparative approach using three beetle species, a single chiltoniid amphipod species and Haloniscus isopods, the latter potentially represented by multiple species (see below). Consideration of several stygobiontic species from a larger sample region permitted the investigation of multispecies population fragmentation. Significantly, our findings generally revealed a shared evolutionary history among the species in this habitat.
The phylogeographic pattern
Comparable phylogeographic patterns among unrelated taxa within an ecological system are strong evidence of a shared spatiotemporal history (Bermingham and Moritz, 1998; Carstens et al., 2005; Sunnucks et al., 2006). At LDC, five unrelated stygobiontic taxa shared a divergent pattern of spatially associated genetic divergence. Each taxon maintained a genetically distinct clade of mtDNA haplotypes that showed up to 5% divergence within each taxon, and much higher in the isopod samples. In subsequent sections, we primarily discuss the results obtained from beetles and amphipods as isopods revealed much larger divergences (∼21%) between lineages, suggesting a different temporal scale to their phylogeographic history. For all taxa, the individuals that yielded divergent haplotypes were sampled from the southernmost MW borefield (that is, the MW-only clade). In contrast, other individuals sampled from this location shared haplotypes with the northern SW (∼11 km) and QW (∼5 km from SW) borefields (that is, the widespread-LAV clade). The processes that have led to this pattern are possibly twofold.
On first inspection, a model of IBD in which individuals from nearby bores are more closely related to each other than to distant bores may explain our observations. IBD was cited as a key mechanism of diversification at the previously studied SMC aquifer (Guzik et al., 2009) and is expected to be used over the spatial scales examined in the LDC. Results of the Mantel tests for all taxa, excluding isopods, showed that there was evidence of IBD between the southern MW borefield and the two northern SW/QW borefields that could be because of the high divergence levels of MW-only haplotypes. However, there was no evidence of IBD between the two northern borefields SW/QW. The lack of IBD between the two northern sites indirectly suggests that distances of ∼6.5 km are not necessarily major barriers to dispersal in the LDC for these taxa. Therefore, we suggest that the divergence between widespread-LAV and MW-only haplotypes is unlikely the product of a purely IBD process. The results of nucleotide diversity (π=8.7–17.0) and the level of divergence (4%) among different haplotypes in the stygobiontic beetles at the southern MW borefield were substantially higher than those found in the SW/QW borefields (Table 3), as well as those previously seen in beetle species from the SMC aquifer in a similarly small geographic area (3.5 km2; Guzik et al., 2009). High nucleotide and haplotype diversity, such as that observed here, even within bore holes, is indicative of a large stable population with a long evolutionary history, allowing retention of ancient haplotype lineages and/or secondary contact between two differentiated populations (Grant and Bowen, 1998). Because we generally see two major clades of haplotypes in all the taxa rather than multiple divergent clades, we consider the latter scenario most probable.
Under a scenario of historical population fragmentation, individuals would have to be restricted to the southern region of the aquifer and physically separated over time from populations in the north. Given sufficient time for isolation, a clear signature of divergent haplotypes in reciprocal monophyly for MW versus SW/QW borefields would be expected. However, although there is evidence for divergent haplotypes, there is no support for reciprocal monophyly among the geographic locations, with several widespread-LAV haplotypes shared between the MW and SW/QW borefields. Either fragmentation has not persisted long enough to provide sufficient time to achieve reciprocal monophyly or, as suggested earlier, there has been long-term fragmentation of populations with recent secondary contact (Taberlet et al., 1998; Hewitt, 2000). This latter hypothesis of post-isolation colonisation, in which a population is fragmented by a geographic barrier (vicariance) and subsequently rejoined, has been demonstrated in many epigean species (Zamudio and Savage, 2003; Phillips et al., 2004; Hoskin et al., 2005; Pinceel et al., 2005). Such a scenario has also been documented from a broad range of cavernicolous habitats and is a plausible hypothesis for the taxa within the LDC aquifer system (Cobolli Sbordoni et al., 1990; Crouau-Roy and Bakalowicz, 1993; Buhay and Crandall, 2005; Hunter et al., 2008).
Short-term vicariance and allopatric divergence with secondary contact
On the basis of the current data, there appears to be a unidirectional gene flow from the north to the south, as indicated by the presence of widespread-LAV haplotypes found at the southern MW borefield and an absence of divergent MW-only haplotypes in northern sites. We mention this directionality with caution because, although our sampling was rigorous, especially for L. windarraensis and the amphipod in SW bores, there is always a possibility that these haplotypes are rare and remain unsampled from the northern sites. However, in the strict sense, the presence of mechanisms of unidirectional gene flow (for example, fast currents) is probably limited in this type of system, because of the generally flat topology of the desert landscape and the low flow of groundwater through the water table (Humphreys et al., 2009). Instead, we propose a scenario in which a barrier may have existed close to the MW borefield that, when removed, permitted gene flow between MW and the population on the other side of the barrier. This scenario, in which MW is located in a zone of introgression between two genetically divergent populations, may explain the presence of some shared haplotypes between the SW/QW and MW borefields and the absence of MW-only clade individuals at SW/QW borefields, which is outside the zone of introgression. We suggest that for the latter population there has been insufficient time for gene flow to spread divergent MW haplotypes to SW/QW. One possible barrier is a salt lake that lies immediately adjacent to MW (Figure 1).
Salt lakes are typically associated with calcrete formation and can be a source of strong saline stratification for proximate groundwater habitats (Humphreys et al., 2009), and marked salinity gradients have been recorded in a number of calcrete aquifers (Mann and Deutscher, 1978; Humphreys et al., 2009). Individuals at LDC may have been isolated on either side of a salinity cline or else separated by an intrusion of hypersaline water. An alternative hypothesis is that of aquifer drawdown (Culver and Sket, 2000; Culver et al., 2000; Leys et al., 2003), in which part of the water body at the MW borefield may have been partitioned from the rest of the calcrete, leading to isolation of populations in distinct refugia in different regions of the calcrete. The cause of such aquifer drawdown could potentially be external climate change events resulting from aridification of Western Australia during the Pleistocene and/or because of cycles of aridity resulting from ice ages (Byrne et al., 2008). At this stage, we cannot discriminate between these hypotheses but future ecological studies may help to resolve the question.
One additional explanation for the phylogeographic pattern observed here is that the clades may reflect divergences that occurred before, or during colonisation of, the subterranean habitat. In the current study, we made the assumption that, unlike the beetle triplet from the previously studied SMC aquifer (Guzik et al., 2009), the LDC species have evolved from different ancestral surface species that colonised the calcrete independently. The basis for this assumption is that the LDC species do not form a monophyletic clade of sister species in comprehensive phylogenetic analyses of Limbodessus species from Yilgarn calcretes (Leys et al., 2003; Leys, unpublished). Therefore, it is possible that the population differentiation that we observe here may reflect allopatric diversification through multiple colonisation events in different regions of the calcrete by the same ancestral species. These events are thought to occur commonly in cave habitats in which the nexus between subterranean and epigean habitats remains open for extended periods of time (Soulier-Perkins, 2004; Villacorta et al., 2008). However, the finding that multiple taxa shared a similar phylogeographic structure within a ∼15 km transect of the LDC is unlikely to be explained by random colonisation events in different regions of the calcrete. Further, we see an overlap in the intra-specific coalescent estimates of time to most recent common ancestor for haplotypes of the LDC and SMC aquifers. In particular, the oldest intra-specific coalescent times occurred in the last 2–3 mya (total L. windarraensis (LDC, Table 6) and in total Paroster mesosturtensis (SMC, Table 4, Guzik et al., 2009)), with the majority of intra-specific coalescent times <1.5 mya (for example, L. lapostae and L. palmulaoides; SMC: Paroster macrosturtensis and Paroster microsturtensis). Major differences in the scale of sampling in the current study and that in Guzik et al. (2009) are likely to have led to considerable variation in the intra-specific coalescent times of taxa from the two calcretes. In particular, the colonisation/speciation/demographic history of the taxa from the two aquifers, as well as the stochastic variation in the process of lineage sorting of haplotypes, has probably impacted TMRC estimates. However, despite this variation, the times are generally lower than the predicted timing for the enclosure and isolation of Yilgarn calcretes (that is, 3–10 mya; Leys et al., 2003; Cooper et al., 2007, 2008). Therefore, we believe that the phylogeographic patterns within taxa from the LDC aquifer most likely resulted from intra-calcrete processes (for example, fragmentation events) after colonisation, rather than from processes that occurred before, or during colonisation of, the subterranean habitat.
Overall, our population demographic results are consistent with the observed structure of the haplotype networks and a model of constant population size was not supported. Given that a range of taxa showed signals of expansion for multiple analyses, it is unlikely that selective sweeps (Turelli and Hoffmann, 1991; Ballard and Kreitman, 1995; Ballard and Rand, 2005) and/or stochastic variation in the coalescent time of the mtDNA were responsible for the significance of these test statistics. Demographic estimates and parsimony network analyses suggested that the MW bores incurred older demographic expansions (L. windarraensis and L. palmulaoides MW-only: τ=17.1–26.1) than that estimated for other parts of the aquifer (τ=1.0–9.5). This difference could be due to a bias because of the presence of divergent haplotype groups at MW. The differences in divergence time between species could also be due to sampling bias, as L. windarraensis was more frequently captured than L. lapostae and L. palmulaoides. We consider it to be more likely that multiple isolation and expansion events have occurred at different times within the aquifer (that is, the temporal patterns of expansion are different for each species) as indicated by BEAST analyses and estimates of τ. These observations are consistent with that of Guzik et al. (2009) for the SMC aquifer.
Crustacean population genetic structure and cryptic species
The level of genetic divergence observed for the crustacean taxa was distinctly different from that of beetles. Among the isopods, divergences were deep and suggest species level differences (see below), whereas the amphipods revealed a more recent expansion. In the amphipod species, there was some evidence of genetic divergence (2% between widespread-LAV and the MW-only clade), but it was smaller than that of beetles (4% intra-specific divergence) and isopods (1–20% divergence). These results may represent differences in mobility, life history or behaviour among the taxa, which were discussed in our earlier study of groundwater beetles (Guzik et al., 2009). The current study also includes isopods and amphipods, which are entirely aquatic and have no larval stage and, hence, are likely to have very different dispersal rates, life history traits and environmental tolerances (such as tolerance to different salinity levels). It is thought that the Haloniscus isopods are benthic-dwelling organisms that feed on detritus and are likely to be tolerant of high salinity levels, given that the related surface dwelling species of Haloniscus live on salt lakes (Bayly and Ellis, 1969). The adults are some of the largest of all stygobiontic invertebrates within calcretes (∼5–6 mm in body length and ∼1 mm in body width; Taiti and Humphreys, 2001) and they crawl on the calcrete surface rather than swimming through the groundwater. Therefore, they are likely to have more localised ranges and reduced rates of dispersal compared with the smaller beetle and amphipod species. The amphipod is thought to scavenge and disperse widely, because of its small size (<2–3 mm) and active juvenile and adult stages, which are capable of both swimming and crawling in the calcrete. They are known to inhabit the upper layers of the water column and are probably the most frequently sampled invertebrate in aquifers after copepods (Allford et al., 2008; Eberhard et al., 2009). Their tolerances to physicochemical differences in the groundwater may also vary widely (Berezina et al., 2001) and may potentially lead to distinct niche partitioning among zones within a salinity gradient (Fenchel and Kolding, 1979; Williams, 2003).
It is unlikely that the widespread-LAV and MW-only clades of the beetles and amphipods represent incipient species, and probably reflect historically divergent populations. Conversely, the isopods showed the presence of two highly divergent (19–21% Table 2) lineages (MW-only and ISOLAV1+2) that are most likely separate species (based on proposed species thresholds within crustaceans; Lefébure et al., 2006). Within each putative species were additional divergent (∼9%) lineages (that is, ISOLAV1 and ISOLAV2) or private haplotypes (ILA5 (BES 12005) and ILA6 (BES12087); Table 2). There is the possibility that ILA5 and ILA6 haplotypes may not be true representatives of the cox1 mtDNA gene. In single locus studies, pseudogenes can provide spurious results and for the mtDNA cox1 gene nuclear mitochondrial (numt) pseudogenes have been shown to inflate the number of identified species (Song et al., 2008). However, our data did not reveal any of the typical indicators of numts (that is, in-frame stop codons, high numbers of point mutations and routine multiple PCR product results such as multiple bands on gels and double peaks, background noise and so on); therefore, we consider it unlikely that pseudogenes have influenced our results.
The origins of these isopod lineages are likely to be more complex than our explanations for the beetles and amphipod; however, the near fixation of each ‘species' to either the MW or SW end of the calcrete is still consistent with our hypothesis of a barrier to dispersal within the system. However, either the two ‘species' diverged before their isolation within the calcrete, with the barrier leading to one predominating by chance in the MW region and the other predominating in the SW region, or, alternatively, fragmentation occurred much earlier in the history of the calcrete and led to a complete barrier to gene flow and speciation for the large-bodied isopods, which also may have colonised the LDC during a much earlier time period than the beetles and amphipods. Currently, we are unable to distinguish between these hypotheses, although it is interesting to note that the private haplotype ILA5 (BES 12087 from SW), which groups closely with the MW-only haplotypes, is likely to be a relictual haplotype from this early fragmentation process and has been retained by chance in the northern part of the calcrete. Additional relictual haplotypes and analyses incorporating nuclear gene markers are likely to shed further light on the history of these calcrete populations.
Conclusion
We have now examined the genetic diversity and population genetic structure of subterranean species within two independent calcrete aquifers in the Yilgarn region. It is clear that individual aquifers vary in their faunal composition and the population genetic substructure of species. Interestingly though, a number of generalisations regarding the calcrete habitat can now be made on the basis of the combined results of these two studies. First, we see that large and genetically diverse populations of stygobiontic organisms are maintained, with persistence over millions of years, despite major changes in climate during the Pleistocene (Byrne et al., 2008). The presence of shared haplotypes throughout both calcretes suggests that connectivity through large regions of the calcrete aquifers exists with preferred pockets of habitat that can facilitate genetic differentiation. It is thought that the connectivity within aquifers is complex, as suggested by evidence for IBD over very short spatial scales at both SMC and LDC, with connectivity among regions likely to vary over time depending on the level of groundwater. The implications of our findings for speciation are that there is clear potential for population genetic fragmentation and in turn speciation to occur from a single troglomorphic ancestral species in situ of the aquifer. However, whether this mode of in situ speciation is responsible for a significant proportion of stygofauna species within the Yilgarn calcretes is yet to be determined.
Acknowledgments
Special thanks to Kathy Saint for laboratory assistance, Chris Watts for the morphological examination of the Limbodessus specimens, and Adam Allford and Tessa Bradford for help with specimen collection. The research was funded by an Australian Research Council (ARC) Discovery grant (DP0663675) to SJBC and WFH, and ARC Linkage grants (LP0348753 and LP100200494) with industry partners Newmont Australia, Placer Dome Asia Pacific, Minara Resources Limited, the Western Australian Museum and the South Australian Museum awarded to ADA, SJBC and WFH.
Appendix A
The authors declare no conflict of interest.
References
- Allford A, Cooper SJB, Humphreys WF, Austin AD. Diversity and distribution of groundwater fauna in a calcrete aquifer: does sampling method influence the story. Invertebr Syst. 2008;22:127–138. [Google Scholar]
- Avise JC. Molecular Markers, Natural History and Evolution. Chapman and Hall: New York; 1994. [Google Scholar]
- Ballard WJ, Kreitman M. Is mitochondrial DNA a strictly neutral marker. Trends Ecol Evol. 1995;10:485–488. doi: 10.1016/s0169-5347(00)89195-8. [DOI] [PubMed] [Google Scholar]
- Ballard WJ, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Syst. 2005;36:621–642. [Google Scholar]
- Ballard JW, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- Bayly IAE, Ellis P. Haloniscus searlei Chilton: an aquatic ‘terrestrial' isopod with remarkable powers of osmotic regulation. Comp Biochem Physiol. 1969;31:523–528. [Google Scholar]
- Berezina NA, Khlebovich VV, Panov VE, Zaporozhets NV. Salinity tolerance of the amphipod Gmelinoides fasciatus (Stebb) introduced into the Gulf of Finland Basin (the Baltic Sea) Dokl Biol Sci. 2001;379:366–368. doi: 10.1023/a:1011664500018. [DOI] [PubMed] [Google Scholar]
- Bermingham E, Moritz C. Comparative phylogeography: concepts and applications. Mol Ecol. 1998;7:367–369. [Google Scholar]
- Brower AVZ. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhay JE, Crandall KA. Subterranean phylogeography of freshwater crayfishes shows extensive gene flow and surprisingly large population size. Mol Ecol. 2005;14:4259–4273. doi: 10.1111/j.1365-294X.2005.02755.x. [DOI] [PubMed] [Google Scholar]
- Buhay JE, Moni G, Mann N, Crandall K. Molecular taxonomy in the dark: evolutionary history, phylogeography, and diversity of cave crayfish in the subgenus Aviticambarus, genus Cambarus. Mol Phylogenet Evol. 2007;42:435–448. doi: 10.1016/j.ympev.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Butlin RK, Galindo J, Grahame JW. Sympatric, parapatric or allopatric: the most important way to classify speciation. Philos Trans R Soc B. 2008;363:2997–3007. doi: 10.1098/rstb.2008.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M, Yeates DK, Joseph L, Kearney M, Bowler J, Williams MA, et al. Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota. Mol Ecol. 2008;17:4398–4417. doi: 10.1111/j.1365-294X.2008.03899.x. [DOI] [PubMed] [Google Scholar]
- Caccone A. Gene flow in cave arthropods: a qualitative and quantitative approach. Evolution. 1985;39:1223–1235. doi: 10.1111/j.1558-5646.1985.tb05688.x. [DOI] [PubMed] [Google Scholar]
- Carstens BC, Brunsfeld SJ, Demboski JR, Good JM, Sullivan J. Investigating the evolutionary history of the Pacific Northwest mesic forest ecosystem: hypothesis testing within a comparative phylogeographic framework. Evolution. 2005;59:1639–1652. [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Cobolli Sbordoni M, Mattoccia M, La Rosa G, De Matthaeis E, Sbordoni V. Secondary sympatric occurrence of sibling species of subterranean shrimps in the Karst. Int J Speleol. 1990;19:9–27. [Google Scholar]
- Cooper SJB, Hinze S, Leys R, Watts CHS, Humphreys WF. Islands under the desert: molecular systematics and evolutionary origins of stygobitic water beetles (Coleoptera: Dytiscidae) from central Western Australia. Invertebr Syst. 2002;16:589–598. [Google Scholar]
- Cooper SJB, Bradbury JH, Saint KM, Leys R, Austin AD, Humphreys WF. Subterranean archipelago in the Australian arid zone: mitochondrial DNA phylogeography of amphipods from central Western Australia. Mol Ecol. 2007;16:1533–1544. doi: 10.1111/j.1365-294X.2007.03261.x. [DOI] [PubMed] [Google Scholar]
- Cooper SJB, Saint KM, Taiti S, Austin AD, Humphreys WF. Subterranean archipelago: mitochondrial DNA phylogeography of stygobitic isopods (Oniscidea: Haloniscus) from the Yilgarn region of Western Australia. Invertebr Syst. 2008;122:195–203. [Google Scholar]
- Crouau-Roy B, Bakalowicz M. Genetic structure in cave beetles and past environment changes. Biochem Syst Ecol. 1993;21:549–556. [Google Scholar]
- Culver DC. Analysis of simple cave communities I. Caves as islands. Evolution. 1970a;24:463–474. doi: 10.1111/j.1558-5646.1970.tb01776.x. [DOI] [PubMed] [Google Scholar]
- Culver DC. Analysis of simple cave communities: niche separation and species packing. Ecology. 1970b;51:949–958. [Google Scholar]
- Culver DC, Holsinger JR, Baroody R. Toward a predictive cave biogeography: the Greenbrier Valley as a case study. Evolution. 1973;27:689–695. doi: 10.1111/j.1558-5646.1973.tb00716.x. [DOI] [PubMed] [Google Scholar]
- Culver DC, Master LL, Christman MC, Hobbs HH., III Obligate cave fauna of the 48 contiguous United States. Conserv Biol. 2000;14:386–401. [Google Scholar]
- Culver DC, Sket B. Hotspots of subterranean biodiversity in caves and wells. J Cave Karst Stud. 2000;62:11–17. [Google Scholar]
- Desutter-Grandcolas L, Grandcolas P. The evolution toward troglobitic life: a phylogenetic reappraisal of climatic relict and local habitat shift hypotheses. Mém Biospéol. 1996;23:57–63. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard SM, Halse SA, Williams MR, Scanlon MD, Cocking J, Barron HJ, et al. Exploring the relationship between sampling efficiency and short-range endemism for groundwater fauna in the Pilbara region, Western Australia. Freshw Biol. 2009;54:885–901. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin version 3.1: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fenchel TM, Kolding S. Habitat selection and distribution patterns of five species of the amphipod genus Gammarus. Oikos. 1979;33:316–322. [Google Scholar]
- Folmer O, Black M, Hoen W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Fu X-Y. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie RG. Community assembly through adaptive radiation in Hawaiian spiders. Science. 2004;303:356–359. doi: 10.1126/science.1091875. [DOI] [PubMed] [Google Scholar]
- Grant WS, Bowen BW. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J Hered. 1998;88:415–426. [Google Scholar]
- Guzik MT, Cooper S, Humphreys WF, Austin AD. Fine-scale comparative phylogeography of a sympatric sister species triplet of subterranean diving beetles from a single calcrete aquifer in Western Australia. Mol Ecol. 2009;18:3683–3698. doi: 10.1111/j.1365-294X.2009.04296.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Harvey MS. Short-range endemism among the Australian fauna: some examples from non-marine environments. Invertebr Syst. 2002;16:555–570. [Google Scholar]
- Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hoskin CJ, Higgie M, McDonald MR, Moritz C. Reinforcement drives rapid allopatric speciation. Nature. 2005;437:1353–1356. doi: 10.1038/nature04004. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Humphreys WF.2000Groundwater calcrete aquifers in the Australian arid zone: the context to an unfolding plethora of stygal biodiversityIn: Humphreys WF, Harvey MS (eds).Subterranean Biology in Australia 2000 63–83.
- Humphreys WF. Aquifers: the ultimate groundwater dependent ecosystems. Aust J Bot. 2006;54:115–132. [Google Scholar]
- Humphreys WF. Rising from down under: developments in subterranean biodiversity in Australia from a groundwater fauna perspective. Invertebr Syst. 2008;22:85–101. [Google Scholar]
- Humphreys WF, Watts CHS, Cooper S, Leys R. Groundwater estuaries of salt lakes: buried pools of endemic biodiversity on the western plateau, Australia. Hydrobiologia. 2009;626:79–95. [Google Scholar]
- Hunter RL, Webb MS, Iliffe TM, Bremer JRA. Phylogeny and historical biogeography of the cave-adapted shrimp genus Typhlatya (Atyidae) in the Caribbean Sea and western Atlantic. J Biogeogr. 2008;35:65–75. [Google Scholar]
- Juan C, Emerson BC. Evolution underground: shedding light on the diversification of subterranean insects. J Biol. 2010;9:17. doi: 10.1186/jbiol227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan C, Guzik MT, Jaume D, Cooper SJB. Evolution in caves: Darwin's ‘wrecks of ancient life' in the molecular era. Mol Ecol. 2010;19:3865–3880. doi: 10.1111/j.1365-294X.2010.04759.x. [DOI] [PubMed] [Google Scholar]
- Lefébure T, Douady CJ, Gouy M, Gibert J. Relationship between morphological taxonomy and molecular divergence within Crustacea: proposal of a molecular threshold to help species delimitation. Mol Phylogenet Evol. 2006;40:435–447. doi: 10.1016/j.ympev.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Leys R, Watts CHS. Systematics and evolution of the Australian subterranean hydroporine diving beetles (Dystiscidae), with notes on Carabhydrus. Invertebr Syst. 2008;22:217–225. [Google Scholar]
- Leys R, Watts CHS, Cooper SJB, Humphreys WF. Evolution of subterranean diving beetles (Coleoptera: Dytiscidae: Hydroporini, Bidessini) in the arid zone of Australia. Evolution. 2003;57:2819–2834. doi: 10.1111/j.0014-3820.2003.tb01523.x. [DOI] [PubMed] [Google Scholar]
- Lunt DH, Zhang D-X, Szymura JM, Hewitt GM. The insect cytochrome oxidase I gene: evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol Biol. 1996;5:153–165. doi: 10.1111/j.1365-2583.1996.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Mann AW, Deutscher RL. Hydrogeochemistry of a calcrete-containing aquifer near Lake Way, Western Australia. J Hydrol. 1978;38:357–377. [Google Scholar]
- Miller MP. Alleles in space: computer software for the joint analysis of inter-individual spatial and genetic information. J Hered. 2005;96:722–724. doi: 10.1093/jhered/esi119. [DOI] [PubMed] [Google Scholar]
- Niemiller ML, Fitzpatrick BM, Miller BT. Recent divergence-with-gene-flow in Tennessee cave salamanders (Plethodontidae: Gyrinophilus) inferred from gene genealogies. Mol Ecol. 2008;17:2258–2275. doi: 10.1111/j.1365-294X.2008.03750.x. [DOI] [PubMed] [Google Scholar]
- Peck SB, Finston TL. Galapagos islands troglobites: the questions of tropical troglobites, parapatric distributions with eyed-sister-species, and their origin by parapatric speciation. Mém Biospéol. 1993;20:19–37. [Google Scholar]
- Phillips BL, Baird SJE, Moritz C. When vicars meet: a narrow contact zone between morphologically cryptic phylogeographic lineages of the rainforest skink, Carlia rubrigularis. Evolution. 2004;58:1536–1548. doi: 10.1111/j.0014-3820.2004.tb01734.x. [DOI] [PubMed] [Google Scholar]
- Pinceel J, Jordaens K, Backeljau T. Extreme mtDNA divergences in a terrestrial slug (Gastropoda, Pulmonata, Arionidae): accelerated evolution, allopatric divergence and secondary contact. J Evolution Biol. 2005;18:1264–1280. doi: 10.1111/j.1420-9101.2005.00932.x. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Poulson TL, Culver DC. Diversity in terrestrial cave communities. Ecology. 1969;50:153–158. [Google Scholar]
- Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Mol Biol Evol. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Ribera I, Bilton DT, Vogler AP. Mitochondrial DNA phylogeography and population history of Meladema diving beetles on the Atlantic Islands and in the Mediterranean basin (Coleoptera, Dytiscidae) Molecular Ecology. 2003;12:153–167. doi: 10.1046/j.1365-294x.2003.01711.x. [DOI] [PubMed] [Google Scholar]
- Ribera I, Fresneda J, Bucur R, Izquierdo A, Vogler AP, Salgado JM, et al. Ancient origin of a Western Mediterranean radiation of subterranean beetles. BMC Evol Biol. 2010;10:29. doi: 10.1186/1471-2148-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Savolainen V, Anstett M-C, Lexer C, Hutton I, Clarkson JJ, Norup MV, et al. Sympatric speciation in palms on an oceanic island. Nature. 2006;441:210–213. doi: 10.1038/nature04566. [DOI] [PubMed] [Google Scholar]
- Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P, et al. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701. [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and nonequilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Soulier-Perkins A. Phylogenetic evidence for multiple invasions and speciation in caves: the Australian planthopper genus Solonaima (Hemiptera: Fulgoromorpha: Cixiidae) Syst Entomol. 2004;30:281–288. [Google Scholar]
- Song H, Buhay JE, Whiting MF, Crandall KA. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci USA. 2008;105:13486–13491. doi: 10.1073/pnas.0803076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnucks P, Blacket MJ, Taylor JM, Sands CJ, Ciavaglia SA, Garrick RC, et al. A tale of two flatties: different responses of two terrestrial flatworms to past environmental climatic fluctuations at Tallaganda in montane southeastern Australia. Mol Ecol. 2006;15:4513–4531. doi: 10.1111/j.1365-294X.2006.03107.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL.2002PAUP*: Phylogenetic Analysis using ParsimonyVersion 4.0b,Sinauer Associates: Sunderland, MA [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy A-C, Cosson J-F. Comparative phylogeography and post-glacial colonisation routes in Europe. Mol Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Taiti S, Humphreys WF.2001New aquatic Oniscidea (Crustacea, Isopoda) from groundwater calcretes of Western AustraliaIn: Humphreys WF, Harvey MS (eds).Subterranean biology in Australia 2000 63–83.
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EB, McPhail JD. Historical contingency and ecological determinism interact to prime speciation in sticklebacks. P Roy Soc Lond B. 2000;267:2375–2384. doi: 10.1098/rspb.2000.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Hoffmann AA. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 1991;353:440–442. doi: 10.1038/353440a0. [DOI] [PubMed] [Google Scholar]
- Villacorta C, Jaume D, Orom P, Juan C. Under the volcano: phylogeography and evolution of the cave-dwelling Palmorchestia hypogaea (Amphipoda, Crustacea) at La Palma (Canary Islands) BMC Biol. 2008;7 doi: 10.1186/1741-7007-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villesen P. FaBox: an online toolbox for fasta sequences. Mol Ecol Notes. 2007;7:965–968. [Google Scholar]
- Watts CHS, Humphreys WF. Three new genera and five new species of Dytiscidae (Coleoptera) from underground waters in Australia. Records of the South Australian Museum. 1999;32:121–142. [Google Scholar]
- Watts CHS, Humphreys WF. Six new species of Nirridessus and Tjirtudessus (Dytiscidae; Coleoptera) from underground waters in Australia. Records of the South Australian Museum. 2000;33:127–144. [Google Scholar]
- Watts CHS, Humphreys WF. A new genus and six new species of Dytiscidae (Coleoptera) from underground waters in the Yilgarn palaeodrainage system of Western Australia. Records of the South Australian Museum. 2001;34:99–114. [Google Scholar]
- Watts CHS, Humphreys WF. Twenty-five new Dytiscidae (Coleoptera) of the genera Tjirtudessus Watts & Humphreys, Nirripirti Watts & Humphreys and Bidessodes Regimbart, from underground waters in Australia. Records of the South Australian Museum. 2003;36:135–187. [Google Scholar]
- Watts CHS, Humphreys WF. Thirteen new Dytiscidae (Coleoptera) of the genera Boongurrus Larson, Tjirtudessus Watts & Humphreys and Nirripirti Watts and Humphreys, from underground waters in Australia. T Roy Soc South Aust. 2004;128 [Google Scholar]
- Watts CHS, Humphreys WF. Twenty-six new Dytiscidae (Coleoptera)of the genera Limbodessus Guignot and Nirripirti Watts & Humphreys, from underground waters in Australia. T Roy Soc South Aust. 2006;1:123–185. [Google Scholar]
- Watts CHS, Humphreys WF. Fourteen new Dytiscidae (Coleoptera) of the genera Limbodessus Guignot, Paroster Sharp and Exocelina Broun, from underground waters in Australia. T Roy Soc South Aust. 2009;133:62–107. [Google Scholar]
- Williams DD. The brackish water hyporheic zone: invertebrate community structure across a novel ecotone. Hydrobiologia. 2003;510:153–173. [Google Scholar]
- Zamudio KR, Savage WK. Historical isolation, range expansion, and secondary contact of two highly divergent mitochondrial lineages in spotted salamanders (Ambystoma maculatum) Evolution. 2003;57:1631–1652. doi: 10.1554/02-342. [DOI] [PubMed] [Google Scholar]