Abstract
HMG-CoA reductase (HMGR), a highly conserved, membrane-bound enzyme, catalyzes a rate-limiting step in sterol and isoprenoid biosynthesis and is the primary target of hypocholesterolemic drug therapy. HMGR activity is tightly regulated to ensure maintenance of lipid homeostasis, disruption of which is a major cause of human morbidity and mortality. HMGR regulation takes place at the levels of transcription, translation, post-translational modification and degradation. In this review, we discuss regulation of mammalian, Saccharomyces cerevisiae and Schizosaccharomyces pombe HMGR and highlight recent advances in the field. We find that the general features of HMGR regulation, including a requirement for the HMGR-binding protein Insig, are remarkably conserved between mammals and ascomycetous fungi, including S. cerevisiae and S. pombe. However the specific details by which this regulation occurs differ in surprising ways, revealing the broad evolutionary themes underlying both HMGR regulation and Insig function.
Keywords: HMG-CoA reductase, sterol, Insig, AMPK, ERAD, HRD
Introduction
Cholesterol biosynthesis is one of the most intensively studied biochemical pathways due to its well-known relevance to human health and disease [1]. Cholesterol is a 27-carbon, tetracyclic molecule that is essential for the structure and function of eukaryotic lipid bilayers [2]. It also has multiple, essential functions in pathways as diverse as bile acid synthesis and Hedgehog signaling [3]. Excessive cholesterol supply results in significant human morbidity and mortality, especially from atherosclerosis, leading to myocardial infarction or stroke [4]. Because animal cells obtain cholesterol by a combination of de novo synthesis and uptake from the bloodstream, the need for end-product feedback inhibition of this biosynthetic pathway is paramount.
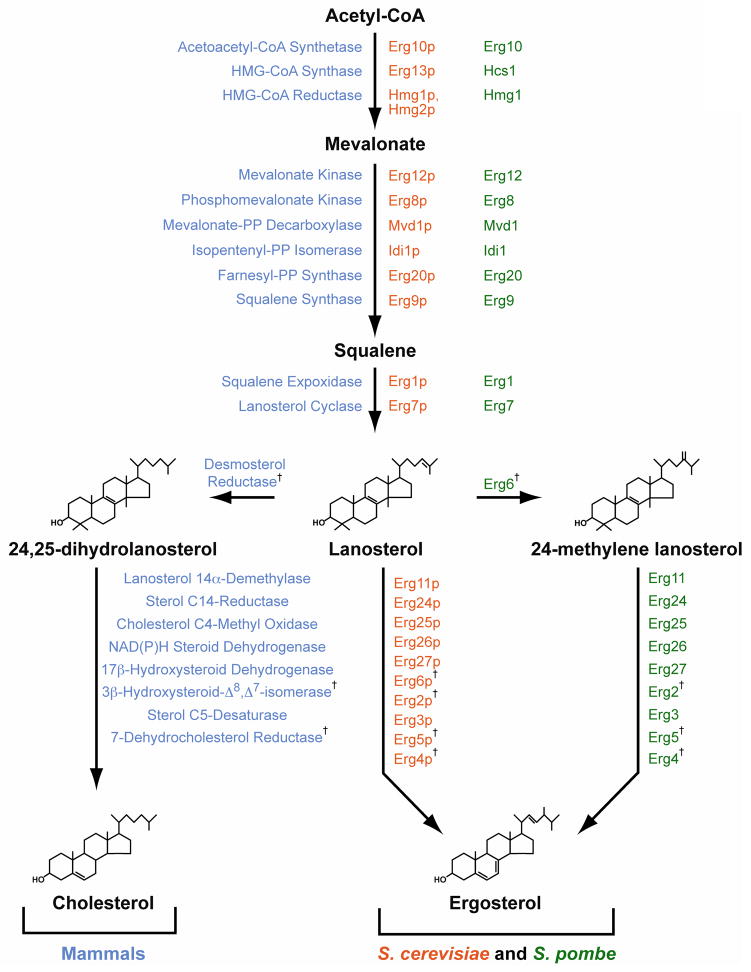
Like mammalian cells, yeast cells require sterol as a structural component of their membranes. In yeast, ergosterol fills this role. Ergosterol is structurally similar to cholesterol, but ergosterol contains double bonds between carbons 7–8 of the B ring and carbons 22–23 of the side chain, and methylation of carbon 24. The similarity between these molecules is enough to allow cholesterol to substitute for ergosterol in the membranes of S. cerevisiae cells [5, 6]. Correspondingly, yeast produce ergosterol in a biosynthetic pathway that shares most enzymatic steps with its mammalian counterpart (Fig. 1)[7].
Figure 1. The sterol biosynthetic pathway in mammals, Saccharomyces cerevisiae and Schizosaccharomyces pombe.
Mammalian enzymes (blue) are listed by their full names; S. pombe enzymes (green) are named based on homology to S. cerevisiae enzymes (orange). † indicates enzymes not conserved between mammals and yeast.
Feedback regulation of cholesterol biosynthesis was discovered by Rudolf Schoenheimer and Fritz Breusch, who observed that mice produce cholesterol in inverse proportion to the amount in their diet [8]. Building on this finding, Marvin Siperstein and M. Joanne Guest determined the target of cholesterol feedback inhibition to be the four-electron reduction of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) to mevalonate by the enzyme HMG-CoA reductase (HMGR)[9]. Subsequent advances revealed that HMGR is regulated at the levels of transcription, translation, post-translational modification and degradation [10, 11]. In addition to sterols, HMGR is required for synthesis of isoprenoids. Assembled from the 5-carbon sterol biosynthesis intermediates isopentenyl pyrophosphate and dimethylallyl pyrophosphate, isoprenoids are precursors in the production of dolichol, heme A, tRNA, ubiquinone and prenylated proteins [10].
Competitive inhibitors of HMGR, colloquially known as statins, are the primary treatment for hypercholesterolemia. Mevastatin, a natural product of Penicillium citrinum, was discovered by Akira Endo and Masao Kuroda in 1976 [12]. This was the first in a remarkably effective and successful line of drugs; annual sales exceed $7.5 billion US dollars for atorvastatin alone [13]. Because of its relevance to human disease, HMGR has been studied more than any other enzyme in the sterol biosynthetic pathway. These studies have yielded a detailed understanding of the molecular mechanisms underlying lipid sensing and have in turn led to significant discoveries in diverse fields such as protein quality control and enzymology. In this review, we highlight recent advances in the regulation of mammalian and yeast HMGRs. These divergent organisms use similar systems to regulate HMGR activity and promote sterol homeostasis.
Structure and enzymology of HMG-CoA reductase
HMGR is a highly conserved enzyme with sequence homologs in eukaryotes, prokaryotes and archaea. Based on sequence alignment, HMGRs can be categorized into two classes [14]. Class I enzymes are highly similar to human HMGR and utilize NADPH as the electron donor. Class II enzymes are found primarily in eubacteria and utilize NADH. Archaeal HMGRs vary widely in sequence with enzymes belonging to each class. The primary sequence element distinguishing the two classes of HMGR is the cis-loop, a strictly conserved feature of Class I HMGRs corresponding to amino acids 682–694 of human HMGR [14, 15].
Eukaryotic HMGRs are typically endoplasmic reticulum (ER)-resident integral membrane proteins consisting of two distinct domains: a hydrophobic NH2-terminal membrane anchor consisting of 2 – 8 transmembrane segments, and a COOH-terminal catalytic domain that extends into the cytoplasm [16]. This hydrophobic NH2-terminus is poorly conserved; there is generally less than 25% amino acid identity between the NH2-termini of fungal and mammalian HMGRs. The NH2-terminus of HMGR usually includes a recognizable domain consisting of 5 consecutive transmembrane spans called a sterol-sensing domain (SSD). The SSD, which is found in other proteins such as the SREBP cleavage-activating protein (Scap)[11] and Neimann-Pick C1-like 1 [17], may bind lipids and plays an important role in HMGR regulation [10, 18, 19].
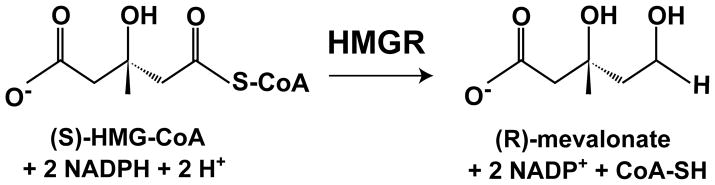
The COOH-terminal catalytic domain of Class I HMGRs forms a dimer that comprises the active enzyme and each monomer contributes catalytic residues to form the active site [20, 21]. Catalysis takes place in two sequential hydride transfers from NADPH, in which HMG-CoA is reduced to mevalonate (Fig. 2). A protonated histidine residue plays a critical role in catalysis by donating a proton to the thiol anion after the first reduction [15, 22]. Substrate binding induces closure of the COOH-terminal helix, moving the catalytic histidine into position and completing the active site [23, 24].
Figure 2. Reaction catalyzed by HMG-CoA reductase.
HMGR catalyzes the reduction of 3-hydroxy-3-methylglutaryl-CoA to mevalonate, thereby oxidizing two molecules of NADPH.
Statins bind directly to the HMGR active site and are competitive inhibitors of the enzyme with respect to HMG-CoA [25]. All statins have structural similarity to the 3-hydroxy-3-methylglutarate moiety of HMG-CoA and occupy the HMG-CoA binding pocket of HMGR [26]. The remainder of the statin molecule is rigid, hydrophobic and highly variable among different statins. This part of the molecule makes contact with residues in the active site, but does not occupy the NADPH binding site [24, 26]. Accordingly, statins do not interfere with NADPH binding to HMGR [25].
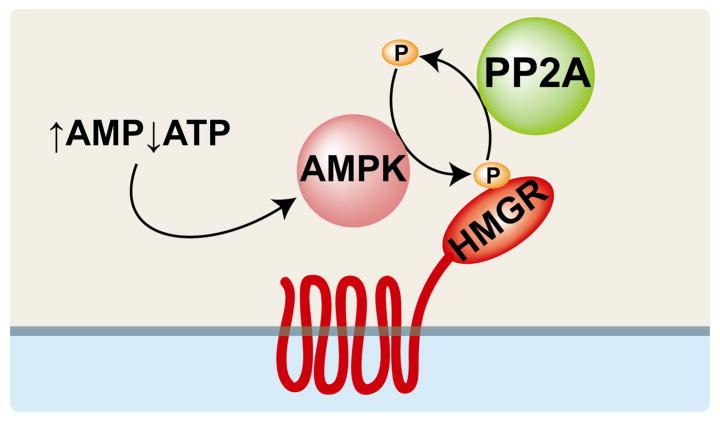
A conserved serine residue corresponding to human HMGR S872 is located near the active site [27] and S872 phosphorylation reversibly decreases enzyme efficiency [15]. S872 is primarily phosphorylated by the AMP-activated protein kinase (AMPK) in response to a high AMP:ATP ratio (Fig. 3)[27, 28]. HMGR is dephosphorylated by protein phosphatase 2A (PP2A)[29]. The residue corresponding to human HMGR S872 is not present in Class II HMGRs, nor is it conserved in Saccharomyces cerevisiae [14]. However human HMGR S872 is conserved in many other fungi, including the fission yeast Schizosaccharomyces pombe [30, 31].
Figure 3. Mammalian HMGR catalytic activity is regulated by AMP kinase.
A high AMP:ATP ratio stimulates AMP-activated protein kinase (AMPK), which phosphorylates HMGR at a conserved serine in the active site, thus inhibiting HMGR activity. Protein phosphatase 2A (PP2A) dephosphorylates HMGR, restoring enzyme activity.
Regulation of mammalian HMG-CoA reductase
Cholesterol biosynthesis is a complex pathway, requiring 20 enzymes to assemble 30 carbons from acetyl-CoA into a 27-carbon structure that includes four rings [4]. Of the 20 enzymes in the pathway, HMGR is uniquely suited to serve as the pathway’s primary point of regulation because it catalyzes an irreversible reaction at the beginning of the pathway [9]. Thus, a decrease in HMGR activity can regulate the output of the overall pathway without accumulating unusable intermediates.
Mammalian HMGR was purified from rat liver by Michael Brown in the laboratory of Marvin Siperstein in 1973 [32]. Advances in molecular biology have since allowed the molecular cloning of the HMGR gene and a detailed dissection of its regulation [33, 34]. HMGR is regulated by sterol-mediated feedback inhibition at the levels of transcription and degradation, ensuring that sterol synthesis meets but does not exceed cellular requirements [10].
The membrane-bound transcription factor sterol regulatory element-binding protein (SREBP) controls HMGR transcription [11]. SREBP forms a complex in the ER membrane with another integral membrane protein, Scap. In cholesterol-replete cells, the inactive SREBP-Scap complex resides in the ER membrane. When cholesterol is depleted, the complex is transported to the Golgi apparatus in COPII vesicles. ER-to-Golgi transport of SREBP requires Scap, which has a COPII recognition site in its NH2-terminal domain. In the Golgi, SREBP is activated by two sequential proteolytic events that cleave the NH2- terminal transcription factor domain from the membrane, allowing it to enter the nucleus and activate HMGR transcription [35].
A major advance in understanding the molecular basis for HMGR sterol feedback inhibition was the discovery of an ER-resident protein called Insig. Insig was originally identified as an mRNA induced in H35 hepatoma cells upon insulin treatment; its name is derived from the description, “insulin induced gene” [36, 37]. Insig contains 6 transmembrane spans and regulates HMGR both transcriptionally and post-translationally [11]. Insig regulates HMGR transcription by inhibiting SREBP activation [11]. Insig binds to Scap in sterol-replete conditions, altering Scap structure and rendering its COPII-recognition sequence inaccessible. This prevents the SREBP-Scap complex from being loaded into COPII vesicles, thereby preventing proteolytic activation of SREBP in the Golgi [38, 39]. Insig dissociates from Scap when sterols are depleted, allowing transport of SREBP-Scap to the Golgi and proteolytic SREBP activation. This results in transcriptional upregulation of cholesterol biosynthetic enzymes, including HMGR, and restoration of cholesterol homeostasis [35].
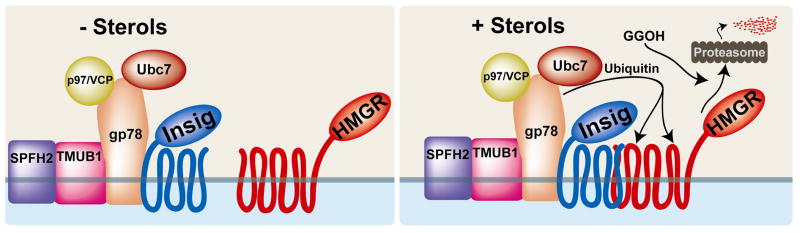
In addition to its role in regulating HMGR transcription through SREBP, Insig also regulates HMGR degradation (Fig. 4)[34]. Because it regulates both HMGR production and degradation, Insig has a striking influence on HMGR levels; liver HMGR activity in mice lacking Insig is 85-fold higher than in control mice [40]. Insig regulates HMGR degradation through a sterol-responsive feedback inhibition system. In sterol-replete conditions, Insig binds to the membrane-embedded NH2-terminal region of HMGR and recruits enzymes that conjugate ubiquitin to HMGR on K89 and K248 (Fig. 4)[41, 42]. The effect of sterols on the half-life of HMGR is dramatic; its half-life decreases from greater than 12 hours in sterol-depleted cells to less than 1 hour in sterol-replete cells [11].
Figure 4. Mammalian HMGR enzyme levels are regulated post-translationally by Insig-dependent and sterol-accelerated degradation.
In sterol-replete cells, Insig binds HMGR and recruits gp78, a ubiquitin E3 ligase. gp78, in concert with the E2 conjugating enzyme Ubc7, ubiquitinates HMGR on K89 and K248, a process which also requires SPFH2 and TMUB1. After membrane extraction, which is enhanced by geranylgeraniol (GGOH), the hexameric ATPase p97/VCP allows proteasomal degradation of HMGR. Insig dissociates from HMGR when sterols are depleted, thus stabilizing HMGR by preventing enzyme ubiquitination.
Insig-mediated, sterol-accelerated degradation of HMGR is accomplished through binding between Insig and gp78 (Fig. 4)[42]. gp78 is a membrane-bound ubiquitin E3 ligase; its NH2-terminal domain has between 5 and 7 transmembrane spans and mediates interaction with Insig [43, 44]. The cytoplasmic face of gp78 recruits Ubc7, an E2 ubiquitin conjugating enzyme that supplies gp78 with activated ubiquitin. gp78 also recruits p97/VCP, a hexameric ATPase [11, 42, 45]. Furthermore, gp78 associates with two ER-localized membrane proteins that contribute to HMGR ubiquitination: SPFH2 and TMUB1 [46]. TMUB1 binds gp78; SPFH2 binds gp78 through TMUB1. After being ubiquitinated by gp78, HMGR is extracted from the membrane and degraded [44]. p97/VCP is required for HMGR degradation, but not for membrane extraction, suggesting that p97/VCP acts by making HMGR accessible to the proteasome [47].
Insig-dependent HMGR degradation is stimulated by two lipid signals: sterols and the 20-carbon isoprenoid geranylgeraniol (GGOH). Each lipid signal acts at a different step in HMGR degradation [44]. Sterols, including certain oxysterols and the cholesterol biosynthetic intermediate 24,25-dihydrolanosterol, stimulate Insig-HMGR binding. Insig recruits the gp78 complex to HMGR, allowing HMGR ubiquitination (Fig. 4)[34, 44]. GGOH is not required for HMGR ubiquitination, but enhances HMGR degradation.
HMGR is also regulated by cellular metabolic state through an Insig-independent mechanism that is thought to help the cell optimize ATP expenditure during metabolic stress (Fig. 3)[28, 40]. AMP-activated protein kinase (AMPK), originally known as HMG-CoA reductase kinase [28], is a heterotrimeric complex consisting of two regulatory subunits (β and γ) and one catalytic subunit (α)[48]. The γ subunit contains four tandem cystathionine β-synthase (CBS) domains, which bind adenosine nucleotides and are thought to participate in energy sensing [49]. AMP allosterically activates AMPK; ATP does not [50]. Because ATP and AMP compete for the same binding site, AMPK is able to respond to a high AMP:ATP ratio by increasing its catalytic activity. Once activated, AMPK phosphorylates HMGR at a conserved residue in the enzyme active site corresponding to serine 872 of human HMGR [51]. The mechanism by which phosphorylation inhibits HMGR activity has not been conclusively determined, but phosphorylation of S872 may either decrease HMGR affinity for NADPH or interfere with closure of the COOH-terminal flap over the active site [21, 23].
Dephosphorylation of HMGR fully restores enzyme activity (Fig. 3). Protein phosphatase 2A (PP2A), the enzyme primarily responsible for dephosphorylating HMGR in vivo [29], has diverse functions including cell cycle control, viral infection, cell morphology and development [52]. Along with AMPK, PP2A may play an important role in regulating HMGR activity through phosphorylation. However, there is no known physiological role for PP2A regulation in controlling HMGR activity.
HMGR is also regulated at the level of translation by a non-sterol isoprenoid [10, 53]. Translational control of HMGR may involve the complex 5′-untranslated region of the HMGR gene. However, this aspect of HMGR regulation has received considerably less investigation than other forms of regulation and thus its mechanism is not clear.
Regulation of Saccharomyces cerevisiae HMG-CoA reductase
The budding yeast Saccharomyces cerevisiae encodes two HMGR genes, designated HMG1 and HMG2. Presumed to be derived from a single ancestral HMGR by gene duplication [54], Hmg1p and Hmg2p have 62% overall amino acid identity to each other. Their NH2-terminal membrane domains are 44% identical, and their COOH-terminal catalytic domains are 95% identical. Either gene can supply the essential HMGR activity when the other is deleted [55]. Hmg1p is a stable protein, whereas Hmg2p is rapidly degraded [56]. Although both enzymes are controlled by feedback inhibition, they are regulated in different ways [54].
Hmg1p is the primary source of HMGR activity during aerobic growth of S. cerevisiae [54]. Aerobic growth promotes synthesis of heme, which activates the transcription factor Hap1p [57]. Hap1p activates HMG1 transcription, resulting in a 10-fold increase in HMGR activity [58]. Simultaneously, aerobic growth represses HMG2 expression by an unknown mechanism [54]. Hmg1p is also regulated at the level of translation by a negative feedback system. Mevalonate-starved cells accumulate Hmg1p protein and show an increase in HMGR activity, even as HMG1 mRNA transcript levels remain unchanged [59]. Translational control of HMG1 expression requires the HMG1 5′-untranslated region insomuch as a lacZ reporter gene was similarly regulated when controlled by the HMG1 promoter. The molecular signal regulating Hmg1p translational control may be mevalonate itself, although the mechanism by which it acts awaits discovery.
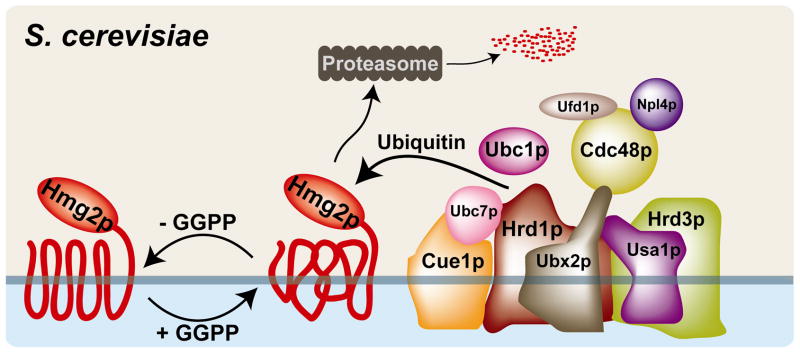
Like mammalian HMGR, S. cerevisiae Hmg2p is regulated by protein turnover through endoplasmic reticulum-associated degradation (ERAD), utilizing the machinery of the HMG-CoA reductase degradation (HRD) pathway [54]. Indeed, regulated ubiquitination of HMGR was initially described in yeast [60]. Hmg2p is recognized and ubiquitinated by the multi-subunit, membrane-associated HRD complex (Fig. 5)[61]. The membrane-spanning E3 ligase Hrd1p, together with the E2 ubiquitin conjugating enzyme Ubc7p, ubiquitinates Hmg2p. Hmg2p is then extracted from the membrane and degraded by the proteasome.
Figure 5. Hmg2p levels are controlled post-translationally by geranylgeranyl pyrophosphate and the HRD complex in S. cerevisiae.
Geranylgeranyl pyrophosphate alters the conformation of the Hmg2p N-terminal membrane domain, promoting recognition and ubiquitination by the membane-bound, multi-subunit HMG-CoA reductase degradation (HRD) complex. Hrd1p, a membrane-spanning ubiquitin E3 ligase with homology to the mammalian E3 ligase gp78, utilizes two E2 ubiquitin conjugating enzymes, Ubc7p and Ubc1p. Ubc7p, a soluble protein that interacts with the membrane through the integral membrane protein Cue1p, is the primary E2 for Hrd1. Hrd3 is involved in substrate recognition and delivery to the HRD complex. Usa1p plays a role in Hrd1p function and Hrd1p self-regulation. Ubx2p is an integral membrane protein that recruits the Cdc48p/Npl4p/Ufd1p complex. Cdc48p, a hexameric ATPase and homolog of mammalian p97/VCP, acts in retrotranslocation of both luminal and membrane-bound HRD substrates. The HRD complex contains other components, including Der1p, Kar2p and Yos9p, that are not required for Hmg2p degradation and are not shown.
Two lipid signals control the rate of Hmg2p degradation by the HRD pathway: a non-sterol isoprenoid and a sterol [62]. The non-sterol signal is geranylgeranyl pyrophosphate (GGPP), a 20-carbon isoprenoid that increases the susceptibility of Hmg2p to HRD-mediated degradation. GGPP may act by altering the conformation of Hmg2p in the membrane. High HMGR activity increases GGPP synthesis, which in turn alters Hmg2p folding and stimulates HRD-mediated Hmg2p degradation, thereby maintaining lipid homeostasis (Fig. 5). The sterol signal that regulates Hmg2p degradation is derived from oxysterols, which can be produced by cyclization of dioxidosqualene [63]. However, unlike GGPP, the oxysterol-derived signal is not required for Hmg2p degradation. Rather, the oxysterol signal enhances isoprenoid-stimulated degradation. Interestingly, dioxidosqualene-derived oxysterols also act as negative regulators of mammalian sterol synthesis [64].
Nsg1p, a homolog of mammalian Insig, binds Hmg2p and regulates Hmg2p protein levels [65]. However, rather than promoting HMGR degradation as in mammals, S. cerevisiae Nsg1p stabilizes Hmg2p by altering its folding and decreasing its susceptibility to HRD-mediated degradation. Nsg1p also interacts with Hmg1p, although the significance of this interaction is not known.
Regulation of Schizosaccharomyces pombe HMG-CoA reductase
The fission yeast Schizosaccharomyces pombe encodes one HMGR enzyme, called Hmg1. The Hmg1 catalytic domain is highly similar to other eukaryotic HMGRs with 58% identity to human HMGR. The NH2-terminal membrane domain has approximately equal similarity to human and S. cerevisiae HMGRs, with amino acid identity less than 25%. Like mammalian HMGR, the NH2-terminus of Hmg1 is predicted to contain 8 transmembrane segments [30]. Notably, the serine corresponding to the residue phosphorylated by AMPK in mammals is conserved in S. pombe Hmg1 at position 1024 [30, 31].
The S. pombe Insig homolog, called ins1+, was identified by sequence homology searches. Unlike mammalian Insig, Ins1 does not regulate the S. pombe SREBP-Scap pathway [31, 66]. Ins1 has low sequence identity with both S. cerevisiae Nsg1p and human Insig, but shares predicted membrane topology with its mammalian homolog [31]. As in mammals and S. cerevisiae, Hmg1 and Ins1 form a stable complex. But in contrast to both mammals and S. cerevisiae, Ins1 does not positively or negatively regulate turnover of Hmg1, which is a stable protein [30, 31].
Instead of controlling HMGR activity by regulating Hmg1 turnover, Ins1 controls HMGR activity through a non-degradative mechanism [31]. Ins1-Hmg1 binding promotes phosphorylation of Hmg1 S1024 and T1028; S1024 corresponds to human HMGR S872, and T1028 is not conserved in mammals. Phosphorylation of these residues inhibits the enzymatic activity of Hmg1. Ins1- mediated inhibition of Hmg1 activity increases the Km for NADPH, consistent with the idea that HMGR phosphorylation interferes with NADPH binding [21].
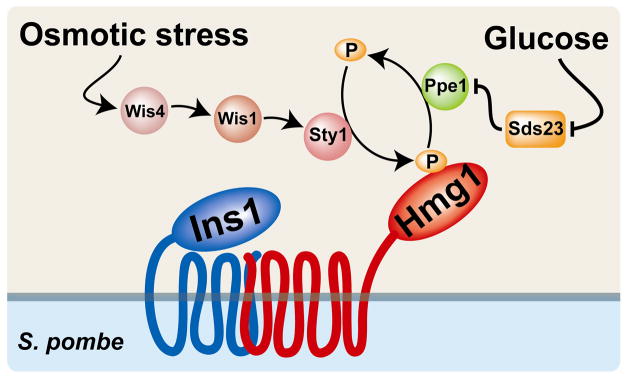
Ins1-dependent Hmg1 phosphorylation is induced by multiple stimuli including growth in minimal medium, osmotic stress and low glucose (Fig. 6)[31, 67]. Hmg1 phosphorylation in minimal medium and osmotic stress requires the stress-responsive mitogen-activated protein kinase (MAPK) Sty1 and its upstream activators Wis4 and Wis1 [31, 68, 69]. In contrast, low glucose phosphorylation of Hmg1 is Sty1-independent (Fig. 6). Low glucose strongly induces phosphorylation of Hmg1 S1024 and T1028, decreasing Hmg1 activity 3-fold [67]. Low glucose-stimulated Hmg1 phosphorylation requires the PP2A-related phosphatase Ppe1 and its negative regulator Sds23 [67, 70]. In cells lacking the Ppe1 phosphatase, Hmg1 is constitutively phosphorylated at S1024 and shows no further increase in S1024 phosphorylation in low glucose [67]. The opposite is true of cells lacking sds23+; sds23Δ cells show no Hmg1 phosphorylation, even in low glucose. Thus, the phosphatase Ppe1 is required for dephosphorylation of Hmg1, and Sds23 is required to prevent Ppe1 from acting when glucose levels are high (Fig. 6). Although phosphorylation of mammalian HMGR is activated by glucose depletion through AMPK [71], the S. pombe AMPK homolog is not essential for low glucose-dependent phosphorylation of Hmg1 [67]. However, like the AMPK γ subunit, Sds23 contains CBS domains, raising the intriguing possibility that S. pombe Hmg1 is regulated by adenosine nucleotide energy sensing like its mammalian counterpart [67, 70].
Figure 6. Hmg1 activity is regulated post-translationally by phosphorylation under the control of the Insig homolog, Ins1, in S. pombe.
Phosphorylation of Hmg1 decreases enzyme activity. Extracellular glucose suppresses activity of the phosphatase Ppe1 through Sds23 thereby preventing Hmg1 dephosphorylation. Osmotic stress stimulates Hmg1 phosphorylation through the MAP kinase Sty1. Ins1 is strictly required for Hmg1 phosphorylation. The enzymes that directly phosphorylate and dephosphorylate Hmg1 have not been identified.
Ins1-dependent Hmg1 phosphoregulation is essential for S. pombe cells to maintain sterol homeostasis, inasmuch as cells lacking ins1+ accumulate sterol pathway intermediates including lanosterol, 24-methylenelanosterol and squalene [31]. Ins1-mediated control of Hmg1 activity is especially important as cells enter stationary phase [67]. Despite low glucose concentrations, ins1Δ cells entering stationary phase continue to produce large quantities of sterol biosynthetic intermediates even as cell growth stops. Thus, Ins1 links nutrient sensing and sterol biosynthesis, allowing the cell to coordinate its anabolism with carbon availability.
Outstanding questions
The past few decades have yielded remarkable progress in our understanding of the molecular details underlying HMGR regulation, but several major questions remain unanswered. In particular, knowledge of how HMGR is dislocated from ER membranes is only beginning to emerge. Because HMGR is degraded by a quality control pathway, it may acquire properties of misfolded proteins. More details regarding HMGR membrane extraction and degradation will be forthcoming as HMGR is a model substrate for understanding ERAD in both yeast and mammalian cells [46, 61].
Many molecular signals that regulate HMGR have been characterized, but our understanding of their action is incomplete (Table 1). 24,25-dihydrolanosterol promotes Insig-HMGR interaction, but whether the lipid binds directly to HMGR as cholesterol does to Scap is unknown [18]. GGOH, which enhances mammalian HMGR membrane dislocation, does not have a defined role and may be required to modify an as-yet unidentified protein [44]. Similarly, GGPP promotes Hmg2p degradation in S. cerevisiae, but its mode of action is unclear [62]. In S. pombe, any mevalonate-derived signals that regulate Hmg1 await discovery.
Table 1.
Signals for HMGR regulation in mammals, S. cerevisiae and S. pombe
| Type of regulation | ||||
|---|---|---|---|---|
| Transcription | Translation | Phosphorylation | Degradation | |
| Mammalian HMGR | cholesterol oxysterols | isoprenoid | ↑AMP/ATP ratio | 24,25-dihydrolanosterol GGOH oxysterols |
| Sc Hmg1p | heme | mevalonate | ||
| Sc Hmg2p | oxygen | GGPP oxysterols | ||
| Sp Hmg1 | glucose osmotic stress | |||
AMPK is thought to be the primary component of mammalian HMGR phosphoregulation [28]. However, the recent finding that phosphoregulation of S. pombe Hmg1 involves multiple signals and regulation of both kinases and phosphatases will stimulate a re-examination of this assumption [31, 67]. While AMPK is clearly a major regulator of mammalian HMGR phosphorylation [28], other kinases may play an equally important role. HMGR phosphorylation is also likely regulated by signals conveyed though PP2A-mediated dephosphorylation.
Conclusions
Here, we summarize the known mechanisms by which HMGR is regulated in mammals, Saccharomyces cerevisiae and Schizosaccharomyces pombe (Tables 1 and 2). Interestingly, S. cerevisiae and S. pombe each employ mechanisms found in the regulation of human HMGR: phosphoregulation and regulated degradation. But, neither yeast employs both. Notably, the signals for each mode of regulation are remarkably consistent across species. Regulated degradation of HMGR is associated with a lipid signal: 24,25-dihydrolanosterol and GGOH in mammals, and GGPP and oxysterols in S. cerevisiae (Figs. 4–5)[34, 62]. Phosphoregulation of HMGR activity is linked to cellular energy state and can be manipulated by altering glucose concentrations in mammals and S. pombe (Figs. 3 and 6)[67, 71].
Table 2.
Insig function in mammals, S. cerevisiae and S. pombe
| Insig homolog | Binds HMGR | Regulated binding | Insig function | |
|---|---|---|---|---|
| Mammalian HMGR | Insig-1 | Yes | Yes | HMGR degradation |
| Sc Hmg1p | Nsg1p | Yes | ? | ? |
| Sc Hmg2p | Nsg1p | Yes | ? | Hmg2p stabilization |
| Sp Hmg1 | Ins1 | Yes | ? | Hmg1 phosphorylation |
Glucose depletion induces HMGR phosphorylation in both S. pombe and mammals, but the mechanisms by which HMGR phosphorylation is regulated are different. S. pombe HMGR phosphorylation requires Insig and occurs in the absence of AMPK (Fig. 6)[67, 71], whereas mammalian HMGR phosphorylation requires AMPK but does not require Insig (Fig. 3)[28, 40]. The fact that the phosphorylation site is conserved suggests that energy-responsive HMGR phosphorylation may be an ancient regulatory system and that the mechanism by which it occurs has diverged over time. Alternatively, HMGR phosphorylation may have developed separately in mammals and S. pombe by convergent evolution.
Insig regulates HMGR in mammals, S. cerevisiae and S. pombe. However, Insig acts in strikingly different ways in each system: it promotes HMGR degradation in mammals, stabilizes Hmg2p in S. cerevisiae and facilitates Hmg1 phosphoregulation in S. pombe (Table 2)[31, 34, 65]. The one common feature of Insig in all three systems is its ability to bind HMGR. Thus, Insig may be thought of as an adaptor protein, evolved primarily to bind SSD-containing proteins and able to facilitate a wide variety of processes.
Acknowledgments
We extend a sincere apology to those whose work was not discussed or cited in this review due to limitations in space and scope. This work was supported by a grant from the National Institutes of Health (HL-077588). PJE is an Established Investigator of the American Heart Association.
The abbreviations used are
- HMGR
HMG-CoA reductase
- ER
endoplasmic reticulum
- SREBP
sterol regulatory element binding protein
- AMPK
AMP-activated protein kinase
- PP2A
protein phosphatase 2A
- CBS
cystathionine β-synthase
- ERAD
endoplasmic reticulum-associated degradation
- HRD
HMG-CoA reductase degradation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldstein JL. A Receptor-mediated pathway for cholesterol homeostasis. In: Frängsmyr T, Lindsten J, editors. Nobel Lectures, Physiology or Medicine 1981–1990. Singapore: World Scientific Publishing Co; 1993. [Google Scholar]
- 2.Yeagle PL. Lipid regulation of cell membrane structure and function. FASEB J. 1989;3:1833–42. [PubMed] [Google Scholar]
- 3.Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274:255–9. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 4.Vance DE, Van den Bosch H. Cholesterol in the year 2000. Biochim Biophys Acta. 2000;1529:1–8. doi: 10.1016/s1388-1981(00)00133-5. [DOI] [PubMed] [Google Scholar]
- 5.Andreasen AA, Stier TJB. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J Cell Physiol. 1953;41:23–36. doi: 10.1002/jcp.1030410103. [DOI] [PubMed] [Google Scholar]
- 6.Nes WR, Sekula BC, Nes WD, Adler JH. The functional importance of structural features of ergosterol in yeast. J Biol Chem. 1978;253:6218–25. [PubMed] [Google Scholar]
- 7.Lees ND, Bard M, Kirsch DR. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1999;34:33–47. [PubMed] [Google Scholar]
- 8.Schoenheimer R, Breusch F. Synthesis and destruction of cholesterol in the organism. J Biol Chem. 1933;103:439–48. [Google Scholar]
- 9.Siperstein MD, Guest MJ. Studies on the site of the feedback control of cholesterol synthesis. J Clin Invest. 1960;39:642–52. doi: 10.1172/JCI104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–30. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 12.Endo A, Kuroda M, Tsujita Y. ML-236A, ML-236B, and ML-236C, new inhibitors of cholesterogenesis produced by Penicillium citrinium. J Antibiot. 1976;29:1346–8. doi: 10.7164/antibiotics.29.1346. [DOI] [PubMed] [Google Scholar]
- 13.Lindsley C. The top prescription drugs of 2009 in the US: CNS therapeutics rank among highest grossing. ACS Chem Neurosci. 2010;1:407–8. doi: 10.1021/cn1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Istvan ES. Bacterial and mammalian HMG-CoA reductases: related enzymes with distinct architectures. Curr Opin Struct Biol. 2001;11:746–51. doi: 10.1016/s0959-440x(01)00276-7. [DOI] [PubMed] [Google Scholar]
- 15.Istvan ES, Deisenhofer J. The structure of the catalytic portion of human HMG-CoA reductase. Biochimica Biophysica Acta. 2000;1529:9–18. doi: 10.1016/s1388-1981(00)00134-7. [DOI] [PubMed] [Google Scholar]
- 16.Bochar DA, Stauffacher CV, Rodwell VW. Sequence comparisons reveal two classes of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol Genet Metab. 1999;66:122–7. doi: 10.1006/mgme.1998.2786. [DOI] [PubMed] [Google Scholar]
- 17.Yu L. The structure and function of Niemann-Pick C1-like 1 protein. Curr Opin Lipidol. 2008;19:263–9. doi: 10.1097/MOL.0b013e3282f9b563. [DOI] [PubMed] [Google Scholar]
- 18.Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. Proc Natl Acad Sci USA. 2007;104:6511–8. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skalnik DG, Narita H, Kent C, Simoni RD. The membrane domain of 3-hydroxy-3-methylglutaryl-coenzyme A reductase confers endoplasmic reticulum localization and sterol-regulated degradation onto beta-galactosidase. J Biol Chem. 1988;263:6836–41. [PubMed] [Google Scholar]
- 20.Frimpong K, Rodwell VW. The active site of hamster 3-hydroxy-3-methylglutaryl-CoA reductase resides at the subunit interface and incorporates catalytically essential acidic residues from separate polypeptides. J Biol Chem. 1994;269:1217–21. [PubMed] [Google Scholar]
- 21.Istvan ES, Palnitkar M, Buchanan SK, Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 2000;19:819–30. doi: 10.1093/emboj/19.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frimpong K, Rodwell VW. Catalysis by Syrian hamster 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proposed roles of histidine 865, glutamate 558, and aspartate 766. J Biol Chem. 1994;269:11478–83. [PubMed] [Google Scholar]
- 23.Tabernero L, Bochar DA, Rodwell VW, Stauffacher CV. Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci USA. 1999;96:7167–71. doi: 10.1073/pnas.96.13.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001;292:1160–4. doi: 10.1126/science.1059344. [DOI] [PubMed] [Google Scholar]
- 25.Endo A, Kuroda M, Tanzawa K. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett. 1976;72:323–6. doi: 10.1016/0014-5793(76)80996-9. [DOI] [PubMed] [Google Scholar]
- 26.Istvan ES. Structural mechanism for statin inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Am Heart J. 2002;144:S27–32. doi: 10.1067/mhj.2002.130300. [DOI] [PubMed] [Google Scholar]
- 27.Clarke PR, Hardie DG. Regulation of HMG-CoA reductase: identification of the site phosphorylated by the AMP-activated protein kinase in vitro and in intact rat liver. EMBO J. 1990;9:2439–46. doi: 10.1002/j.1460-2075.1990.tb07420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardie DG, Hawley SA, Scott JW. AMP-activated protein kinase--development of the energy sensor concept. J Physiol. 2006;574:7–15. doi: 10.1113/jphysiol.2006.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friesen JA, Rodwell VW. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome biology. 2004;5:248. doi: 10.1186/gb-2004-5-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lum PY, Edwards S, Wright R. Molecular, functional and evolutionary characterization of the gene encoding HMG-CoA reductase in the fission yeast, Schizosaccharomyces pombe. Yeast. 1996;12:1107–24. doi: 10.1002/(SICI)1097-0061(19960915)12:11%3C1107::AID-YEA992%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 31.Burg JS, Powell DW, Chai R, Hughes AL, Link AJ, Espenshade PJ. Insig regulates HMG-CoA reductase by controlling enzyme phosphorylation in fission yeast. Cell Metab. 2008;8:522–31. doi: 10.1016/j.cmet.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown MS, Dana SE, Dietschy JM, Siperstein MD. 3-Hydroxy-3-methylglutaryl coenzyme A reductase. Solubilization and purification of a cold-sensitive microsomal enzyme. J Biol Chem. 1973;248:4731–8. [PubMed] [Google Scholar]
- 33.Reynolds GA, Basu SK, Osborne TF, Chin DJ, Gil G, Brown MS, et al. Hmg Coa Reductase - A negatively regulated gene with unusual promoter and 5′ untranslated regions. Cell. 1984;38:275–85. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- 34.DeBose-Boyd RA. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 2008;18:609–21. doi: 10.1038/cr.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet. 2007;41:401–27. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 36.Mohn KL, Laz TM, Hsu JC, Melby AE, Bravo R, Taub R. The immediate-early growth response in regenerating liver and insulin-stimulated H-35 cells: comparison with serum-stimulated 3T3 cells and identification of 41 novel immediate-early genes. Mol Cell Biol. 1991;11:381–90. doi: 10.1128/mcb.11.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diamond RH, Du K, Lee VM, Mohn KL, Haber BA, Tewari DS, et al. Novel delayed-early and highly insulin-induced growth response genes. Identification of HRS, a potential regulator of alternative pre-mRNA splicing. J Biol Chem. 1993;268:15185–92. [PubMed] [Google Scholar]
- 38.Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, et al. Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 39.Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci USA. 2007;104:6519–26. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engelking LJ, Liang G, Hammer RE, Takaishi K, Kuriyama H, Evers BM, et al. Schoenheimer effect explained--feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J Clin Invest. 2005;115:2489–98. doi: 10.1172/JCI25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sever N, Song BL, Yabe D, Goldstein JL, Brown MS, DeBose-Boyd RA. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J Biol Chem. 2003;278:52479–90. doi: 10.1074/jbc.M310053200. [DOI] [PubMed] [Google Scholar]
- 42.Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–40. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin Cell Dev Biol. 2007;18:770–9. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jo Y, Debose-Boyd RA. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit Rev Biochem Mol Biol. 2010;45:185–98. doi: 10.3109/10409238.2010.485605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2001;98:14422–7. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jo Y, Sguigna PV, DeBose-Boyd RA. Membrane-associated Ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl coenzyme a reductase. J Biol Chem. 2011;286:15022–31. doi: 10.1074/jbc.M110.211326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartman IZ, Liu P, Zehmer JK, Luby-Phelps K, Jo Y, Anderson RG, et al. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J Biol Chem. 2010;285:19288–98. doi: 10.1074/jbc.M110.134213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–55. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 49.Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, et al. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–84. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corton JM, Gillespie JG, Hawley SA, Hardie DG. 5-aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur J Biochem. 1995;229:558–65. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- 51.Omkumar RV, Darnay BG, Rodwell VW. Modulation of Syrian hamster 3-hydroxy-3-methylglutaryl-CoA reductase activity by phosphorylation. Role of serine 871. J Biol Chem. 1994;269:6810–4. [PubMed] [Google Scholar]
- 52.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–39. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakanishi M, Goldstein JL, Brown MS. Multivalent control of 3-hydroxy-3-methylglutaryl coenzyme-A reductase - mevalonate-derived product inhibits translation of messenger-RNA and accelerates degradation of enzyme. J Biol Chem. 1988;263:8929–37. [PubMed] [Google Scholar]
- 54.Hampton R, Dimster-Denk D, Rine J. The biology of HMG-CoA reductase: the pros of contra-regulation. Trends Biochem Sci. 1996;21:140–5. [PubMed] [Google Scholar]
- 55.Basson ME, Thorsness M, Rine J. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci USA. 1986;83:5563–7. doi: 10.1073/pnas.83.15.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hampton RY, Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol. 1994;125:299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Hach A. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell Mol Life Sci. 1999;56:415–26. doi: 10.1007/s000180050442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorsness M, Schafer W, D’Ari L, Rine J. Positive and negative transcriptional control by heme of genes encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:5702–12. doi: 10.1128/mcb.9.12.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dimster-Denk D, Thorsness MK, Rine J. Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:655–65. doi: 10.1091/mbc.5.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hampton RY, Bhakta H. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci USA. 1997;94:12944–8. doi: 10.1073/pnas.94.24.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hampton RY, Garza RM. Protein quality control as a strategy for cellular regulation: lessons from ubiquitin-mediated regulation of the sterol pathway. Chem Rev. 2009;109:1561–74. doi: 10.1021/cr800544v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garza RM, Tran PN, Hampton RY. Geranylgeranyl pyrophosphate is a potent regulator of HRD-dependent 3-Hydroxy-3-methylglutaryl-CoA reductase degradation in yeast. J Biol Chem. 2009;284:35368–80. doi: 10.1074/jbc.M109.023994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gardner RG, Shan H, Matsuda SP, Hampton RY. An oxysterol-derived positive signal for 3-hydroxy- 3-methylglutaryl-CoA reductase degradation in yeast. J Biol Chem. 2001;276:8681–94. doi: 10.1074/jbc.M007888200. [DOI] [PubMed] [Google Scholar]
- 64.Brown AJ. 24(S),25-epoxycholesterol: a messenger for cholesterol homeostasis. Int J Biochem Cell Biol. 2009;41:744–7. doi: 10.1016/j.biocel.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 65.Flury I, Garza R, Shearer A, Rosen J, Cronin S, Hampton RY. INSIG: a broadly conserved transmembrane chaperone for sterol-sensing domain proteins. EMBO J. 2005;24:3917–26. doi: 10.1038/sj.emboj.7600855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–42. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Burg JS, Espenshade PJ. Glucose controls phosphoregulation of HMG-CoA reductase through the PP2A-related phosphatase Ppe1 and Insig in fission yeast. J Biol Chem. 2011 doi: 10.1074/jbc.M111.233452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Millar JB, Buck V, Wilkinson MG. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–30. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- 69.Zhou X, Ma Y, Sugiura R, Kobayashi D, Suzuki M, Deng L, et al. MAP kinase kinase kinase (MAPKKK)-dependent and -independent activation of Sty1 stress MAPK in fission yeast. J Biol Chem. 2010;285:32818–23. doi: 10.1074/jbc.M110.135764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanyu Y, Imai KK, Kawasaki Y, Nakamura T, Nakaseko Y, Nagao K, et al. Schizosaccharomyces pombe cell division cycle under limited glucose requires Ssp1 kinase, the putative CaMKK, and Sds23, a PP2A-related phosphatase inhibitor. Genes Cells. 2009;14:539–54. doi: 10.1111/j.1365-2443.2009.01290.x. [DOI] [PubMed] [Google Scholar]
- 71.Sato R, Goldstein JL, Brown MS. Replacement of serine-871 of hamster 3-hydroxy-3-methylglutaryl-CoA reductase prevents phosphorylation by AMP-activated kinase and blocks inhibition of sterol synthesis induced by ATP depletion. Proc Natl Acad Sci USA. 1993;90:9261–5. doi: 10.1073/pnas.90.20.9261. [DOI] [PMC free article] [PubMed] [Google Scholar]