Abstract
Background
Due to the structural and biochemical similarities between the anti-tumor p53 and p73 proteins, we hypothesized that individuals who carry high risk genotypes of p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms have a higher risk of developing second primary malignancy (SPM) in patients after an index squamous cell carcinomas of the head and neck (SCCHN).
Methods
A cohort of 1,269 patients with index cases of SCCHN was recruited between May 1995 and January 2007 at M.D. Anderson Cancer Center and followed for SPM development. Patients were genotyped for p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms. A log-rank test and Cox proportional hazard models were used to compare SPM-free survival and SPM risk among different risk groups with the combined risk genotypes of the two polymorphisms.
Results
Our data demonstrated that patients with p53 WP + PP and p73 GC/GC genotypes had a worse SPM-free survival and an increased SPM risk compared with the corresponding p53 WW and p73 GC/AT +AT/AT genotypes. After combining the two polymorphisms, a borderline significantly or significantly reduced SPM-free survival and increased SPM risk were observed in medium-risk group (p53 WW and p73 GC/GC or p53 P carrier and p73 AT carriers) and high-risk group (p53 P carriers and p73 GC/GC) compared with low-risk group (p53 WW and p73 AT carriers), respectively.
Conclusions
Our results suggest an increased risk of SPM after index SCCHN with both p53 and p73 polymorphisms individually and in combination.
Keywords: p53, p73, Polymorphisms, Squamous cell carcinoma of the head and neck, Second primary malignancy
Introduction
Squamous cell carcinoma of the head and neck (SCCHN) is one of the most common cancers worldwide 1. SCCHN is characterized by highly aggressive tumor growth and results in significant morbidity, commonly in the form of disfigurement and loss or impairment in the ability to speak and swallow 2. These medical and psychosocial consequences are exacerbated by relatively stagnant survival rates over the last 30 years despite advances in treatment 3. The survival advantages afforded by new treatment modalities are undermined by poor prognosis of SCCHN which is to some extent due to the increased likelihood of developing second primary malignancies (SPM) 4.
SPMs are estimated to occur in about 15% of SCCHN patients and are a significant cause of post-treatment morbidity and mortality 4. Although continued use of alcohol and tobacco 5, 6, as well as some cancer treatments 7, 8, has been determined to play a role in the development of SPM, these factors alone do not explain the risk of SPM. Many patients, including smokers and drinkers, never develop SPM, suggesting that genetic susceptibility may also contribute to SPM etiology 9. Determining a genetically susceptible risk group would allow better identification of high-risk SPM subgroups from cancer survivors. By identifying markers of risk for SPM, improved initial treatment management, increased secondary prevention, and currently limited to basic clinical post-treatment screenings, would be possible.
Cell cycle control is paramount in maintaining normal growth and differentiation of cells. Both p53 and p73 are important tumor suppressor genes that regulate the cell cycle via apoptosis and cell cycle arrest. The p53 protein plays an important role in the prevention of carcinogenesis in that upon DNA damage from various agents it mediates pathways leading to DNA repair, cell cycle arrest, and apoptosis 10. Downregulation of p53 leads to diminished DNA repair and poor cell cycle control, ultimately resulting in cellular malignancy 11. Furthermore, p53 has been shown to be mutated in most cancers and approximately half of all SCCHN exhibit such mutations 12, 13. Although the p73 protein does not function as a traditional tumor suppressor gene, its high level of sequence homology with the DNA-binding domains of p53 enables p73 to transactivate p53-response genes, resulting in cell cycle arrest, DNA repair, and apoptosis. Thus, the two proteins, p53 and p73, are interrelated and are considered members of the same family 14-16. In human malignancies involving p53 mutations, p73 expression has been found to be increased, proposing an additional role for p73 as a compensator for p53 in the event of dysfunctional p53 mutations 17-20.
A polymorphism of the p53 consisting of either proline or arginine at amino-acid position 72 has been found in a proline-rich domain necessary for full induction of apoptosis 21. Of the two amino acids, the Arg72 type has been shown to induce apoptosis with faster kinetics and suppresses transformation more efficiently than the p53 Pro72 variant 22. It has been proposed that this increased apoptotic ability is due to an increased ability of Arg72 to localize to the mitochondria resulting in cytochrome c release into the cytosol and subsequent apoptosis 21. Research has suggested an association with the p53 codon 72 polymorphism with risk of several cancers and survival outcomes 23-26. While findings suggest that p73 mutations are rare 17, 27, it is possible that genetic variation of p73 may lead to differences in susceptibility to cancer. Specifically, it is believed that the two linked, noncoding polymorphisms at exon 2 of p73 at positions 4 (G→A) and 14 (C→T) (the p73 G4C14-to-A4T14 polymorphism) affect p73 function by altering gene expression 28. Previous studies have documented the role of this polymorphism on risk of several cancers including SCCHN and survival outcomes 29-33.
More recently, we have reported that each of p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms alone was associated with risk of SPM in patients after index SCCHN 34, 35. However, since these proteins do not function in isolation from one another, a combined analysis of both p53 and p73 polymorphisms has not been performed to determine the joint effects on risk of SPM in patients with index SCCHN. To test whether individuals who carry a higher number of risk genotypes of both p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms have a higher risk of SPM after index SCCHN, we analyzed the combined effect of these two polymorphisms in a cohort of 1,269 index cases of SCCHN to compare SPM-free survival and SPM risk between different risk groups with the combined risk genotypes.
Materials and Methods
Study Subjects
This research was approved by the institutional review board of the University of Texas M. D. Anderson Cancer Center. Details and response rate for this study have been previously published 34, 35. For this combined analysis, the cases with index SCCHN were recruited through the Head and Neck Clinic at the University of Texas M. D. Anderson Cancer Center between May 1995 and January 2007 as part of an ongoing molecular epidemiological study.
At our institution, SCCHN patients are typically followed and monitored through their treatment and post-treatment courses with regularly scheduled clinical and radiographic examinations. Based on modified criteria of Warren and Gates 36, SPMs were considered if the second lesions were different histopathologic type, or if they occurred more than 5 years following treatment for the index tumor, and/or clearly separated by normal epithelium based on clinical and radiographic assessment. Pulmonary lesions were considered as a SPM if they had a non-squamous histology; or if they were isolated squamous lesions greater than 5 years from initial SCCHN and felt to be SPM by the thoracic oncologist and thoracic surgeon. If there was discrepancy or differing of opinions regarding the origin of the tumor (i.e., recurrence vs. SPM), the second lesion was classified as a local recurrence rather than a SPM.
Genotype analysis
Genomic DNA was isolated from patients' peripheral leukocyte pellets according to manufacturer's instructions (QIAGEN Inc., Valencia, CA). Genotyping of p53 and p73 polymorphisms was performed as previously described 30, 34. More than 10% of the samples were randomly selected and retested, and the results were 100% concordant.
Statistical Analysis
Software utilized for analysis was Statistical Analysis System software (SAS version 9.1.3; SAS Institute). Statistical significance was set at p<0.05 and all tests were two-sided. Chi-squared tests were used to assess differences in demographic and clinical variables, as well as genotype distributions between the groups of patients who developed SPM and those who remained SPM free.
Kaplan-Meier methods were used to determine if there were significant differences (p < 0.05) in SPM-free survival between different risk groups with the combined genotypes. Both univariate and multivariable Cox proportional hazards regression model were used for assessment as previously published 34, 35. Details in building the multivariable proportional hazards model was described previously 34, 35. After a stepwise search strategy was used in building the multivariable proportional hazards model, the final, fully adjusted Cox regression models included age, sex, ethnicity, and smoking and alcohol status.
Results
Patient Characteristics
The demographics and clinical variables for the study patients are shown in Table 1. Overall, a total of 1,269 SCCHN patients were included in the study. Of those recruited, 1,160 patients remained SPM free and 109 developed SPMs. SPM-free patients and those who developed SPM appeared to be no significant differences in sex, ethnicity, and alcohol drinking status) (P = 0.704, P = 0.100, and P = 0.124, respectively); however, patients with SPM were more likely to be older (P < 0.001) and smokers (P = 0.021). Compared with the SPM-free group, patients who developed SPM had similar characteristics with respect to index cancer site (P = 0.220), index cancer stage (P = 0.866), and treatment (P = 0.910).
Table 1. Distribution of selected characteristics of the patient cohort (n = 1269).
| Variable | Total | SPM/Total | SPM-Free | SPM | P-values* | |||
|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |||
| Total patients | 1269 | 100 | 8.6 | 1160 | 91.4 | 109 | 8.6 | |
| Age | ||||||||
| ≤ median (57 years) | 664 | 52.3 | 5.3 | 629 | 54.2 | 35 | 32.1 | < 0.001 |
| > median (57 years) | 605 | 47.7 | 12.2 | 531 | 45.8 | 74 | 67.9 | |
| Sex | 0.704 | |||||||
| Male | 959 | 75.6 | 8.8 | 875 | 75.4 | 84 | 77.3 | |
| Female | 310 | 24.4 | 8.1 | 285 | 24.6 | 25 | 22.7 | |
| Ethnicity | 0.100 | |||||||
| Non-Hispanic White | 1084 | 85.4 | 7.8 | 999 | 86.1 | 85 | 78.0 | |
| Other | 185 | 14.6 | 13.0 | 161 | 13.8 | 24 | 22.0 | |
| Smoking | 0.021 | |||||||
| Never | 340 | 26.8 | 5.6 | 321 | 27.7 | 19 | 17.4 | |
| Ever | 929 | 73.2 | 9.7 | 839 | 72.3 | 90 | 82.6 | |
| Alcohol | 0.124 | |||||||
| Never | 335 | 26.4 | 6.6 | 313 | 27.0 | 22 | 20.2 | |
| Ever | 934 | 73.6 | 9.3 | 847 | 73.0 | 87 | 79.8 | |
| Index Cancer Site | 0.220 | |||||||
| Oral cavity | 405 | 31.9 | 8.4 | 371 | 32.0 | 34 | 31.2 | |
| Oropharynx | 573 | 45.2 | 7.5 | 530 | 45.7 | 43 | 39.4 | |
| Larynx/Hypopharynx | 291 | 22.9 | 11.0 | 259 | 22.3 | 32 | 29.4 | |
| Index Cancer Stage | 0.866 | |||||||
| 1 or 2 | 329 | 25.9 | 8.8 | 300 | 25.9 | 29 | 26.6 | |
| 3 or 4 | 940 | 74.1 | 8.5 | 860 | 74.1 | 80 | 73.4 | |
| Treatment | 0.910 | |||||||
| Surgery only | 225 | 17.7 | 8.4 | 206 | 17.8 | 19 | 17.4 | |
| Surgery + Adjuvant Tx1 | 309 | 24.4 | 9.4 | 280 | 24.1 | 29 | 26.6 | |
| XRT2 | 334 | 26.3 | 7.8 | 308 | 26.6 | 26 | 23.9 | |
| XRT + Chemotherapy | 401 | 31.6 | 8.7 | 366 | 31.5 | 35 | 32.1 | |
Adjuvant Tx: adjuvant radiotherapy and/or chemotherapy
XRT: radiotherapy
P values were calculated from chi-square test
Combined effects of the p53 and p73 polymorphisms on risk of SPM
In this study, we examined the distribution of the combined p53 and p73 genotypes among the patients who developed SPM, those who remained SPM free, and the associations with risk of SPM (Table 2). Because both p53 and p73 variant homozygous genotypes were relatively uncommon, we combined the variant homozygous with the heterozygous genotypes for the final analyses. As previously reported 34, 35, the p53 WP + PP variant genotypes were more common among patients with SPM than among patients who remained SPM free (P = 0.008) and was associated with approximately 60% increased risk for SPM compared with the WW genotype (adjusted HR, 1.58; 95% CI, 1.07-2.34), while the p73 GC/GC genotype was more common in patients who developed SPM (P = 0.019) and was associated with approximately 70% increased risk for SPM compared with the GC/AT + AT/AT variant genotypes (HR, 1.68; 95% CI, 1.12-2.52). Because p53 and p73 share a common pathway, we used meaningful combination of the two polymorphisms to determine whether the combined risk genotypes modified the risk of SPM. The patients carrying p53 WW and p73 GC/AT + AT/AT genotypes were placed in low-risk group; the patients with p53 WW and p73 GC/GC or p53 WP + PP and p73 GC/AT + AT/AT were placed in medium-risk group; and the patients with p53 WP + PP and p73 GC/GC were placed in high-risk group. Our data demonstrated that the patients had significant differences in SPM-free survival among the three different risk groups (overall log-rank: P = 0.0008, specifically, P = 0.0004 for high-risk to low-risk; P = 0.0860 for medium-risk to low-risk; and P = 0.0096 for high-risk to medium-risk, respectively) (Figure 1). After adjusting for age, sex, ethnicity, tobacco smoking and alcohol drinking, the patients in medium-risk and high-risk groups had an approximately 1.7- and 2.7-fold elevated risk for developing a SPM compared with those in low-risk group (adjusted HR, 1.66; 95% CI, 1.00-3.06 for medium-risk group and adjusted HR, 2.69; 95% CI, 1.44-5.00 for high-risk group). Furthermore, a dose-response relationship was observed among the three risk groups with different numbers of risk genotypes of the two polymorphisms (Ptrend = 0.0007).
Table 2. SPM risk associated with p53 and p73 polymorphisms after index SCCHN.
| Genotypes and no. of variant alleles | Total (No. = 1269) |
SPM/Total | SPM-free (No. = 1160) |
SPM (No. = 109) |
Pa | HR(95% CI)b | |||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | % | No. | % | No. | % | |||
| p53 | 0.008 | ||||||||
| WW | 655 | 51.6 | 6.6 | 612 | 52.8 | 43 | 39.4 | 1.00 (Reference) | |
| WP+PP | 614 | 48.4 | 10.7 | 548 | 47.2 | 66 | 60.6 | 1.58 (1.07-2.34) | |
| p73 | 0.019 | ||||||||
| GA+ AA | 530 | 41.8 | 6.4 | 496 | 42.8 | 34 | 31.2 | 1.00 (Reference) | |
| GG | 739 | 58.2 | 10.1 | 664 | 57.2 | 75 | 68.8 | 1.68 (1.12-2.52) | |
| Combined risk genotypes | 0.002 | ||||||||
| Low risk group | 279 | 22.0 | 4.7 | 266 | 22.9 | 13 | 11.9 | 1.00 (Reference) | |
| Medium risk group | 627 | 49.4 | 8.1 | 576 | 49.7 | 51 | 46.8 | 1.66 (1.00-3.06) | |
| High risk group | 363 | 28.6 | 12.4 | 318 | 27.4 | 45 | 41.3 | 2.69 (1.44-5.00) | |
| Trend test | 0.0007 | ||||||||
χ2 test for differences in the distribution of p73 genotypes between the patients who developed SPM and the patients who did not.
Adjusted for age, sex, ethnicity, tobacco smoking and alcohol drinking in a Cox model.
Fig. 1.
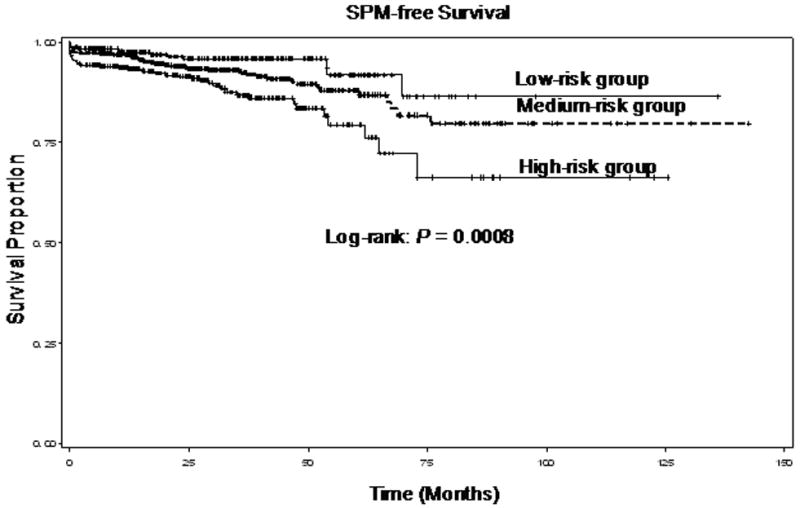
SPM-free survival of patients with SCCHN by the combined risk genotypes of the p53 and p73 polymorphisms (overall log-rank: P = 0.0008, specifically, P = 0.0004 for high-risk to low-risk; P = 0.0860 for medium-risk to low-risk; and P = 0.0096 for high-risk to medium-risk, respectively)
Stratification Analysis of the combined p53 and p73 genotypes with risk of SPM
To further evaluate risk of SPM for specific subgroups, the data was further stratified by age, sex, ethnicity, smoking status, drinking status, treatment, index tumor stage, and tumor site (Table 3). In each subgroup except females, the patients in medium-risk group had an increased risk for SPM compared with those in low-risk group, although the increased risk was only statistically significant for males (adjusted HR, 2.50; 95% CI, 1.12-5.56). While there was an increased risk of SPM for all subgroups in high-risk group and the increased risk was statistically significant for patients older than 57 years (adjusted HR, 3.46; 95% CI, 1.52-7.86), males (adjusted HR, 3.32; 95% CI, 1.47-7.52), non-Hispanic whites (adjusted HR, 2.56; 95% CI, 1.28-5.13), smokers (adjusted HR, 3.0; 95% CI, 1.49-6.06), and drinkers (adjusted HR, 2.53; 95% CI, 1.28-4.99). Furthermore, the patients with late stage index SCCHN (3 or 4) at time of diagnosis, those with DNA damaging treatments (radiotherapy or chemotherapy), and those with index non-oropharyngeal cancer had a significantly pronounced SPM risk (adjusted HR, 2.79; 95% CI, 1.37-5.70, adjusted HR,3.17; 95%CI, 1.57-6.39, and adjusted HR, 2.92; 95% CI, 1.33-6.41, respectively).
Table 3. Stratification analysis of combined association of p53 and p73 polymorphisms with SPM risk.
| Low risk group (Ref.) | Medium risk group | High risk group | ||||||
|---|---|---|---|---|---|---|---|---|
| SPM-free No. % | SPM No. % | SPM-free No. % | SPM No. % | HR (95%CI)* | SPM-free No. % | SPM No. % | HR (95%CI)* | |
| Age(years) | ||||||||
| ≤57 | 156 (58.6) | 6 (46.2) | 297(51.6) | 19(37.3) | 1.65 (0.65-4.21) | 176(55.3) | 10(22.2) | 1.50 (0.54-4.21) |
| >57 | 110 (41.4) | 7 (53.8) | 279(48.4) | 32(62.7) | 1.69 (0.74-3.84) | 142(44.7) | 35(77.8) | 3.46 (1.52-7.86) |
| Sex | ||||||||
| Male | 192(72.2) | 7(53.8) | 437(75.9) | 44(86.3) | 2.50 (1.12-5.56) | 246(77.4) | 33(73.3) | 3.32 (1.47-7.52) |
| Female | 74 (27.8) | 6 (46.2) | 139(24.1) | 7(13.7) | 0.53 (0.18-1.59) | 72(22.6) | 12(26.7) | 1.95 (0.69-5.47) |
| Ethnicity | ||||||||
| Non-HW | 245(92.1) | 11(84.6) | 497(86.3) | 44(86.3) | 1.81 (0.93-3.51) | 257(80.8) | 30(66.7) | 2.56 (1.28-5.13) |
| Others | 21(7.9) | 2(15.4) | 79(13.7) | 7(13.7) | 1.30 (0.27-6.33) | 61(19.2) | 15(33.3) | 3.50 (0.77-15.9) |
| Smoking Status | ||||||||
| Ever | 197(74.1) | 10(76.9) | 405(70.3) | 42(82.4) | 1.88 (0.94-3.77) | 237(74.5) | 38(84.4) | 3.00 (1.49-6.06) |
| Never | 69(25.9) | 3(23.1) | 171(29.7) | 9(17.6) | 1.23 (0.32-4.65) | 81(25.5) | 7(15.6) | 1.43 (0.34-6.00) |
| Drinking Status | ||||||||
| Ever | 200(75.2) | 11(84.6) | 415(72.1) | 39(76.5) | 1.49 (0.76-2.93) | 232(73.0) | 37(82.2) | 2.53 (1.28-4.99) |
| Never | 66(24.8) | 2(15.4) | 161(27.9) | 12(23.5) | 2.25 (0.49-10.3) | 86(27.0) | 8(17.8) | 2.84 (0.57-14.1) |
| Treatment | ||||||||
| Surgery only | 51(19.2) | 3(23.1) | 92(16.0) | 10(19.6) | 1.22 (0.32-4.59) | 63(19.8) | 6(13.3) | 1.08 (0.26-4.39) |
| DNA damaging | 215(80.8) | 10(76.9) | 484(84.0) | 41(80.4) | 1.73 (0.86-3.46) | 255(80.2) | 39(86.7) | 3.17 (1.57-6.39) |
| Stage | ||||||||
| Early (1 or 2) | 66(24.8) | 3(23.1) | 156(27.1) | 15(29.4) | 1.55 (0.44-5.49) | 78(24.5) | 11(24.4) | 2.22 (0.61-8.06) |
| Late (3 or 4) | 200(75.2) | 10(76.9) | 420(72.9) | 36(70.6) | 1.75 (0.86-3.53) | 240(75.5) | 34(75.6) | 2.79 (1.37-5.70) |
| Tumor site | ||||||||
| Oropharnx | 126(47.4) | 5(38.5) | 263(45.7) | 24(47.1) | 2.08 (0.79-5.48) | 141(44.3) | 14(31.1) | 2.30 (0.82-6.47) |
| Non-oropharynx | 140(52.6) | 8(61.5) | 313(54.3) | 27(52.9) | 1.44 (0.65-3.18) | 177(55.7) | 31(68.9) | 2.92 (1.33-6.41) |
Adjusted for age, sex, ethnicity, tobacco smoking and alcohol drinking in a Cox model.
TA: Tobacco-Associated
Discussion
The role of both p53 and p73 proteins in modulating carcinogenesis has been well established; and there is an apparent difference between the different polymorphic forms. In our study of 1,269 SCCHN patients, we analyzed the two well known polymorphisms, p53 codon 72 and p73 G4C14-to-A4T14, and their associations with the risk of SPM. Our previous studies have shown that the p53 and p73 polymorphisms individually modify the risk of SPM 34, 35, however, no study has been done to assess the joint effect of the two polymorphisms on risk of SPM. In this study, we found that the p53 and p73 polymorphisms jointly borderline significantly (medium-risk group) or significantly (high-risk group) increased the risk of SPM, and such joint effect on risk of SPM was more pronounced in certain subgroups, suggesting that the p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms may jointly modify the risk of SPM after index SCCHN.
It is biologically plausible that p53 and p73 play a role in the development of SPM since these two proteins have similar biological properties and each may play similar roles in the regulation of cell cycle control, DNA repair, and apoptosis. Furthermore, members of the p53 family, including p53 and p73, have been shown to interact in development of human cancers. In malignancies associated with loss of p53 expression, an increased expression of p73 has been observed in malignant tissues compared with adjacent normal tissues, providing evidence that p73 may compensate for the loss of p53 function 17-20. Thus, these two proteins may also have a combined effect on SPM risk.
The two polymorphic forms of p53 may result in a marked alteration of the primary structure of the protein, thereby modifying its biochemical properties and effects 36. The Pro72 variant interacts more effectively with elements of the transcriptional machinery and is capable of inducing higher levels of transcriptional activity than the Arg72 form. It also induces G1 arrest and more effectively activates DNA repair system 21, 22, 37, 38. However, Arg72 has demonstrated apoptotic induction with faster kinetics and suppresses transformation more efficiently than the Pro72 variant 22, 37. Thus, the differences in these biological activities caused by each of two polymorphic variants may result in a different effect modification of SPM risk. On the other hand, the p73 protein, similar to p53, also plays a role in DNA repair, cell cycle regulation, and apoptosis, and therefore influences tumor development and progression. The location of the p73 G4C14-to-A4T14 polymorphism is upstream of the initiating AUG of exon 2 and may have a role in the formation of a stem-loop structure, which may result in an alteration of gene expression by altering the initiation of translation 39, thereby modifying the risk of human cancer including SPM. However, further studies are needed to confirm these biological functions of the two polymorphisms.
Either p53 or p73 polymorphism has been reported to be associated with risk for several human malignancies, including SCCHN 30, 32, 40. Recently, we also found that each of the p53 codon 72 and the p73 G4C14-to-A4T14 polymorphisms moderately modified the risk of SPM after an index SCCHN 34, 35. Since there is considerably biological interaction between p53 and p73 proteins, a recent case-control study found a combined effect of p53 and p73 polymorphisms on risk of head and neck cancer in an Italian population 41. Similarly, we, therefore, undertook the current study with a combined analysis for the p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms and their association with risk for SPM. We did find that the results from current study were consistent with the notion that these two polymorphisms may jointly increase risk of SPM. Moreover, the risk of the combined risk genotypes of the two polymorphisms was more pronounced in several subgroups, including older patients, males, non-Hispanic whites, smokers, drinkers, patients with late stage index SCCHN, and those with DNA damaging treatments, and patients with non-oropharyngeal cancer, in each of which we found the similar results for each of the two polymorphisms 34, 35. It is possible that these polymorphisms may affect the DNA repair capacity of damage induced by tobacco and alcohol carcinogens, DNA damaging therapy, or reduced DNA repair capacity by aging. Although how ethnicity affects the SPM risk is not clear, it is possible that certain behaviors and other genetic factors may play a role in development of SPM. Additionally, the late-stage patients with more extensive treatment modalities including chemotherapy and/or radiotherapy may have more extensive DNA damage.
Our study has some limitations. First, although the study was performed in a large cohort of SCCHN patients, approximately 85% of the patients were non-Hispanic white. Nevertheless, ethnicity was adjusted for in the multivariable analyses. Secondly, while demographics, exposure, and clinical data for the cohort were collected prospectively, the clinical outcomes such as SPM were collected retrospectively. Therefore, follow-up time was limited and patients may not have had enough time to develop SPM or could have been lost to follow-up. Also, the prevalence of never-smokers, late stage index cancer patients, and our strict criteria for determining SPM, resulted in an SPM rate that was lower than expected. Therefore, the low rate of SPM limited statistical power for the analysis, particularly for the stratified analysis. Finally, data on HPV status, one of the major risk factors for SCCHN, was not taken into account. Although the major risk factor for SCCHN is the exposure to tobacco and/or alcohol, currently sufficient evidence concludes that there is strong and consistent association between oncogenic human papillomavirus (HPV) (principally type 16 and occasionally type 18) and a distinct subset of head and neck cancers (i.e., soft palate, palatine tonsil, and base of tongue / lingual tonsil) 42-44. Despite declining smoking rates in the United States, the rising incidence of oral cavity and pharyngeal cancer within certain sites, particularly the base of tongue, tonsil, and oropharynx, among white men born since the mid-1940s appears attributed to the increasingly prevalent infection of oncogenic subtypes of HPV and may reflect changes in sexual practices since the mid-1960s 45. Molecular studies have shown that oncogenic E6 and E7 proteins of HPV have a high binding affinity for p53 and RB promoting the ubiquitination and complete degradation of these tumor suppressor genes, leading to the deregulation of cell cycle control and subsequent tumor development 42-44. Studies also have shown that HPV-positive patients appear to be a distinct epidemiologic, clinical, and molecular subgroup which exhibits unique clinical behaviors and treatment responses compared with HPV-negative patients 43, 44. In current study, the absence of HPV status did not allow us to evaluate its potential influence on the development of SPMs in patients with index SCCHN. Thus, we will closely monitor the role of HPV in the outcomes of SCCHN patients in our future studies when a much larger patient cohort with HPV-associated tumor becomes available.
In conclusion, our results show that p73 and p53 polymorphisms jointly significantly increase the risk of SPM development following an index SCCHN, such risk was more pronounced in several subgroups. This study provides evidence that simultaneous presence of the p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms may have joint effects on increased risk of SPM, and the combination of the two polymorphisms may provide more comprehensive and accurate estimates of the risk of SPM than the single polymorphism alone.
Acknowledgments
The authors wish to thank Ms. Margaret Lung, Ms. Angeli Fairley, Ms. Liliana Mugartegui, and Ms. Kathryn Tipton with assistance with patient recruitment, and Dr. Chong Zhao and Ms. Yingdong Li for laboratory assistance.
Funded by: Research Training Award, The American Laryngological, Rhinological, and Otological Society (to E.M.S.); U.T. M.D. Anderson Cancer Center Start-up Funds (to E.M.S.); N.I.H. Grant K-12 88084 (to E.M.S., faculty trainee; to R.C. Bast, P.I.); National Institute of Environmental Health Sciences Grant R01 ES-11740 (to Q.W.); CA135679-01 (G.L.); and CA133099-01A1 (G.L.).
Abbreviations
- CI
confidence interval
- HR
hazard ratio
- SCCHN
squamous cell carcinoma of the head and neck
- SPM
second primary malignancy
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010 Jun 17; doi: 10.1002/ijc.25516. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 2.Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328:184–194. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society, Inc. Cancer Facts and Figures. American Cancer Society; Atlanta: 2007. [Google Scholar]
- 4.Sturgis EM, Miller RH. Second primary malignancies in the head and neck cancer patient. Ann Otol Rhinol Laryngol. 1995;104:946–954. doi: 10.1177/000348949510401206. [DOI] [PubMed] [Google Scholar]
- 5.Day GL, Blot WJ, Shore RE, et al. Second cancers following oral and pharyngeal cancers: role of tobacco and alcohol. J Natl Cancer Inst. 1994;86:131–137. doi: 10.1093/jnci/86.2.131. [DOI] [PubMed] [Google Scholar]
- 6.Do KA, Johnson MM, Lee JJ, et al. Longitudinal study of smoking patterns in relation to the development of smoking-related secondary primary tumors in patients with upper aerodigestive tract malignancies. Cancer. 2004;101:2837–2842. doi: 10.1002/cncr.20714. [DOI] [PubMed] [Google Scholar]
- 7.Ng AK, Travis LB. Second primary cancers: an overview. Hematol Oncol Clin North Am. 2008;22:271–289. doi: 10.1016/j.hoc.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert ES, Stovall M, Gospodarowicz M, et al. Lung cancer after treatment for Hodgkin's disease: focus on radiation effects. Radiat Res. 2003;159:161–173. doi: 10.1667/0033-7587(2003)159[0161:lcatfh]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Zafereo ME, Sturgis EM, Liu Z, Wang L, Wei Q, Li G. Nucleotide excision repair core gene polymorphisms and risk of second primary malignancy in patients with index squamous cell carcinoma of the head and neck. Carcinogenesis. 2009;30:997–1002. doi: 10.1093/carcin/bgp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helton ES, Chen X. p53 modulation of the DNA damage response. J Cell Biochem. 2007;100:883–896. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- 11.Millau JF, Bastien N, Drouin R. P53 transcriptional activities: a general overview and some thoughts. Mutat Res. 2009;681:118–133. doi: 10.1016/j.mrrev.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Brachman DG, Graves D, Vokes E, et al. Occurrence of p53 gene deletions and human papilloma virus infection in human head and neck cancer. Cancer Res. 1992;52:4832–4836. [PubMed] [Google Scholar]
- 13.Bénard J, Douc-Rasy S, Ahomadegbe JC. TP53 family members and human cancers. Hum Mutat. 2003;21:182–191. doi: 10.1002/humu.10172. [DOI] [PubMed] [Google Scholar]
- 14.Melino G. p73, the “assistant” guardian of the genome? Ann N Y Acad Sci. 2003;1010:9–15. doi: 10.1196/annals.1299.002. [DOI] [PubMed] [Google Scholar]
- 15.Tomasini R, Mak TW, Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol. 2008;18:244–252. doi: 10.1016/j.tcb.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Flores ER, Tsai KY, Crowley D, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 17.Yokomizo A, Mai M, Tindall DJ, et al. Overexpression of the wild type p73 gene in human bladder cancer. Oncogene. 1999;18:1629–1633. doi: 10.1038/sj.onc.1202474. [DOI] [PubMed] [Google Scholar]
- 18.Nimura Y, Mihara M, Ichimiya S, et al. p73, a gene related to p53, is not mutated in esophageal carcinomas. Int J Cancer. 1998;78:437–440. doi: 10.1002/(sici)1097-0215(19981109)78:4<437::aid-ijc8>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 19.Zaika AI, Kovalev S, Marchenko ND, Moll UM. Overexpression of the wild type p73 gene in breast cancer tissues and cell lines. Cancer Res. 1999;59:3257–3263. [PubMed] [Google Scholar]
- 20.Kang MJ, Park BJ, Byun DS, et al. Loss of imprinting and elevated expression of wild-type p73 in human gastric adenocarcinoma. Clin Cancer Res. 2000;6:1767–1771. [PubMed] [Google Scholar]
- 21.Dumont P, Leu JI, Della Pietra AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 22.Thomas M, Kalita A, Labrecque S. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almeida PS, Manoel WJ, Reis AA, et al. TP53 codon 72 polymorphism in adult soft tissue sarcomas. Genet Mol Res. 2008;7:1344–1352. doi: 10.4238/vol7-4gmr497. [DOI] [PubMed] [Google Scholar]
- 24.Han J, Cox DG, Colditz GA, Hunter DJ. The p53 codon 72 polymorphism, sunburns, and risk of skin cancer in US Caucasian women. Mol Carcinog. 2006;45:694–700. doi: 10.1002/mc.20190. [DOI] [PubMed] [Google Scholar]
- 25.Yi SY, Lee WJ. A p53 genetic polymorphism of gastric cancer: difference between early gastric cancer and advanced gastric cancer. World J Gastroenterol. 2006;12:6536–6539. doi: 10.3748/wjg.v12.i40.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ezzikouri S, El Feydi AE, Chafik A. The Pro variant of the p53 codon 72 polymorphism is associated with hepatocellular carcinoma in Moroccan population. Hepatol Res. 2007;37:748–754. doi: 10.1111/j.1872-034X.2007.00126.x. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi H, Ichimiya S, Nimura Y, et al. Mutation, allelotyping, and transcription analyses of the p73 gene in prostatic carcinoma. Cancer Res. 1998;58:2076–2077. [PubMed] [Google Scholar]
- 28.Chen X, Sturgis EM, Etzel CJ, Wei Q, Li G. p73 G4C14-to-A4T14 polymorphism and risk of human papillomavirus-associated squamous cell carcinoma of the oropharynx in never smokers and never drinkers. Cancer. 2008;113:3307–3314. doi: 10.1002/cncr.23976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li G, Wang LE, Chamberlain RM, Amos CI, Spitz MR, Wei Q. p73 G4C14-to-A4T14 polymorphism and risk of lung cancer. Cancer Res. 2004;64:6863–6866. doi: 10.1158/0008-5472.CAN-04-1804. [DOI] [PubMed] [Google Scholar]
- 30.Li G, Sturgis EM, Wang LE, et al. Association of a p73 exon 2 G4C14-to-A4T14 polymorphism with risk of squamous cell carcinoma of the head and neck. Carcinogenesis. 2004;25:1911–1916. doi: 10.1093/carcin/bgh197. [DOI] [PubMed] [Google Scholar]
- 31.Ryan BM, McManus R, Daly JS, et al. Common p73 polymorphism is associated with a reduced incidence of oesophageal carcinoma. Br J Cancer. 2001;85:1499–1503. doi: 10.1054/bjoc.2001.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeifer D, Arbman G, Sun XF. Polymorphism of the p73 gene in relation to colorectal cancer risk and survival. Carcinogenesis. 2005;26:103–107. doi: 10.1093/carcin/bgh305. [DOI] [PubMed] [Google Scholar]
- 33.Niwa Y, Hamajima N, Atsuta Y, et al. Genetic polymorphisms of p73 G4C14-to-A4T14 at exon 2 and p53 Arg72Pro and the risk of cervical cancer in Japanese. Cancer Lett. 2004;205:55–60. doi: 10.1016/j.canlet.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Li F, Sturgis EM, Zafereo M, Wei Q, Li G. Association of p53 codon 72 polymorphism with risk of second primary malignancy in patients with squamous cell carcinoma of the head and neck. Cancer. 2010;116:2350–2359. doi: 10.1002/cncr.25072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, Sturgis EM, Zafereo M, et al. p73 G4C14-to-A4T14 polymorphism and risk of second primary malignancy after index squamous cell carcinoma of the head and neck. Int J Cancer. 2009;125:2660–2665. doi: 10.1002/ijc.24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren S, Gates O. Multiple primary malignant tumors: a survey of the literature and statistical study. Am J Cancer. 1932;51:1358. [Google Scholar]
- 37.Pim D, Banks L. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer. 2004;108:196–199. doi: 10.1002/ijc.11548. [DOI] [PubMed] [Google Scholar]
- 38.Siddique M, Sabapathy K. Trp53-dependent DNA-repair is affected by the codon 72 polymorphism. Oncogene. 2006;25:3489–3500. doi: 10.1038/sj.onc.1209405. [DOI] [PubMed] [Google Scholar]
- 39.Kaghad M, Bonnet H, Yang A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 40.De Feo E, Persiani R, La Greca A, et al. A case-control study on the effect of p53 and p73 gene polymorphisms on gastric cancer risk and progression. Mutat Res. 2009;675:60–65. doi: 10.1016/j.mrgentox.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Gallì P, Cadoni G, Volante M, et al. A case-control study on the combined effects of p53 and p73 polymorphisms on head and neck cancer risk in an Italian population. BMC Cancer. 2009;9:137. doi: 10.1186/1471-2407-9-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14:467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 43.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 44.Gillison ML, Lowy DR. A causal role for human papillomavirus in head and neck cancer. Lancet. 2004;363:1488–1489. doi: 10.1016/S0140-6736(04)16194-1. [DOI] [PubMed] [Google Scholar]
- 45.Brown LM, Check DP, Devesa SS. Oropharyngeal cancer incidence trend: diminishing racial disparities. Cancer Cause Control. 2011 doi: 10.1007/s10552-011-9748-1. epub 3-05-2011. [DOI] [PubMed] [Google Scholar]


