Abstract
We tested different fitness components on a series of conspecific mtDNA haplotypes, detected by RFLPs in Drosophila subobscura. Additionally, haplotype VIII, endemic to the Canary Islands, was tested upon its own native nuclear DNA background and upon that of the rest of mtDNAs tested herein. We found that both nuclear and mitochondrial DNA can have a significant effect upon their hosts' fitness, and that negative selection is one of the mechanisms that can intervene in this species' mtDNA haplotype pattern. We discuss the importance of this mechanism in relation to genetic drift, in the form of periodic population bottlenecks, and how the latter can enhance the former. We also detected a significant positive effect of haplotype VIII upon fitness that could explain in part the dominance of this endemic haplotype on some of the Canary Islands, and a mitochondrial heterosis involving this haplotype when on a foreign nuclear DNA background.
Keywords: RFLPs, mtDNA haplotypes, fitness components, Drosophila subobscura
Introduction
Drosophila subobscura is a Palaearctic species of the obscura subgroup of Drosophila. It is distributed over most Europe, the Middle East, northern Africa and the Atlantic islands of the Azores, Madeira and the Canaries. In addition, in recent times it has colonized in the American continent (Ayala et al., 1989; Rozas et al., 1990).
A consistent observation in studies of mtDNA evolution in Old and New World populations of this species, is the high prevalence of two widespread and almost equally frequent RFLP haplotypes (I and II), and the sporadic appearance of rare ones at low frequencies generally present in never more than a single locality (Afonso et al., 1990; Latorre et al., 1992; Moya et al., 1993; González et al., 1994; Castro et al., 1999; Oliver et al., 2002; Christie et al., 2010). The exception is on the Canary Islands, where an endemic haplotype (VIII) is the most frequent on some islands (Pinto et al., 1997).
Different studies have tried to determine which evolutionary forces account for this species' mtDNA haplotype pattern in nature. The maintenance of the widespread equidistribution of the two main haplotypes has been thoroughly studied, but not so the case of the local less common haplotypes. The equidistribution of haplotypes I and II has been mainly explained by population expansion, but not ruling out the action of natural selection (García-Martínez et al., 1998; Oliver et al., 2005; Castro et al., 2010). Recently, Christie et al. (2010) studied monthly samples in a single geographic population of Drosophila subobscura over a 2 year period. Results indicated that natural populations have not yet reached a drift-selection equilibrium, because of periodic bottlenecks as well as some kind of selection, due to environmental heterogeneity (seasonal changes), possibly acting upon nuclear inversion polymorphism frequencies associated with mtDNA haplotypes. Many studies concerning the genetic dynamics of mtDNA in D. subobscura indicate the existence of cytonuclear interactions between the mtDNA and nuclear markers (Fos et al., 1990; García-Martínez et al., 1998; Oliver et al., 2002; Castro et al., 2003; Christie et al., 2004).
In relation to the nucleotide composition of the mtDNA in Drosophila subobscura, up to date only partial sequencing has been carried out. Moya et al. (1993) sequenced 15% of the genome and only found three silent substitutions between haplotypes I and II, one of which corresponded to the HaeIII restriction site that distinguishes haplotypes I and II, which is located on the ND5 gene. These authors supported the idea that haplotypes I and II were selectively neutral and the individuals phenotypically equivalent. Further sequencing by Castro et al. (2010) analysed nucleotide diversity in a 942-bp fragment of the mtDNA ND5 gene in haplotypes I, II and some rare ones. Results revealed some non-synonymous substitutions, although synonymous substitutions were far more frequent, so most of the rare haplotypes should also be considered a priori as neutral variants; even though, it is reported that a substantial proportion of intraspecific diversity in mtDNA results from slightly deleterious mutations (Rand and Kann, 1996). In relation to the presence of rare haplotypes in nature, Christie et al. (2010) found significant D values of the Tajima test for non-neutral evolution, which reflected an excess of rare haplotypes. This population dynamics was explained mainly because of periodic population bottlenecks followed by expansions but, at the same time, without discarding the possibility of purifying (negative) selection when non-silent changes at the protein level are involved. Thus, the competence of the rare mtDNA haplotypes of Drosophila subobscura in nature in comparison to that of haplotypes I and II might be an interesting issue.
In the present research, we focus in particular on the effect of mtDNA on its host's fitness. We tested the fitness of different conspecific mtDNA variants upon a same common nuclear DNA background with the following aims: (i) to investigate the fitness of the rare mtDNA haplotypes of Drosophila subobscura in comparison to haplotypes I and II and (ii) to determine the importance of selection in the populational dynamics of the mtDNA haplotypes. We demonstrate that mtDNAs have a significant effect upon their host's fitness, and that purifying (negative) selection is one of the mechanisms that can intervene in this species' mtDNA haplotype pattern in nature.
Materials and methods
mtDNA haplotypes
A total of 11 mtDNA RFLP haplotypes were used in this paper. In all, 10 of the haplotypes were sampled at a pine forest in Calvià, on Majorca (Spain). These were the two main haplotypes present in D. subobscura, I and II, and eight local rare haplotypes: III, IV, V, VI, IX, XI, XIII and XVI, are described in Christie et al. (2010). The eleventh was haplotype VIII, described in Latorre et al. (1992), endemic to the Canary Islands, and sampled at La Laguna, on Tenerife (Spain). The flies were collected with conventional traps containing fermented bananas, and classified individually under a microscope. Extraction and digestion of mtDNA was obtained by the method described by Latorre et al. (1986). mtDNA haplotypes were named following the notation used by Latorre et al. (1986, 1992).
Construction of isofemale lines
A total of 12 isofemale lines were established. In all, 10 corresponded to the haplotypes from Calvià with their own native nuclear background. Additionally, two haplotype VIII isofemale lines (derived from the same female fly) were established: one with its native Tenerife nuclear background (VIIIt) and another with that of Calvià (VIIIc). In order to uniform the nuclear DNA variability, introgression of nuclear background was carried out in all the isofemale lines by backcrossing for eight generations. A total of 20 virgin females (except generation zero, with one) from all lines were mated to twice as many wild males (from generations zero, one or two from established population cages; see below). In theory, 99.61% of the original nuclear genome was replaced with that of the wild populations. Each isofemale line's haplotype was re-confirmed after backcrossing concluded to discard mtDNA contamination. Isofemale lines for experiments were maintained by serial transfer in five 500 ml bottles. All flies were maintained in the laboratory at 19 °C, 70% relative humidity, 1:1 day–night cycle and fed with corn-meal food.
Population cages
Additionally, two population cages were established for the wild-stock nuclear backgrounds of Calvià and Tenerife used for backcrossing, consisting of boxes with 12 jars of corn-meal food, with a mean number of approximately 2000 individuals per generation. These cages were found with at least 50 wild females plus corresponding males. The cage with the Calvià nuclear background was re-found every three generations, thanks to the proximity of the local population.
Presence of Wolbachia
An incompatibility system in D. subobscura promoted by the presence of Wolbachia was excluded. PCR assays using 16S rDNA Wolbachia-specific primers on local populations were carried out in the recent past (García-Martínez et al., 1998; Christie et al., 2004), giving negative results. Original Drosophila stocks from Tenerife were treated with tetracycline, and PCR assayed. The cure was carried out at least 10 generations before experiments, avoiding, in this way, the negative consequences indicated by Ballard and Melvin (2007). Moreover, no kind of cytoplasmic incompatibility was detected, such as embryonic death or sex ratio distortion in crosses with different isofemale lines.
Experiments
Larva–adult viability and developmental time without competition (VDT)
Groups of approximately 15 adult couples per haplotype were transferred for 24 h from the 500 ml maintaining bottles to 250 ml bottles, with fresh food in order to provide sufficient adult offspring reared without competition. At 2 or 3 days after eclosion, offsprings (males and females together) were transferred to new 250 ml bottles with fresh food, where the flies could mate, feed and lay eggs freely for 1 week. These flies with 9–10 days of age were used for egg laying. They were transferred for 2 h to ‘oviposition vials', which contained a watch glass with agar, water, acetic acid, ethyl alcohol and a few milligrams of live yeast. The eggs were kept in petri dishes for 40 h at 19 °C until the larvae hatched. Then 50 larvae of the same age were seeded in 10 × 3 cm tubes with 10 ml of fresh food. Larvae were picked up one by one directly from the watch glasses, with a lancet under a microscope. The adults that emerged were counted daily, and a total of 13–15 replicates were carried out for each isofemale line. Viability (V) was expressed as following:
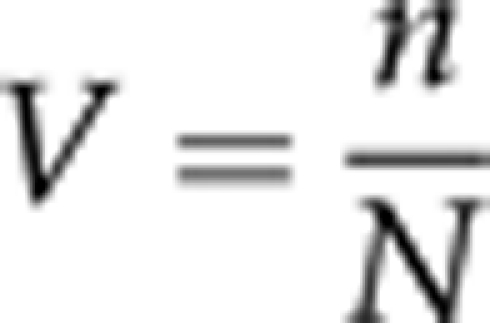 |
where N is the input number of larvae and n is the output number of adults emerging from these N larvae.
Developmental time (DT) was measured in days by the formula:
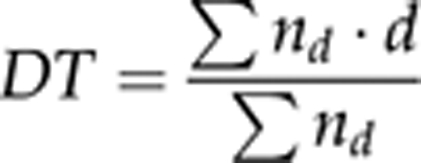 |
where nd is the number of flies emerging d days after the larvae were placed on the medium.
Longevity
The flies tested for longevity were obtained as explained in the previous experiment (reared without competition), and taken as they hatched from their pupae. In all, 50 individuals of each isoline and sex were maintained individually in 10 × 3 cm vials, with 10 ml of food medium containing active yeast on the surface. Flies were checked daily. Vials were changed every fortnight, before dehydration of food was evident. All the flies that escaped, were damaged or died during the experiment were included in the analyses.
Fertility
The flies tested for fertility were taken at eclosion from pupae, and were obtained as in the experiments above (reared without competition). Females were mated individually to two males and were transferred daily to vials with fresh food for 21 consecutive days. The emerged offsprings were counted and sexed.
Statistics
Results from each experiment were analysed globally (all 12 isofemale lines), considering only those isolines with Calvià nuclear DNA (11 isofemale lines) and only those haplotypes native to Calvià (10 isofemale lines). Additionally, direct comparisons were done of haplotype I and II, and of both the lines with haplotype VIII.
Statistical analyses were performed by means of the SPSS package (IBM SPSS Statistics, New York, NY, USA). One-way ANOVA was used to test for differences among the several independent groups in the fertility and VTD experiments. Viabilities were subjected to the arcsine-square root transformation for statistical analyses, but the results with transformed data were qualitatively indistinguishable from analyses with untransformed data, so only the latter are presented. The longevity was analysed by means of the Survival Analysis Procedure (included in the SPSS package). The method was that of Kaplan–Meier survival curves. The analysis gives, among others parameters, the cumulative survival with time and the mean with standard error (s.e.) for each curve. The comparison of the several curves was made with the log-rank test, so called because it can be shown to be related to a test that uses the logarithms of the ranks of the data.
E-statistic (Moya et al., 1986; Christie et al., 2004) is a fitness parameter that combines data of viabilities and developmental times, and follows the optimality principle that the largest number of adults emerged in the shortest possible time gives the best fitness. The characteristic of this parameter is that it simultaneously maximises viability, V, and the reciprocal of the development time, DT, as follows:
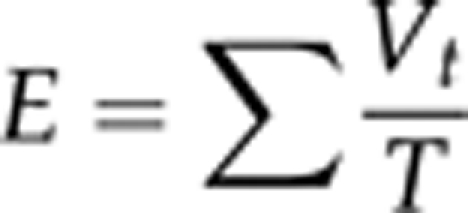 |
where Vt=nt/N and T=t/toptimal; nt is the number of individuals emerged at time t; N is the input number; and toptimal is a conventional minimum developmental time (here considered 17 days). Hence,
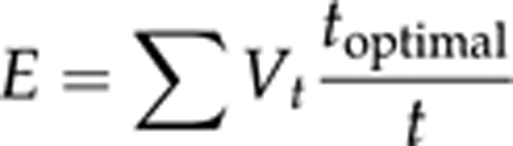 |
For computational purposes this formula can also be expressed as
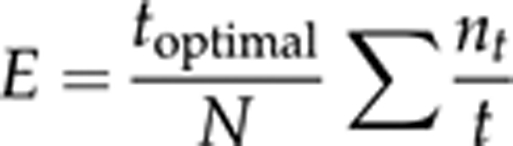 |
E-statistic is only a combination of viabilities and developmental times, and it does not include other fitness components (such as population growth rates or fecundities).
Results
Longevity
Average longevities with standard errors calculated with Kaplan–Meier for males and females of each haplotype are presented on Table 1. In general, females of each haplotype lived significantly longer than their corresponding males. Survival analyses showed significant differences in the dynamics of the different haplotypes. Considering only those haplotypes native to Calvià, haplotypes I and II had intermediate values, and log-rank tests between them were not significant; haplotypes XI and XIII stood out for having the lowest longevities in both sexes, whereas haplotypes IV and V had the longest longevities for females and males, respectively. With respect to haplotype VIII, log-rank tests between both isofemale lines (VIIIc and VIIIt) indicated a superiority of the flies with the Calvià nuclear DNA background. Furthermore, when comparing all groups, this line was the longest lived, whereas haplotype VIII upon its own native nuclear background was the shortest lived, in both males and females.
Table 1. Average adult longevities with standard errors of males and females of each isofemale line, calculated with Kaplan–Meier.
| Isofemale line |
Adult longevity (days) |
||
|---|---|---|---|
| Males | Females | Log-rank | |
| I | 78.86±3.78 | 91.21±3.19 | 2.82 NS |
| II | 70.44±3.46 | 93.64±3.83 | 17.56*** |
| VIIIt | 38.36±2.58 | 52.76±3.68 | 10.50** |
| VIIIc | 83.76±2.62 | 94.44±4.49 | 12.74*** |
| III | 72.68±3.43 | 82.82±3.60 | 6.01* |
| IV | 77.16±3.55 | 94.44±4.09 | 13.75*** |
| V | 82.42±3.17 | 81.36±3.60 | 0.03 NS |
| VI | 63.46±4.75 | 82.94±3.65 | 6.06* |
| IX | 67.49±3.48 | 86.57±3.89 | 12.79*** |
| XI | 58.24±3.25 | 78.28±2.94 | 12.26*** |
| XIII | 59.39±3.28 | 77.74±4.47 | 20.29*** |
| XVI | 68.58±3.35 | 87.02±4.01 | 13.63*** |
|
Comparisons |
Sex |
Log-rank |
|
| Global | Females | 117.39*** (11) | |
| Males | 167.49*** (11) | ||
| Calvià nDNA | Females | 30.68*** (10) | |
| Males | 52.98*** (10) | ||
| Without VIII | Females | 23.22** (9) | |
| Males | 43.00*** (9) | ||
| I vs II | Females | 0.85 NS (1) | |
| Males | 3.06 NS (1) | ||
| VIIIt vs VIIIc | Females | 45.81*** (1) | |
| Males | 75.83*** (1) | ||
Abbreviation: NS, not significant.
Comparisons are calculated with log-rank. Comparisons are done globally (all 12 lines), with those lines with Calvià nuclear DNA (11 lines), excluding haplotype VIII (10 lines), haplotype I vs haplotype II, and between lines with haplotype VIII (VIIIt, with Tenerife nuclear DNA, and VIIIc, with Calvià nuclear DNA).
*P<0.05, **P<0.01, ***P<0.001; degrees of freedom are within parentheses.
Fertility
Average offspring with standard errors per female and haplotype is presented on Table 2. ANOVA analyses considering all isofemale lines, those with Calvià nuclear DNA, and only those with mtDNA native to Calvià, showed significant differences. On the other hand, t-tests between haplotypes I and II, and between both lines with haplotype VIII, were not significant. Haplotypes I and II, and both isolines with haplotype VIII, had intermediate fertility values. Considering those haplotypes native to Calvià, haplotypes XI and XIII, as in the case of longevity, stood out for having the lowest average fertility, whereas haplotypes IV and V had the most offspring.
Table 2. Average fertility (offspring per female) with standard errors of each isofemale line.
| Isofemale lines | Fertility (average offspring per female) |
|---|---|
| I | 522.14±36.11 |
| II | 570.33±49.65 |
| VIIIt | 450.93±37.91 |
| VIIIc | 506.29±37.17 |
| III | 485.00±41.78 |
| IV | 593.71±57.48 |
| V | 575.14±29.84 |
| VI | 468.86±51.14 |
| IX | 509.93±38.83 |
| XI | 364.54±45.30 |
| XIII | 411.14±39.95 |
| XVI | 442.93±44.28 |
|
Comparisons |
ANOVA/t-test |
| Global | 2.520** (11) |
| Calvià nDNA | 2.615** (10) |
| Without VIII | 2.822** (9) |
| I vs II | −0.776 NS (27) |
| VIIIt vs VIIIc | −1.043 NS (26) |
Abbreviations: ANOVA, analysis of variance; NS, not significant.
Comparisons are calculated with ANOVA (more than two lines) or with t-test (two lines). Comparisons are done globally (all 12 lines), with those lines with Calvià nuclear DNA (11 lines), excluding haplotype VIII (10 lines), haplotype I vs haplotype II, and between lines with haplotype VIII (VIIIt, with Tenerife nuclear DNA, and VIIIc, with Calvià nuclear DNA).
*P<0.05, **P<0.01, ***P<0.001; degrees of freedom are within parentheses.
Larval viability
Larva-to-adult viabilities with standard errors are shown on Table 3. ANOVA analyses showed significant differences between haplotypes as long as both haplotype VIII isofemale lines were included; these lines obtained the highest viabilities and were significantly different to the Calvià haplotypes. t-tests between haplotypes I and II and between lines with haplotype VIII were not significant.
Table 3. Larval developmental times, viabilities and resulting E statistic with standard errors of each isofemale line.
| Isofemale line |
Larval developmental time (days)a |
Viability (%) | E statistic | ||
|---|---|---|---|---|---|
| Males | Females | Both sexes | Both sexes | Both sexes | |
| I | 17.48±0.30 | 17.75±0.32 | 17.63±0.31 | 80.80±1.75 | 0.74±0.02 |
| II | 17.46±0.22 | 17.78±0.23 | 17.63±0.22 | 76.27±1.91 | 0.69±0.02 |
| VIIIt | 19.06±0.38 | 19.35±0.43 | 19.22±0.40 | 88.93±1.74 | 0.74±0.01 |
| VIIIc | 17.05±0.17 | 17.36±0.19 | 17.22±0.17 | 87.43±2.38 | 0.81±0.02 |
| III | 17.70±0.31 | 17.92±0.35 | 17.82±0.33 | 83.57±2.21 | 0.75±0.02 |
| IV | 17.97±0.23 | 18.33±0.28 | 18.14±0.25 | 85.47±2.25 | 0.76±0.02 |
| V | 17.46±0.11 | 17.77±0.14 | 17.62±0.12 | 80.15±1.75 | 0.73±0.01 |
| VI | 17.75±0.27 | 18.06±0.30 | 17.90±0.28 | 80.93±2.08 | 0.73±0.02 |
| IX | 18.17±0.17 | 18.44±0.17 | 18.31±0.17 | 80.80±2.87 | 0.71±0.02 |
| XI | 17.78±0.13 | 17.94±0.13 | 17.86±0.13 | 79.20±1.69 | 0.71±0.01 |
| XIII | 17.87±0.19 | 18.27±0.21 | 18.06±0.19 | 78.40±2.02 | 0.70±0.02 |
| XVI | 17.49±0.29 | 17.72±0.30 | 17.60±0.29 | 81.08±2.18 | 0.74±0.02 |
|
Comparisons |
Sex |
ANOVA/t-test |
Comparisons |
ANOVA/t-test |
ANOVA/t-test |
| Global | Females | 3.585*** (11) | Global | 3.228** (11) | 2.905** (11) |
| Males | 4.241*** (11) | Calvià nDNA | 2.222* (10) | 3.034** (10) | |
| Calvià nDNA | Females | 1.637 NS (10) | Without VIII | 1.504 NS (9) | 1.353 NS (9) |
| Males | 1.815 NS (10) | I vs II | 1.747 NS (28) | 1.573 NS (28) | |
| Without VIII | Females | 1.122 NS (9) | VIIIt vs VIIIc | 0.515 NS (27) | −2.657* (27) |
| Males | 1.122 NS (9) | ||||
| I vs II | Females | −0.096 NS (28) | |||
| Males | 0.06 NS (28) | ||||
| VIIIt vs VIIIc | Females | 4.143*** (27) | |||
| Males | 4.680*** (27) | ||||
Abbreviations: ANOVA, analysis of variance; NS, not significant.
Comparisons are calculated with ANOVA (more than two lines) or with t-test (two lines). Comparisons are done globally (all 12 lines), with those lines with Calvià nuclear DNA (11 lines), excluding haplotype VIII (10 lines), haplotype I vs haplotype II, and between lines with haplotype VIII (VIIIt, with Tenerife nuclear DNA, and VIIIc, with Calvià nuclear DNA).
*P<0.05, **P<0.01, ***P<0.001; degrees of freedom are within parentheses.
The t-test was not significant in any case when comparing males and females.
Developmental time
Average developmental times with standard errors of each haplotype are also presented on Table 3. ANOVA analyses were only significant when isoline VIIIt was included. t-tests between haplotypes I and II were not significant. t-tests between haplotypes VIII were significant, and similarly to that found in the longevity experiment, males and females had the lowest fitness results when upon their own native nuclear background, and had the best fitness results for males and females when upon the nuclear DNA from Calvià.
E-statistic
E-statistic is a fitness parameter that combines data of viabilities and developmental times. Results are shown on Table 3. Results depend heavily upon each haplotype's viability. ANOVA analyses showed significant differences when considering all isolines and when considering those with Calvià nuclear DNA. ANOVA analyses were not significant when both isolines with haplotype VIII were excluded. t-tests between haplotypes I and II were not significant, but were so between isolines with haplotypes VIII.
Discussion
In this study, we tested different fitness components on a series of isofemale lines of Drosophila subobscura that differed in their mtDNAs. The main aim was to study the effects of those conspecific mtDNAs upon their hosts' fitness. Nuclear DNA background variability was uniform due to backcrossing. In doing so, we attempted to: (1) resolve the competence of the rare mtDNA haplotypes of Drosophila subobscura in nature in comparison to that of haplotypes I and II and (2) determine if selection is one of the mechanisms that intervene in this species' mtDNA haplotype pattern. Additionally, in order to observe the effect of nuclear DNA, haplotype VIII, endemic to the Canary Islands, was tested upon its own native nuclear DNA and upon that of Calvià. Our results demonstrate that mtDNA can compromise fitness in D. subobscura. Significant differences in fitness were detected in adult longevities, larval viabilities, fertilities and statistic E. Although single parameters cannot be directly used to estimate global fitness because of the possibility of antagonistic pleiotropy (Prout, 1971a, 1971b), in the present work we can affirm that as a general trend some haplotypes were more efficient than others. Considering only those haplotypes native to Calvià, haplotypes XI and XIII seemed to have a negative effect upon their hosts' fitness, as they obtained the worst results in various experiments. These disadvantageous haplotypes would surely tend to disappear from the population unless they only affect males, while having apparently little or no effect on females (Ruiz-Pesini et al., 2000; Rand et al., 2001; Sackton et al., 2003). Recent work indicates that mutations in mtDNA are mainly expected to affect males, having an important role in sperm function, male fertility and male fitness (Gemmell et al., 2004). Our results support these facts as differences between haplotypes were always higher among males than among females. Although in some experiments we have failed to distinguish between male and female fitness, as in the fertility experiment, we have observed that these mtDNAs can clearly affect female fitness too, as seen in the longevity experiment.
The significant differences in fitness that we have detected in the different experiments, could indicate the implication of non-synonymous mutations in the mtDNA with deleterious or near-neutral effects, and be the consequence of a gradual accumulation of point mutations in mtDNA rather than by a single specific point mutation, as indicated by other authors (Nachman, 1998; Holyoake et al., 2001; St John et al., 2001). Consequently, the mean fitness of each isofemale line could reflect the total number of deleterious mutations in that one line (Bergstrom and Pritchard, 1998), although evidently selective effects of deleterious mutations will vary, with some being effectively neutral and others strongly deleterious. The accumulation of these non-beneficial mutations in mtDNA, would be caused mainly by the bottlenecking of germline mitochondria during transmission between generations (Bergstrom and Pritchard, 1998; Neiman and Taylor, 2009).
The accumulation of deleterious mutations and irreversible decline in fitness in obligatory asexual lineages, known as Muller's Ratchet, has been described in organelle genomes like mitochondria (Lynch, 1996). This process within each particular mtDNA lineage would presumably proceed undetected by RFLPs, until a chance mutation would determine a novel haplotype, but a larger accumulation of mutations implies a higher chance of being detected as a novel haplotype and of also having an impaired fitness. As a result, negative selection can be expected to purge from populations these novel or rare haplotypes, reducing their frequency and number instead of them accumulating over time, as would be expected if they were all selectively neutral variants. Therefore, we consider that negative selection can be considered one of the mechanisms that contribute to this species' mtDNA haplotype pattern in nature. Although there is recent accumulated evidence of recombination in mtDNA and that it may occur regularly (Piganeau et al., 2004; Tsaousis et al., 2005), reparation of the fixed deleterious mutations in a host's mtDNA through recombination is not possible unless the individuals be heteroplasmic through paternal leakage. Reports in D. subobscura of heteroplasmy are scarce (Afonso et al., 1990; Volz-Lingenhöhl et al., 1992; Morel et al., 2006). Moreover, in Christie et al. (2010) heteroplasmy was not detected in none of the 607 isofemale lines characterized. Consequently, it would be reasonable to suppose that the role of recombination in mitochondrial haplotype survival would not be relevant in Drosophila through infrequency, but its importance should not be underestimated if very little recombination is required to counter mutation accumulation (Neiman and Taylor, 2009).
Previous studies have mainly indicated genetic drift as the main mechanism implicated in this dilemma, in part because population bottlenecks in D. subobscura are known to be frequent and to have important effects (González et al., 1994; Christie et al., 2010), with a lack of a stable population equilibrium, reflected in an excess of rare haplotypes, indicated by the negative Tajima′s D values. The results of the present work also support genetic drift because many of the novel haplotypes studied in this paper have similar fitnesses to haplotypes I and II, and so can be considered selectively neutral variants. But which of the two mechanisms would mainly determine this species' mtDNA haplotype pattern? Hedrick (1970) argued that changes in haplotype frequency would be determined primarily by genetic drift rather than by natural selection when the product of the effective population size and the selection coefficient is less than one, because chance effects outweigh those of selection. In this case, we consider that the extinction of rare haplotypes would be mainly due to genetic drift, as the result of seasonal population bottlenecks (with a decrease in the effective population size), having selection when implicated a secondary role. During the seasonal population expansions, disadvantageous mtDNAs would relatively reduce their hosts' offspring production in comparison to other mtDNAs. This phenomenon would increment the probability of extinction of those mtDNA lineages (already at low frequencies as them being novel) upon the event of the annually occurring population bottlenecks. Further still, it is interesting to note the close relationship between bottlenecks and selection. The event of a bottleneck itself has been proved to enhance selection among mtDNA lineages (Bergstrom and Pritchard, 1998). These authors determined that population size bottlenecks between host generations increase the efficiency of selection among mtDNA lineages against deleterious mutations by increasing the variance in fitness among their eukaryotic hosts.
With respect to haplotype VIII, it is interesting to note that this mtDNA may have a positive effect upon the fitness of its hosts independently to the nuclear DNA background considered, because both isofemale lines with this haplotype obtained the best results in the larval viability test and comparisons among lineages lost signification when both were excluded. This result could partially explain why haplotype VIII is the most frequent haplotype on some of the Canary Islands. Additionally, comparisons between haplotypes I and II did not give significative differences, contrarily to Castro et al. (2003) and Christie et al. (2004). These present results confirm that the fitness differences observed in these two haplotypes were surely due to cytonuclear interactions and not to direct selection upon the mtDNA.
On the other hand, our results also confirm that nuclear DNA has an important effect upon fitness too in D. subobscura. In this case, significant differences were observed when comparing haplotype VIII isolines. Against what was expected, haplotype VIII upon a foreign nuclear DNA (isofemale line VIIIc) not only provided better results than upon its own native nuclear DNA (isofemale line VIIIt) but scored better fitness results than the local haplotypes too. Line VIIIc enjoyed of some sort of hybrid vigour that indicates a strong nuclear x mtDNA epistatic interaction effect. We find this mitochondrial heterosis hard to explain because disruption of nuclear-mitochondrial coadaptation is generally expected to lower fitness (Fos et al., 1990), although it has been observed in interspecific Drosophila mtDNA strains (Rand et al., 2006). On the other hand, we consider the poor results of line VIIIt due to the adaptation of the species to the particular conditions found on the Canary Islands and not to inbreeding in the population cages: although the expectation of life at eclosion of inbred flies is approximately half that of non inbred flies (Clarke and Maynard Smith, 1955), fertility results were normal and viability results were the highest indicating that these flies were not inbred (Hollingsworth and Maynard Smith, 1955).
In conclusion, both nuclear and mitochondrial DNA can affect fitness in D. subobscura and negative selection can be considered one of the mechanisms that contribute to this species' mtDNA haplotype pattern in nature. The importance of genetic drift, in the form of seasonal population bottlenecks, must also be considered (Castro et al., 2010; Christie et al., 2010).
Acknowledgments
This work was supported by grants PB96-0793 and BOS2000-1000 from the Dirección General de Enseñanza Superior (Ministerio de Educación y Cultura, Spain).
The authors declare no conflict of interest.
References
- Afonso JM, Volz A, Hernández M, Ruttkay H, González AM, Larruga JM, et al. Mitochondrial DNA variation and genetic structure in Old-World populations of Drosophila subobscura. Mol Biol Evol. 1990;7:123–142. doi: 10.1093/oxfordjournals.molbev.a040590. [DOI] [PubMed] [Google Scholar]
- Ayala FJ, Serra LL, Prevosti A. A grand experiment in evolution: the Drosophila subobscura colonisation of the Americas. Genome. 1989;31:246–255. [Google Scholar]
- Ballard JWO, Melvin RG. Tetracycline treatment influences mitochondrial metabolism and mtDNA density two generations after treatment in Drosophila. Insect Mol Biol. 2007;16:799–802. doi: 10.1111/j.1365-2583.2007.00760.x. [DOI] [PubMed] [Google Scholar]
- Bergstrom CT, Pritchard J. Germline bottlenecks and the evolutionary maintenance of mitochondrial genomes. Genetics. 1998;149:2135–2146. doi: 10.1093/genetics/149.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro JA, Barrio E, González A, Picornell A, Ramon MM, Moya A. Nucleotide diversity of a ND5 fragment confirms that population expansion is the most suitable explanation for the mtDNA haplotype polymorphism of Drosophila subobscura. Genetica. 2010;138:819–829. doi: 10.1007/s10709-010-9464-x. [DOI] [PubMed] [Google Scholar]
- Castro JA, Oliver P, Christie JS, Picornell A, Ramon M, Moya A. Assortative mating and fertility in two Drosophila subobscura strains with different mitochondrial DNA haplotypes. Genetica. 2003;119:295–301. doi: 10.1023/b:gene.0000003656.19330.ba. [DOI] [PubMed] [Google Scholar]
- Castro JA, Ramon M, Picornell A, Moya A. The genetic structure of Drosophila subobscura populations from the islands of Majorca and Minorca (Balearic Islands, Spain) based on allozymes and mitochondrial DNA. Heredity. 1999;83:271–279. doi: 10.1038/sj.hdy.6885500. [DOI] [PubMed] [Google Scholar]
- Christie JS, Castro JA, Oliver P, Picornell A, Ramon MM, Moya A. Fitness and life-history traits of the two major mitochondrial DNA haplotypes of Drosophila subobscura. Heredity. 2004;93:371–378. doi: 10.1038/sj.hdy.6800513. [DOI] [PubMed] [Google Scholar]
- Christie JS, Picornell A, Moya A, Ramon MM, Castro JA. Dynamics of the mtDNA haplotype variability in a Drosophila subobscura population over a two-year period. The Open Evolution Journal. 2010;4:23–30. [Google Scholar]
- Clarke JM, Maynard Smith J. The genetics and cytology of Drosophila suboscura. XI. Hybrid vigour and longevity. J Genetics. 1955;53:172. [Google Scholar]
- Fos M, Domínguez MA, Latorre A, Moya A. Mitochondrial DNA evolution in experimental populations of Drosophila subobscura. Proc Natl Acad Sci USA. 1990;87:4198–4201. doi: 10.1073/pnas.87.11.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martínez J, Castro J.A, Ramón M, Latorre A, Moya A. Mitochondrial DNA haplotype frequencies in natural and experimental populations of Drosophila subobscura. Genetics. 1998;149:1377–1382. doi: 10.1093/genetics/149.3.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell NJ, Metcalf VJ, Allendorf FW. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol Evol. 2004;19:238–244. doi: 10.1016/j.tree.2004.02.002. [DOI] [PubMed] [Google Scholar]
- González A, Carrió R, Fernández-Pedrosa V, Moya A. Lack of seasonal changes in mitochondrial DNA variability of a Drosophila subobscura population. J Evol Biol. 1994;7:29–38. [Google Scholar]
- Hedrick PW. Selection in finite populations. I. The probability of fixation and rate of response using transition matrix iteration. Genetics. 1970;65:157–173. doi: 10.1093/genetics/65.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth MJ, Maynard Smith J. The effects of inbreeding on rate of development and on fertility in Drosophila subobscura. J Genet. 1955;53:295–314. [Google Scholar]
- Holyoake AJ, McHugh P, Wu M, O'Carroll S, Benny P, Sin IL, et al. High incidence of single nucleotide substitutions in the mitochondrial genome is associated with poor semen parameters in men. Int J Androl. 2001;24:175–182. doi: 10.1046/j.1365-2605.2001.00292.x. [DOI] [PubMed] [Google Scholar]
- Latorre A, Hernández C, Martínez D, Castro J.A, Ramón M, Moya A. Population structure and mitochondrial DNA gene flow in old World populations of Drosophila suboscura. Heredity. 1992;68:15–24. doi: 10.1038/hdy.1992.2. [DOI] [PubMed] [Google Scholar]
- Latorre A, Moya A, Ayala FJ. Evolution of mitochondrial DNA in Drosophilasubobscura. Proc Natl Acad Sci USA. 1986;83:8649–8653. doi: 10.1073/pnas.83.22.8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Mutation accumulation in Transfer RNAs: molecular evidence for muller's ratchet in mitochondrial genomes. Mol Biol Evol. 1996;13:209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]
- Morel F, Renoux M, Alziari S. Mitochondrial biochemical activities and heteroplasmy evolution in established D. subobscura cell line. In Vitro Cell Dev Biol Anim. 2006;42:201–217. doi: 10.1290/0601003.1. [DOI] [PubMed] [Google Scholar]
- Moya A, González-Candelas F, Ayala FJ. Intra- and intergenotypic competition in Drosophila melanogaster: effects of density on larval survival and rate of development. Genetica. 1986;70:59–67. [Google Scholar]
- Moya A., Barrio E, Martínez D, Latorre A, González-Candelas F, Ramón M, et al. Molecular characterisation and cytonuclear disequilibria of two Drosophila subobscura mitochondrial haplotypes. Genome. 1993;36:890–898. doi: 10.1139/g93-117. [DOI] [PubMed] [Google Scholar]
- Nachman MW. Deleterious mutations in animal mitochondrial DNA. Genetica. 1998;103:61–69. [PubMed] [Google Scholar]
- Neiman M, Taylor DR. The causes of mutation accumulation in mitochondrial genomes. Proc R Soc B. 2009;276:1201–1209. doi: 10.1098/rspb.2008.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P, Balanyà J, Ramon MM, Picornell A, Serra L, Moya A, et al. Population dynamics of the two major mitocondrial DNA haplotypes in experimental populations of Drosophila subobscura. Genome. 2005;48:1010–1018. doi: 10.1139/g05-077. [DOI] [PubMed] [Google Scholar]
- Oliver P, Castro JA, Picornell A, Ramon MM, Solé E, Balanyà J, et al. Linkage disequilibria between mtDNA haplotypes and chromosomal arrangements in a natural population of Drosophila subobscura. Heredity. 2002;89:133–138. doi: 10.1038/sj.hdy.6800116. [DOI] [PubMed] [Google Scholar]
- Piganeau G, Gardner M, Eyre-Walker A. A Broad Survey of Recombination in Animal Mitochondria. Mol Biol Evol. 2004;21:2319–2325. doi: 10.1093/molbev/msh244. [DOI] [PubMed] [Google Scholar]
- Pinto FM, Brehm A, Hernandez M, Larruga JM, González AM, Cabrera VM. Population genetic structure and colonisation sequence of Drosophila subobscura in the Canaries and Madeira Atlantic islands as inferred by autosomal, sex-linked and mtDNA traits. J Hered. 1997;88:108–114. doi: 10.1093/oxfordjournals.jhered.a023067. [DOI] [PubMed] [Google Scholar]
- Prout T. The relation between fitness components and population prediction in Drosophila. I: the estimation of fitness components. Genetics. 1971a;68:127–149. doi: 10.1093/genetics/68.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prout T. The relation between fitness components and population prediction in Drosophila. II: population prediction. Genetics. 1971b;68:151–167. doi: 10.1093/genetics/68.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Clark AG, Kann LM. Sexually antagonistic cytonuclear fitness interaction in Drosophila melanogaster. Genetics. 2001;159:173–187. doi: 10.1093/genetics/159.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Fry A, Sheldahl L. Nuclear–mitochondrial epistasis and Drosophila aging: Introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics. 2006;172:329–341. doi: 10.1534/genetics.105.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:737–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Rozas JM, Hernandez M, Cabrera VM, Prevosti A. Colonisation of America by Drosophila subobscura: effect of the founder event on the mitochondrial DNA polymorphism. Mol Biol Evol. 1990;7:103–109. doi: 10.1093/oxfordjournals.molbev.a040584. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Lapena AC, Diez-Sanchez C, Perez-Martos A, Montoya J, Alvarez E, et al. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am J Hum Genet. 2000;67:682–696. doi: 10.1086/303040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Haney RA, Rand DM. Cytonuclear coadaptation in Drosophila: disruption of cytochrome c oxidase activity in backcross genotypes. Evolution. 2003;57:2315–2325. doi: 10.1111/j.0014-3820.2003.tb00243.x. [DOI] [PubMed] [Google Scholar]
- St John JC, Jokhi RP, Barratt CLR. Men with oligoasthenoteratazoospermia harbour higher numbers of multiple mitochondrial DNA deletions in their spermatozoa, but individual deletions are not indicative of overall aetiology. Mol Hum Reprod. 2001;7:103–111. doi: 10.1093/molehr/7.1.103. [DOI] [PubMed] [Google Scholar]
- Tsaousis AD, Martin DP, Ladoukakis ED, Posada D, Zouros E. Widespread Recombination in Published Animal mtDNA Sequences. Mol Biol Evol. 2005;22:925–933. doi: 10.1093/molbev/msi084. [DOI] [PubMed] [Google Scholar]
- Volz-Lingenhöhl A, Solignac M, Sperlich D. Stable heteroplasmy for a large-scale deletion in the coding region of Drosophila subobscura mitochondrial DNA. Proc Natl Acad Sci USA. 1992;89:11528–11532. doi: 10.1073/pnas.89.23.11528. [DOI] [PMC free article] [PubMed] [Google Scholar]


