Abstract
Despite the advances in immunosuppression, renal allograft attrition over time remains unabated due to chronic allograft dysfunction (CAD) with interstitial fibrosis (IF) and tubular atrophy (TA). We aimed to evaluate microRNA (miRNA) signatures in CAD with IF/TA and appraise correlation with paired urine samples and potential utility in prospective evaluation of graft function. MicroRNA signatures were established between CAD with IF/TA vs. normal allografts by microarray. Validation of the microarray results and prospective evaluation of urine samples was performed using RT-qPCR. Fifty-six miRNAs were identified in samples with CAD-IF/TA. Five miRNAs were selected for further validation based on: array fold change, p-value and in silico predicted mRNA targets. We confirmed the differential expression of these 5 miRNAs by RT-qPCR using an independent set of samples. Differential expression was detected for miR-142-3p, miR-204, miR-107, and miR-211 (P<0.001) and miR-32 (p<0.05). Furthermore, differential expression of miR-142-3p (p<0.01), miR-204 (p<0.01) and miR-211 (p<0.05) was also observed between patient groups in urine samples. A characteristic miRNA signature for IF/TA that correlates with paired urine samples was identified. These results support the potential use of miRNAs as non-invasive markers of IF/TA and for monitoring graft function.
Keywords: miRNA, chronic allograft dysfunction, IF/TA
INTRODUCTION
Kidney transplantation (KT) is the treatment of choice for end-stage kidney disease (1). Despite the advances in immunosuppression, allograft attrition over time remains unabated due to chronic allograft dysfunction (CAD) (1). As defined by the Banff criteria, CAD remains a non-specific pathologic entity encompassing histological changes of interstitial fibrosis and tubular atrophy (IF/TA), glomerulosclerosis, splitting of glomerular capillary basement membranes and/or vascular intimal hyperplasia (1,2).
A major obstacle in the management of transplant recipients is a lack of accurate and specific tests for immune monitoring as well as predicting long-term graft function. Currently available and accessible methods for evaluating kidney function are either ineffective and inaccurate or highly invasive, such as tissue biopsies (3,4). Measurements of serum creatinine, estimated glomerular filtration rate (eGFR) and proteinuria as well as biopsies remain the current gold standards for the evaluation of renal allografts (5-11). These tests have significant limitations in predicting which patients are destined for immune tolerance or immune-mediated graft loss, and assisting in the management of long-term immunosuppression (5-11). Biomarkers allowing sensitive and accurate monitoring of graft function, early and specific diagnosis of rejection, assessment of long-term outcome allowing for a tailored immunosuppressive therapy in a non-invasive, cost-effective manner are critically needed (12).
MicroRNAs (miRNAs) are a family of noncoding 22-nucleotide RNAs expressed in a tissue-specific manner that can regulate gene expression levels of target mRNAs. MicroRNAs can repress mRNA translation or reduce mRNA stability of target genes although translational activation mechanisms and transcriptional effects have also been reported in the literature (13-17). MicroRNAs are now shown to be stably expressed in serum, plasma, urine, saliva, and other body fluids. Moreover, the expression patterns of these circulating miRNAs are correlated with human diseases including various types of cancer and represent potentially informative biomarkers. Furthermore, urinary miRNA profiling might offer valuable information in kidney disease (17-19).
In the KT field, to date, there have been two studies evaluating miRNA profiles in kidney allograft with acute cellular rejection (ACR) (20,21). However, miRNA profiling associated with other factors such as monitoring graft function and predicting long-term outcomes has not been evaluated yet.
Here, we have identified a miRNA expression signature for IF/TA and have confirmed the differential expression of five of these miRNAs in an independent set of allograft biopsy samples. Moreover, we have confirmed detection, and differential expression of three of these miRNAs, in urine samples of patients diagnosed with IF/TA. To the best of our knowledge, this is the first study to report a miRNA signature associated with CAD with IF/TA. Furthermore, we prospectively evaluated the identified miRNA signature in kidney transplant recipients (KTRs) identifying several patients with consistent IF/TA-like miRNA changes in concordance with histological findings while their serum creatinine and eGFR measurements indicated normal renal function.
METHODS
Kidney tissue samples
KTRs of unique deceased donor kidneys were included in the study. Written informed consent was obtained from all patients. The Institutional Review Board at Virginia Commonwealth University (VCU-IRB Protocol #HM11454) approved the study protocol. Renal allograft tissue was obtained with an 18-gauge needle and placed in RNAlater (Ambion) immediately after collection. No living donors, HIV positive or re-transplantation patients were included. All samples were classified based on histopathology using Banff criteria (2,22). All normal allograft (NA) samples selected were collected at 9-months or later post-transplantation and had continue eGFR values >60 mL/min/1.73m2. Estimated GFR was calculated using the Modification of Diet in Renal Disease (MDRD) formula (23).
RNA isolation
Total RNA was extracted using Trizol (Life Technologies) from biopsy tissue or urinary cell fractions following the manufacturer’s protocol. Prior to microarray processing, RNA integrity and quality was evaluated as previously described (24).
Illumina BeadChip® Array microarray processing
Total RNA from IF/TA (N=13), and NA (N=5) was processed following the manufacturer’s protocol. The final reaction products were hybridized to Sentrix Universal BeadChip® Arrays. After hybridization, the arrays were imaged using an Illumina BeadArray® Reader and the intensity files analyzed with the BeadStudio® software suite.
Data Analysis
Following the pre-normalization assessment described by Cunningham et al (25), unnormalized intensities without background subtraction were exported from GenomeStudio. Two technical replicates and two hybridization replicates were included in the experimental design to evaluate the reproducibility of the assay. For all hybridization replicates, MA plots were constructed (not shown) and Spearman’s rank correlations calculated (0.936 and 0.921 for the technical replicates and 0.996 and 0.911 for the hybridization replicates). Unsupervised hierarchical clustering using Ward’s method was applied to the miRNA data where 1-∣Pearson’s correlation∣ was the distance measure; all replicates hybridization clustered together (not shown). A detection p-value ≤0.01 was used to identify detected miRNAs. The miRNA dataset was then filtered to include only those miRNAs detected in at least 50% of the samples and having an miRNA standard deviation of expression intensities in the 90th percentile or higher. The miRNA data was then normalized using the quantile normalization method and then the dataset was restricted to the independent sample groups. To identify miRNAs exhibiting significant differential expression between these groups, miRNA level analysis of variance models were fit considering miRNA expression as the response and tissue type as the fixed effect of interest. A group means parameterization of tissue type was included as the fixed effect term to facilitate extraction of linear contrasts of interest. To adjust for multiple comparisons, the q-value method was used; miRNAs with a false discovery rate (FDR) ≤5% were considered significant. The limma package was used to fit the mixed effects models using restricted maximum likelihood procedure; an empirical Bayes method was applied to moderate miRNA standard errors by borrowing information across the entire set of miRNAs.
Validation of miRNA Microarray Results
Real-time (RT) quantitative-PCR (qPCR) reactions were used for quantifying the expression of miR-142-3p (assay ID: 000464), miR-32 (assay ID: 0021090), miR-204 (assay ID: 000508), miR-107 (assay ID: 000443), and miR-211 (assay ID: 000514) using pre-designed TaqMan® miRNA expression assays (Applied Biosystems) following the manufacturer’s protocol. The endogenous control RNU48 (assay ID: 001094) was used for normalization. Threshold cycle (Ct) values were used to calculate relative expression using the ΔΔCt method where: relative expression = 2−ΔΔCt, where ΔΔCt = (ΔCt of IF/TA) – (ΔCt of NA).
MicroRNA/mRNA correlation analyses
Correlation analyses between the miRNA array expression data and an independently generated mRNA gene expression signature previously published (24). Pearson correlation coefficients and p-values were calculated for every miRNA/mRNA pair between the differentially expressed IF/TA miRNAs (n=15) and probesets (n=2,223). A permutation analysis was performed to assess the statistical significance of the observed differentially expressed miRNA/mRNA correlations and determine the null distribution. For each correlation, 1,000 permutations were carried out between each miRNA and a randomly selected sample of 2,223 probesets to determine the null distribution. Empirical p-values were derived by comparing the observed correlation coefficients to the null distribution obtained from the permuted Pearson values.
Interaction Networks and Functional Analysis
Gene ontology and gene interaction analyses were analyzed through the use of ToppGene (http://toppgene.cchmc.org/) (26) and Ingenuity Pathways Analysis (IPA, Ingenuity® Systems, www.ingenuity.com). Gene lists containing Entrez GeneIDs were used as input. Biological functions and pathways with a p-value <0.05 were considered significant.
RESULTS
Patients and samples
A total of 81 KTRs (117 samples: 45 allograft kidney biopsies and 117 urine samples) were included in the present study. As part of the study design, KTRs were categorized in three groups (Figure 1). Demographic and clinical characteristics of the patients and donors are shown in the Table 1.
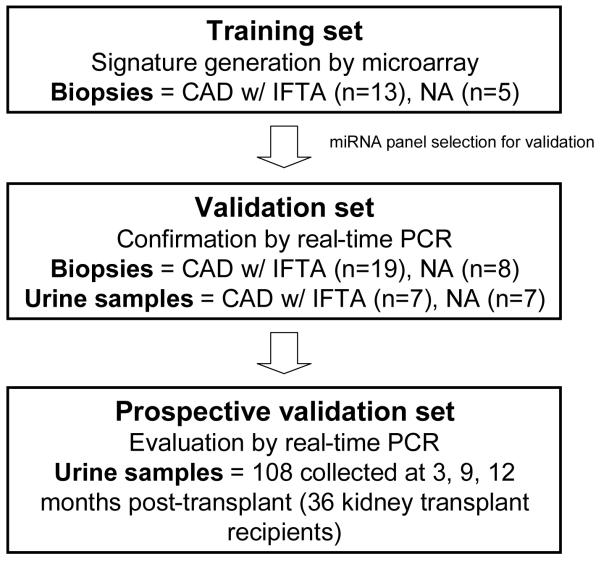
Figure 1.
Study design. Training set: For creating a signature of differentially expressed miRNAs, allograft tissue samples with histological diagnosis of CAD with IF/TA were evaluated using microarrays. Normal allograft (NA) samples (defined as samples obtained from kidney transplant recipients with at least 9 months post-transplantation, normal histology and continue eGFR >60 mL/min/1.73 m20 were used as control group. Validation set: The selected miRNAs for validation (using in silico target prediction and consecutive filtering of the results using our pre-established CAD with IF/TA gene expression signature) were validated in an independent set of patients (CAD with IF/TA and NA) using unique and paired biopsy and urine samples and using a second method (QPCR). Prospective validation set: The miRNAs identified as having correlation between tissue and urine expression were validated in an independent set of urine samples (collected at 3, 9, and 12 months post-transplantation) from 36 kidney transplant recipients.
Table 1.
Demographic and clinical characteristics of patients and donors by group
| Training Set | Validation Set | Prospective Validation Set |
|||
|---|---|---|---|---|---|
| NA | IF/TA | NA | IF/TA | ||
| N | 5 | 13 | 8 | 19 | 36 |
| Donor Type (DDK) | 5 | 13 | 8 | 19 | 36 |
|
Time to collection
(months) |
12.4 ± 3.9 | 82.5 ± 78.0 | 12.8 ± 3.3 | 92.6 ± 66.4 | 3 months, 3.9 ± 1.3 |
| 9 months, 9.5 ± 1.1 |
|||||
| 12 months, 12.1 ± 1.2 |
|||||
| Recipient Age (yrs) | 43.6 ± 12.6 | 39.8 ± 11.3 | 44.8 ± 16.3 | 41.4 ± 11.9 | 50.5 ± 12.2 |
| Recipient Gender (M/F) | 1 / 4 | 7 / 6 | 2 / 6 | 11 / 8 | 17 / 19 |
|
Recipient Race
(C/AA/O) |
0 / 4 / 1 | 1 / 12 / 0 | 5 / 2 / 1 | 2 / 17 / 0 | 5 / 29 / 2 |
| Recipient HCV (pos) | 0 | 1 | 0 | 1 | 3 |
| Recipient CMV (pos) | 3 | 8 | 6 | 11 | 28 |
| CMV Disease (pos) | 0 | 0 | 0 | 0 | 0 |
| Donor Age (yrs) | 42.6 ± 7.3 | 33.2 ± 15 | 37 ± 13 | 37.8 ± 13.8 | 42.4 ± 15.6 |
| Donor Gender (M/F) | 2 / 3 | 7 / 6 | 2 / 6 | 8 / 11 | 19 / 17 |
| Donor Race (C/AA/O) | 0 / 5 / 0 | 7 / 5 / 1 | 5 / 2 / 1 | 12 / 5 / 2 | 21 / 14 / 1 |
| Donor HCV (pos) | 0 | 2 | 0 | 2 | 2 |
| Donor HBV cAb (pos) | 1 | 1 | 1 | 2 | 4 |
| Donor HBV sAb (pos) | 1 | 1 | 1 | 1 | 1 |
| Donor CMV (pos) | 2 | 9 | 3 | 9 | 19 |
| Donor HIV (pos) | 0 | 0 | 0 | 0 | 0 |
| CIT (min) | 1039.2 ± 434.4 |
1097.5 ± 486.2 |
1092.1 ± 240.6 |
976.1 ± 556.9 |
1186.3 ± 416.3 |
| WIT (min) | 28.6 ± 6.7 | 29.5 ± 4.3 | 29.1 ± 6.4 | 32.8 ± 10.5 | 29.1 ± 6.3 |
| PPP (min) | 362.4 ± 418.2 |
793.4 ± 785.1 |
473.5 ± 363.5 |
179.6 ± 462.2 |
550.6 ± 494.7 |
| DGF incidence | 2 | 4 | 2 | 4 | 12 |
|
Acute Rejection
incidence |
1 | 4 | 1 | 6 | 3 |
| HLA MM total | 4 ± 1.4 | 3.25 ± 2.5 | 4.3 ± 1.2 | 3.5 ± 1.6 | 4.3 ± 1.3 |
| PRA at Tx (T-cell) | 67.6 ± 38.6 | 73.5 ± 0.7 | 55.5 ± 38.0 | 50.5 ± 31.8 | 48.0 ± 35.2 |
|
eGFR 1mo
(mL/min/1.73m2) |
>60 | N/A | >60 | N/A | 53.7 ± 22.0 |
|
eGFR 3mo
(mL/min/1.73m2) |
>60 | N/A | >60 | N/A | 56.7 ± 21.1 |
|
eGFR 6mo
(mL/min/1.73m2) |
>60 | N/A | >60 | N/A | 56.0 ± 21.8 |
|
eGFR 9mo
(mL/min/1.73m2) |
>60 | N/A | >60 | N/A | 56.3 ± 25.8 |
|
eGFR 12mo
(mL/min/1.73m2) |
>60 | N/A | >60 | N/A | 54.8 ± 23.2 |
Deceased Donor Kidney (DDK), Caucasian (C), African-American (AA), Other (O), Cold Ischemia Time (CIT), Warm Ischemia Time (WIT), Pump Perfusion Preservation (PPP), Delayed Graft Function (DGF), eGFR* (estimated Glomerular Filtration Rate).
*(GFR (ml/min/1.73 m2) = 175 × (Scr)1.154 × (age in yrs)−0.203 × (0.742 if female) × (1.21 if African-American) where Scr is serum creatinine concentration (mg/dl)).
Identification of miRNA signature
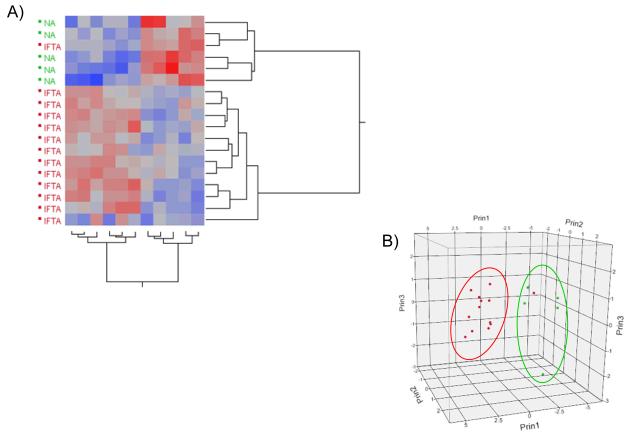
Biopsy samples were classified as either NA (N=5) or IF/TA (N=13) based on histopathological reports using the most current Banff criteria (2,22). MicroRNAs with an FDR ≤5% were considered significant. Fifty-six microRNAs were identified as significantly differentially expressed between the two groups (Table 2). Based on in silico mRNA target prediction and comparison of these targets to an independently generated mRNA IF/TA signature (24), we reduced this list to 15 microRNAs of interest (Table 2, miRNAs in bold). Supervised hierarchical clustering and principal component analysis of the miRNA array data for the reduced miRNA set shows a distinct separation between NA and IF/TA samples (Figure 2).
Table 2.
Differentially expressed miRNAs identified from each pairwise comparison. P-values and q-values are listed for each comparison.
| MicroRNA | Target Mature Sequences on Illumina BeadChip® Array |
p-value | FDR |
|---|---|---|---|
| HS_29 | TGTGCTTGGCTGAGGAGAA | 2.81E-13 | 3.22E-10 |
| HS_22.1 | CCAAGGAAGGCAGCAGGC | 8.33E-08 | 3.18E-05 |
| HS_243.1 | GTGAAGGCCCGGCGGAGA | 5.69E-07 | 1.63E-04 |
| hsa-miR-663 | AGGCGGGGCGCCGCGGGACCGC | 1.92E-06 | 4.39E-04 |
| hsa-miR-891a | TGCAACGAACCTGAGCCACTGA | 5.02E-06 | 9.56E-04 |
| hsa-miR-30c-1* | CTGGGAGAGGGTTGTTTACTCC | 5.85E-06 | 9.56E-04 |
| HS_263.1 | TCCTCCTCCTCCCCCGTC | 1.78E-05 | 2.55E-03 |
| hsa-miR-483-3p | TCACTCCTCTCCTCCCGTCTT | 2.32E-05 | 2.95E-03 |
| hsa-miR-1275 | GTGGGGGAGAGGCTGTC | 2.86E-05 | 3.27E-03 |
| hsa-miR-142-3p | TGTAGTGTTTCCTACTTTATGGA | 3.68E-05 | 3.77E-03 |
| hsa-miR-194* | CCAGTGGGGCTGCTGTTATCTG | 4.29E-05 | 3.77E-03 |
| hsa-miR-603 | CACACACTGCAATTACTTTTGC | 4.67E-05 | 3.77E-03 |
| hsa-miR-663b | GGTGGCCCGGCCGTGCCTGAGG | 5.05E-05 | 3.77E-03 |
| solexa-8048-104 | ATGAATGGATGAACGA | 5.06E-05 | 3.77E-03 |
| hsa-miR-494 | TGAAACATACACGGGAAACCTC | 5.26E-05 | 3.77E-03 |
| HS_197 | CCCGCTCTCCCGAAGCTGTTCG | 6.28E-05 | 4.23E-03 |
| hsa-miR-107 | AGCAGCATTGTACAGGGCTATCA | 1.17E-04 | 7.42E-03 |
| HS_10 | ACTGGAGATATGGAAGAGCTG | 2.23E-04 | 1.29E-02 |
| hsa-miR-32 | TATTGCACATTACTAAGTTGCA | 2.26E-04 | 1.29E-02 |
| hsa-miR-139-3p | GGAGACGCGGCCCTGTTGGAGT | 2.53E-04 | 1.38E-02 |
| hsa-miR-608 | AGGGGTGGTGTTGGGACAGCTCCGT | 3.34E-04 | 1.74E-02 |
| hsa-miR-129* | AAGCCCTTACCCCAAAAAGTAT | 3.93E-04 | 1.96E-02 |
| hsa-miR-34a | TGGCAGTGTCTTAGCTGGTTGT | 4.62E-04 | 2.16E-02 |
| hsa-miR-30c-2* | CTGGGAGAAGGCTGTTTACTCT | 4.72E-04 | 2.16E-02 |
| hsa-miR-572 | GTCCGCTCGGCGGTGGCCCA | 5.18E-04 | 2.28E-02 |
| hsa-miR-526b:9.1 | CTCTTGAGGGAAGCACTTTCTGTT | 5.63E-04 | 2.35E-02 |
| hsa-miR-216b | AAATCTCTGCAGGCAAATGTGA | 5.86E-04 | 2.35E-02 |
| hsa-miR-576-3p | AAGATGTGGAAAAATTGGAATC | 5.95E-04 | 2.35E-02 |
| hsa-miR-1227 | CGTGCCACCCTTTTCCCCAG | 6.15E-04 | 2.35E-02 |
| hsa-miR-148a | TCAGTGCACTACAGAACTTTGT | 6.86E-04 | 2.53E-02 |
| hsa-miR-223 | TGTCAGTTTGTCAAATACCCCA | 8.02E-04 | 2.82E-02 |
| hsa-miR-211 | TTCCCTTTGTCATCCTTCGCCT | 8.14E-04 | 2.82E-02 |
| HS_157 | TGCCTGCCCTCTTCCTCCAG | 1.02E-03 | 3.29E-02 |
| hsa-miR-15a | TAGCAGCACATAATGGTTTGTG | 1.03E-03 | 3.29E-02 |
| hsa-miR-7-2* | CAACAAATCCCAGTCTACCTAA | 1.05E-03 | 3.29E-02 |
| HS_94 | TGGGGAGCGGGAATGGATA | 1.07E-03 | 3.29E-02 |
| hsa-miR-215 | ATGACCTATGAATTGACAGAC | 1.12E-03 | 3.29E-02 |
| hsa-miR-448 | TTGCATATGTAGGATGTCCCAT | 1.14E-03 | 3.29E-02 |
| hsa-miR-548j | AAAAGTAATTGCGGTCTTTGGT | 1.15E-03 | 3.29E-02 |
| hsa-miR-204 | TTCCCTTTGTCATCCTATGCCT | 1.20E-03 | 3.29E-02 |
| hsa-miR-362-3p | AACACACCTATTCAAGGATTCA | 1.21E-03 | 3.29E-02 |
| hsa-miR-1257 | AGTGAATGATGGGTTCTGACC | 1.32E-03 | 3.51E-02 |
| hsa-miR-192* | CTGCCAATTCCATAGGTCACAG | 1.36E-03 | 3.55E-02 |
| hsa-miR-21* | CAACACCAGTCGATGGGCTGT | 1.40E-03 | 3.56E-02 |
| hsa-miR-16-1* | CCAGTATTAACTGTGCTGCTGA | 1.51E-03 | 3.69E-02 |
| hsa-miR-154* | AATCATACACGGTTGACCTATT | 1.51E-03 | 3.69E-02 |
| hsa-miR-1270 | CTGGAGATATGGAAGAGCTGTGT | 1.57E-03 | 3.75E-02 |
| hsa-miR-1249 | ACGCCCTTCCCCCCCTTCTTCA | 1.63E-03 | 3.77E-02 |
| hsa-let-7b* | CTATACAACCTACTGCCTTCCC | 1.65E-03 | 3.77E-02 |
| hsa-miR-767-5p | TGCACCATGGTTGTCTGAGCATG | 1.72E-03 | 3.86E-02 |
| solexa-4793-177 | TGATGATGATGATGATGATG | 2.04E-03 | 4.49E-02 |
| solexa-3793-229 | CAGGGCTGGCAGTGACATGGGT | 2.11E-03 | 4.56E-02 |
| hsa-miR-744 | TGCGGGGCTAGGGCTAACAGCA | 2.24E-03 | 4.74E-02 |
| HS_64 | TCCCTCTCCCTCCTTGCTCC | 2.28E-03 | 4.74E-02 |
| hsa-let-7d* | CTATACGACCTGCTGCCTTTCT | 2.34E-03 | 4.74E-02 |
| hsa-miR-20b* | ACTGTAGTATGGGCACTTCCAG | 2.36E-03 | 4.74E-02 |
Figure 2.
(A) Graphical representation using supervised hierarchical clustering of the reduced set for the differentially expressed miRNAs using Ward’s method. Higher intensity values are colored in red; lower values in blue. (B) Principal component analysis for the miRNA BeadChip® Array data for the 15 miRNAs of interest. NA samples are colored in green; IF/TA samples in red.
Validation of miRNA BeadChip® Array Results
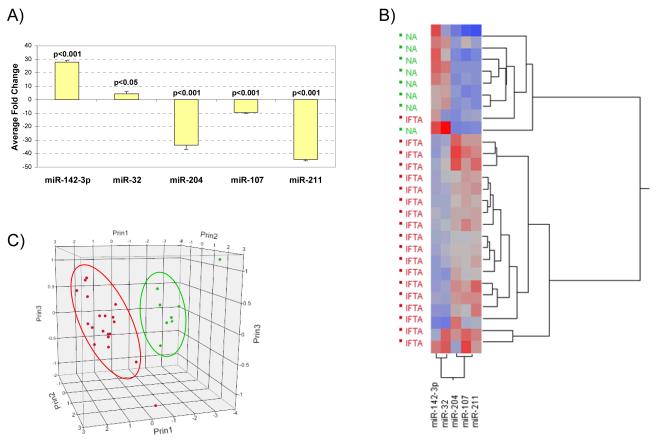
Five miRNAs from the identified signature were selected for further confirmation based on a combination of detected array fold change, statistical significance and in silico target mRNA target prediction. Differential expression of these five miRNAs was confirmed by RT-qPCR in the same RNAs used for the generation of the initial signature and using an independent set of samples from 27 additional deceased donor KTRs (NA=8 and IF/TA=19). A significant change in expression was detected for miR-142-3p, miR-204, miR-107, miR-211 (p<0.001) and miR-32 (p=0.023) (Figure 3A). Unsupervised hierarchical clustering (Figure 3B) and principal component analysis (Figure 3C) of RT-qPCR ΔCt values shows a clear separation between NA and IF/TA samples. Receiver operating characteristics (ROC) were calculated using ΔCt values for each miRNA (Table 3). To derive a multi-miRNA classifier predicting IF/TA versus NA, quadratic discriminant analysis (QDA) was performed using the RT-qPCR ΔCt values for all five miRNAs simultaneously. All NA and IF/TA biopsies were correctly classified (0% misclassification)
Figure 3.
(A) Taqman® MicroRNA Assays RT-qPCR results using tissue samples with histology confirmed IF/TA for 5 of the miRNAs identified in the Illumina Array study. The fold change was calculated using the ΔΔCt method. RNU48 was used as the normalizing endogenous control. (B) Unsupervised hierarchical clustering of all RT-qPCR ΔCt values using Ward’s method. Higher ΔCt values are colored red; lower values are blue. (C) Principal component analysis using the RT-qPCR ΔCt data. NA samples are colored in green; IF/TA samples in red.
Table 3.
Measurement properties of the predictive models for the individual and combined (QDA) miRNAs confirmed by RT-qPCR using the ΔCt data obtained in tissue samples. IF/TA was defined as positive.
| Specificity | Sensitivity | PPV | NPV | ACC | TP | TN | FP | FP | AUC | |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-142- 3p |
1.00 | 0.89 | 1.00 | 0.80 | 0.93 | 17 | 8 | 0 | 2 | 0.974 |
| miR-32 | 0.88 | 0.89 | 0.94 | 0.78 | 0.89 | 17 | 7 | 1 | 2 | 0.822 |
| miR-204 | 1.00 | 0.95 | 1.00 | 0.89 | 0.96 | 18 | 8 | 0 | 1 | 0.967 |
| miR-107 | 0.88 | 1.00 | 0.95 | 1.00 | 0.96 | 19 | 7 | 1 | 0 | 0.954 |
| miR-211 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 19 | 8 | 0 | 0 | 1.000 |
| QDA | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 19 | 8 | 0 | 0 | 1.00 |
PPV: positive predictive value; NPV: negative predictive value; ACC: accuracy; TP: true positive; TN: true negative; FP: false positive; FN: false negative; AUC: area under the curve.
QDA = Quadratic Discriminant Analysis
Detection of miRNAs in urine samples
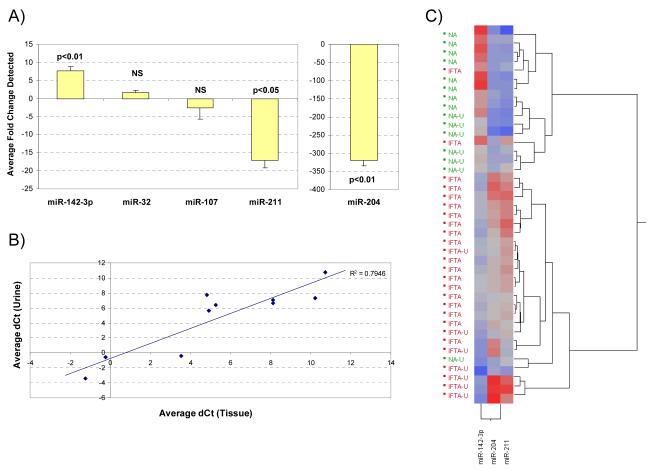
The expression of these five miRNAs was evaluated in paired urine samples from patients with IF/TA confirmed histology (N=7) compared to NA (N=7) samples. Differential expression of miR-142-3p (p=0.005), miR-204 (p=0.007) and miR-211 (p=0.017) was observed between IF/TA and NA urine samples (Figure 4A). Fold changes of 1.5 and 2.6 were calculated for miR-32 and miR107; however they did not meet statistical significance. Pearson correlation coefficient between average ΔCt values in tissue and urine samples was 0.891 (p<0.001, Figure 4B) for all miRNAs tested and 0.914 (p=0.011, not shown) when considering only miRNAs with statistically significant differential expression in both tissue and urine samples. Unsupervised hierarchical clustering of ΔCt values shows urine IF/TA samples clustering with IF/TA biopsies (Figure 4C). One urine NA sample clustered within the IF/TA cluster. However, their ΔCt pattern was not the same as that of other IF/TA samples within the cluster; this may be indicative of early kidney dysfunction and development of IF/TA.
Figure 4.
(A) Taqman® MicroRNA Assays RT-qPCR results using urine samples collected from patients with confirmed IF/TA. The fold change was calculated using the ΔΔCt method. RNU48 was used as the endogenous control. (B) Scatterplot comparing average ΔCt values obtained for NA and IFTA tissue and urine samples for the 5 miRNAs of interest. (C) Unsupervised hierarchical clustering of all RT-qPCR ΔCt values for miR-142-3p, miR-204 and miR-211 using Ward’s method for all tissue and urine samples; urine samples have been designated with the letter U. Higher ΔCt values are colored red; lower values are blue.
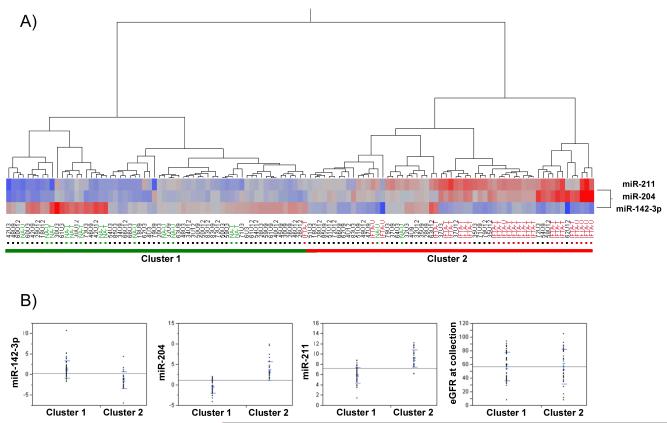
Subsequently, prospective evaluation of miRNA expression in urine samples from a second independent group of KTRs (N=36) undergoing protocol-biopsy was performed. Since only miR-142-3p, miR-204 and miR-211 showed statistically significant differential expression in tissue and urine samples, we focused our examination on these three miRNAs. Samples were collected at 3, 9 and 12 months post-transplantation. MicroRNA expression was examined by RT-qPCR and ΔCt values were calculated as previously described. Hierarchical clustering of the obtained ΔCt values revealed 2 clusters of samples; cluster 1 was composed of samples with ΔCt values similar to those of NA tissue and urine samples whereas cluster 2 was composed of samples with ΔCt values similar to those of IF/TA tissue and urine samples (Figure 5A).
Figure 5.
(A) Hierarchical clustering of RT-qPCR ΔCt values for miR-142-3p, miR-204 and miR-211 using Ward’s method for all urine samples evaluated prospectively. NA and IF/TA tissue (T) and urine (U) samples used in the signature confirmation have been colored in green (NA) and red (IF/TA) for reference; higher ΔCt values are colored red; lower values are blue. (B) Comparison of ΔCt values for miR-142-3p, miR-204, miR-211 and eGFR values for samples in clusters 1 and 2 from (A).
Samples from nineteen patients segregated into cluster 2. Spearman’s rank correlation showed significant correlations between the ΔCt for all three miRNAs (p<0.001) but not between miRNA expression and estimated glomerular filtration rate (eGFR) at the time of collection. When comparing cluster 1 to cluster 2, ΔCt values for miR-142-3p, miR-204 and miR-211 were statistically significantly different (p<0.001); however, eGFR at the time of collection was not significant (p=0.951, Figure 5B) between groups. Furthermore, paired urine samples from allograft biopsies showing histological evidence of IF/TA progression (IF (ci≥1) and TA (ct≥1) involving more than 25% of the cortical area) were localized within cluster 2. Finally, for a selected set of patients’ longitudinal analysis of the urine samples showed separation of the KTRs also in concordance with the previously described cluster 1 and 2 (Supplemental Figure 1).
Identification of miRNA target mRNAs
Our group recently published a gene expression comparative analysis between IF/TA and NA biopsies with the purpose of identifying molecular pathways involved in IF/TA development as well as possible early biomarkers of IF/TA progression (24). Using the publicly available miRecords database (http://mirecords.biolead.org/index.php) a list of possible mRNA targets for each of the miRNAs identified was generated (27). The predicted target tool component of miRecords integrates the prediction algorithms of several published miRNA target prediction tools: DIANA-microT, MicroInspector, miRanda, MirTarget2, miTarget, NBmiRTar, PicTar, PITA, RNA22, RNAhybrid, and TargetScan (27). Only mRNA targets identified through a consensus of three individual databases were considered when generating the list of potential mRNA targets for each of the miRNAs. When comparing predicted consensus target genes to the experimentally identified IF/TA genes (1,729 genes identified from 2,223 probesets), 1148 genes were found to overlap between the two lists (not shown) strongly suggesting that the differentially expressed miRNAs identified here may have a regulatory role in the expression of the IF/TA genes identified in our previous study.
Correlation of miRNA expression changes with gene expression changes
Ten patient samples (5 NA and 5 IF/TA) used in the generation of the miRNA signature were also used in the generation of mRNA signature (24). Therefore, we conducted correlation analyses between the two sets of array expression data to identify promising miRNA/mRNA pairs that could also be used as biomarkers of IFTA and possibly enhance the predictive/diagnostic potential of the miRNA signature.
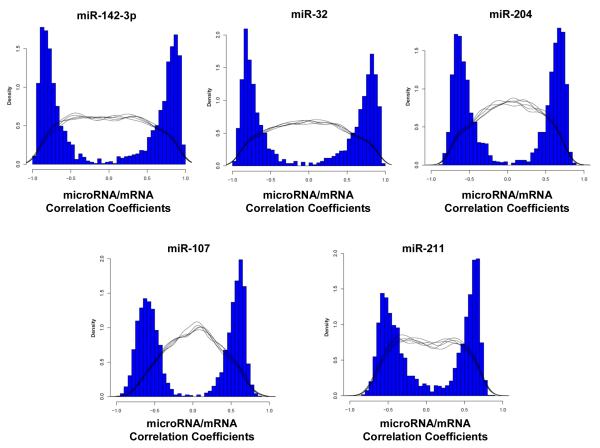
Pearson correlation coefficients and p-values were calculated for every miRNA/mRNA pair as described in the Methods. All calculated Pearson coefficients with significant empirical p-values were >∣0.658∣; 244 predicted mRNA target genes with significant miRNA/mRNA correlations were identified. Pearson correlation coefficients distributions for the five miRNAs confirmed by RT-qPCR are displayed in Figure 6. A summary of the correlation results can be found in Table 4. Pearson coefficients and respective empirical p-value distributions for all significant miRNA/mRNA pairs can be found in the Supplemental Information.
Figure 6.
Distribution of the miRNA/mRNA Pearson correlation values for the five miRNAs confirmed by RT-qPCR. Only Pearson coefficients with associated significant empirical p-values were graphed. The black lines represent five representative repetitions of the null distribution obtained when expression for each of the five miRNAs was compared to randomly selected mRNA expression data.
Table 4.
Summary of correlation values obtained for the 15 miRNAs of interest. Predicted targets refer to mRNA targets predicted by a consensus of at least three database algorithms queried by miRecords (http://mirecords.biolead.org/index.php). The numbers in the table indicate probesets with significant miRNA/mRNA correlations.
| MicroRNA | Number of significant p-values |
Number of significant empirical p-values |
Number of predicted targets |
Number of non- predicted targets |
|---|---|---|---|---|
| hsa-miR-194* | 1731 | 673 | 30 | 634 |
| hsa-miR-142-3p | 1797 | 632 | 40 | 592 |
| hsa-miR-32 | 1731 | 583 | 53 | 530 |
| hsa-miR-148a | 1373 | 566 | 56 | 509 |
| hsa-miR-216b | 1017 | 551 | 4 | 544 |
| hsa-miR-204 | 1062 | 547 | 21 | 525 |
| hsa-miR-107 | 782 | 520 | 46 | 474 |
| hsa-miR-223 | 1475 | 438 | 37 | 401 |
| hsa-miR-15a | 1407 | 398 | 71 | 327 |
| hsa-miR-34a | 1038 | 325 | 26 | 298 |
| hsa-miR-30c-2* | 545 | 265 | 16 | 249 |
| hsa-miR-211 | 512 | 250 | 7 | 243 |
| hsa-miR-30c-1* | 219 | 142 | 17 | 125 |
| hsa-let-7b* | 492 | 52 | 3 | 49 |
| hsa-miR-7-2* | 263 | 13 | 2 | 11 |
Bold highlights the miRNAs confirmed by RT-QPCR
Interaction Networks and Functional Analysis
Gene ontology and gene interaction analyses were carried out through the use of ToppGene (26) and IPA to identify biological functions and pathways over-represented by predicted consensus mRNA target genes with significant miRNA/mRNA correlations. Top biological functions identified from up-regulated genes in this list included regulation of lymphocyte proliferation, B-cell and T-cell activation/differentiation, NK cell differentiation, and positive regulation of apoptosis. Top biological functions identified from genes down-regulated included lipid oxidation/modification and protein dephosphorylation (Supplemental Tables). Primarily up-regulated pathways identified included CD28 signaling in T-helper cells, B-cell development and allograft rejection signaling. Pathways such as butanoate metabolism, valine-leucine-isoleucine degradation, and citrate cycle were composed primarily of down-regulated genes (Supplemental Figure 2 and Supplemental Tables).
DISCUSSION
The identification of biomarkers for monitoring of renal function in KTRs and early detection of progression to CAD with IF/TA is crucial in order to improve long-term outcomes (28-30). Currently, determination of late renal allograft failure due to IF/TA relies mainly on histological evaluation of biopsies taken when physiological parameters indicate possible dysfunction. However, development of IF/TA begins before the appearance of any clinical manifestation resulting in a diagnosis that often occurs after significant damage to the allograft has taken place (30-34). Here, we report the first CAD with IF/TA-related miRNA signature and show that non-invasive detection of this miRNA signature is also possible in urine samples of IF/TA diagnosed patients.
Two recent reports have studied miRNA changes occurring in kidney grafts with ACR (20,21). Of the miRNAs identified in our study, six had been previously reported by Anglicheau et al. (20). Five of these (let-7, miR-30c, miR-204, miR-223 and miR-142-3p) showed similar directional expression changes in our signature compared to the earlier report suggesting a similarity in the underlying processes involved in the two pathologies. Two additional miRNAs, miR-663 and miR-483, reported by Sui et al. (21) as differentially expressed in ACR were also identified in our signature. Our study only included deceased donor kidneys. However, the Anglicheau et al. study included both living and deceased donor transplant kidneys while the Sui et al. study used normal kidneys (not normal allografts) to generate the miRNA signatures. It has been shown that post-transplant gene expression profiles of living and deceased donor kidneys are significantly different (35,36) making a true comparison between these miRNA signatures difficult. Although miRNA signatures have been generated for ACR, to date there has been no report of a miRNA signature for CAD with IF/TA.
Studies of kidney transplant recipients have shown that subclinical acute rejection is associated with an increased risk of graft fibrosis (37-39). Subclinical inflammation occurring in association with IF/TA, even when it is below the threshold for the Banff criteria for acute rejection, has also been previously reported (40). These findings support the overlapping between miRNAs in ACR and CAD with IF/TA. Presence of inflammatory cells into the renal allograft interstitium is the biologic hallmark of alloimmune responses that leads to tubulointerstitial injury and subsequent interstitial fibrosis and chronic allograft failure. Of the miRNAs confirmed by RT-qPCR, miR-142-3p (also identified as significant in ACR by Anglicheau et al. (20)) has been shown to be implicated in the functional regulation of regulatory T-cells and macrophages (41). It has also been shown that Foxp3 mediates transcriptional repression of miR-142-3p; down-regulation of miR-142-3p then confers regulatory T-cells with suppressor functions by increasing levels of adenylyl cyclase 9 and subsequently levels of cAMP (21). Increased levels of Foxp3 have been implicated as a marker of acute rejection (42-44) as well as correlated with the presence of interstitial inflammation, tubulitis, interstitial fibrosis, and tubular atrophy (45). The increased levels of miR-142-3p observed in samples with IF/TA may indicate a decreased ability of regulatory T-cells to suppressed immune related processes occurring within the kidney. The involvement of miR-142-3p in the previously described gene expression regulation also supports the common ground between ACR and CAD with IF/TA. It is expected that the final panel of biomarkers that can be used for graft monitoring will result from the combination of a set of markers instead of unique markers. Progression to CAD with IF/TA is characterized by activation of different components of the immune response and response of the graft to the injury. Consequently, it is anticipated that the resulting biomarker panel will include miRNAs involved in all the different pathways related to the disease progression.
Additionally, miR-142-3p and miR-223, another miRNA identified in our signature, have been shown to be expressed in peripheral blood mononuclear cells (46,47). Expression of miR-142-3p was shown to be present in normal human T-cells and granulocytes but weak in monocytes and B-cells while miR-223 expression was confined to only myeloid lineages (47). Elevated levels of these two miRNAs in IF/TA samples may suggest involvement and/or presence of T- and B-cells.
MicroRNA-32 has been primarily shown to be dysregulated in different types of cancer (48-53). Interestingly, in prostate tumors, elevated levels of C9orf5, the host transcript for miR-32, were reported suggesting that dysregulation of miR-32 was secondary to changes in gene expression associated with the host gene (48). This may be possible for some of the miRNAs identified in our signature since several of them are located within intronic sequences of host genes (not shown).
Several roles have been proposed for miR-204. In HeLa cells inhibition of miR-204, along with seven others, resulted in increased levels of apoptosis; the authors concluded that these miRNAs were part of a miRNA network that played a role in controlling apoptosis (54). We have observed decreased expression of miR-204 in IF/TA samples and consistent with the proposed role in apoptosis, our gene expression IF/TA signature identified elevated expression of several genes involved in induction of apoptosis (24).
Finally, there is evidence that different members of the miR-15/107 group regulate cell cycle related mRNA targets; over-expression of these miRNAs results in G0/G1 arrest in cell lines (55,56). In pancreatic cancer cell lines, transfection of cells with miR-107 leads to decreased expression of cyclin dependent kinase 6 (CDK6) and decreased cell growth overall (57). Both miR-107 and miR-15 were differentially expressed in IF/TA samples. In this context, the observed down-regulation of miR-107 would suggest increased expression of cell cycle progression and cell proliferation genes. Consistent with the proposed roles of each of these miRNAs, we have identified differential expression of several genes involved in lymphocyte activation/differentiation, induction and regulation of apoptosis and cell proliferation in our IF/TA gene mRNA signature (24).
More importantly, using this signature, we were able to identify changes in miRNA expression in urine samples of patients with apparently normal renal function (cluster 2, Figure 4A). Kidney physiology is dynamic, constantly adjusting to body needs. Transplant renal physiology is even more complex as the organ must not only adjust to physiological demands but also to the new host; moreover, there is also damage to the kidney associated with harvesting, transport and transplantation surgery itself. Moreover, the presence of persistent IF/TA-like miRNA expression changes detected in multiple consecutive samples of the same patient may be indicative of early IF/TA-related molecular changes and progression. Further follow up, both clinical and molecular, of these patients is needed to confirm these findings.
It has become evident that miRNA-regulated gene expression plays an important role in renal function and physiology (58-61) and that their dysregulation may contribute to renal diseases. It has been suggested that the miR-30 family of miRNAs contribute to the maintenance of homoeostasis and function of podocytes and that loss of regulation by these miRNAs contributes to the formation of the observed phenotypes (58-61). Two members of this family (miR-30-c1 and miR-30-c2) were found to be differentially expressed in IF/TA samples compared to normal allografts.
MicroRNA repression and activation of translational and transcriptional mechanisms have been reported in the literature (13-17). Consistent with these observations we have identified a number of both positive and negative miRNA/mRNA correlations. Bioinformatic analyses such as the one performed here have identified many miRNA/mRNA pairs. Unfortunately, only a very small proportion of such associations have been confirmed experimentally (19). Despite advances in the sensitivity of target prediction algorithms, the overlap of predicted targets between different platforms remains relatively small. Additionally, these in silico predictions are not cell-type or context specific. To overcome this limitation, our study used a previously established gene expression signature with the established critical pathways to limit the list of statistical significant interaction. Further studies are currently ongoing to validate the possible miRNA/mRNA pairs identified.
The most immediate clinical benefit of miRNAs is their use as biomarkers for diagnosis, prognosis or response to therapy. In kidney transplantation, the use of urine as a non-invasive “surrogate” biosample source is an attractive alternative because it can be easily collected at multiple time points. There is ongoing research to expand our validated signature and evaluate the discovered markers as predictors of disease progression. The association between a miRNA panel and clinical characteristics might conduct to the establishment of a “risk score panel” for monitoring graft function allowing possible early therapeutic interventions.
In summary, we have identified a miRNA expression signature for IF/TA and have confirmed the differential expression of five of these miRNAs in an independent set of tissue biopsy samples. Moreover, we have confirmed detection, and differential expression of three of these miRNAs, in urine samples of patients diagnosed with IF/TA. To the best of our knowledge, this is the first study to report a miRNA signature associated with CAD with IF/TA. Furthermore, we prospectively evaluated the identified miRNA signature in kidney transplant recipients identifying several patients with consistent IF/TA-like miRNA changes while their serum creatinine and eGFR measurements indicated normal renal function. Further studies are currently ongoing incorporating gene expression data to the miRNA expression data to improve the predictive and discriminatory accuracy of the identified miRNA signature.
Supplementary Material
ACKNOWLEDGEMENTS
The research results included in this report were supported by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant, R01DK080074 and the National Institute of Mental Health (MH-20030).
ABBREVIATIONS
- ACR
acute cellular rejection
- CAD
chronic allograft dysfunction
- Ct
Threshold cycle
- eGFR
estimated glomerular filtration rate
- IF
interstitial fibrosis
- IPA
Ingenuity Pathways Analysis
- KT
Kidney transplantation
- KTRs
kidney transplant recipients
- MDRD
Modification of Diet in Renal Disease
- miRNA
microRNAs
- MSO
miRNA-specific oligonucleotide
- QDA
Quadratic discriminant analysis
- ROC
Receiver operating characteristics
- RT-qPCR:
Real-time quantitative-PCR
- TA
tubular atrophy
Footnotes
DISCLOSURE The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
- Supplemental Table S1 – List of significant miRNA/mRNA pairs identified.
- Supplemental Table S2 – Biological processes identified by ToppGene.
- Supplemental Table S3 – Biological pathways identified by ToppGene.
- Supplemental Table S4 – Biological processes identified by IPA.
- Supplemental Table S5 – Biological pathways identified by IPA.
REFERENCES
- (1).Racusen LC, Solez K, Colvin RB, Bonsib SM, Castro MC, Cavallo T, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55(2):713–723. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- (2).Solez K, Colvin RB, Racusen LC, Haas M, Sis B, Mengel M, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am.J.Transplant. 2008;8(4):753–760. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- (3).Schold JD, Kaplan B. The elephant in the room: failings of current clinical endpoints in kidney transplantation. Am.J.Transplant. 2010;10(5):1163–1166. doi: 10.1111/j.1600-6143.2010.03104.x. [DOI] [PubMed] [Google Scholar]
- (4).Poggio ED, Batty DS, Flechner SM. Evaluation of renal function in transplantation. Transplantation. 2007;84(2):131–136. doi: 10.1097/01.tp.0000269108.59275.dc. [DOI] [PubMed] [Google Scholar]
- (5).Manotham K, Booranalertpaisarn V, Eiam-Ong S, Chusil S, Praditpornsilpa K, Tungsanga K. Accurately simple estimation of glomerular filtration rate in kidney transplant patients. Transplant.Proc. 2002;34(4):1148–1151. doi: 10.1016/s0041-1345(02)02788-4. [DOI] [PubMed] [Google Scholar]
- (6).Mariat C, Alamartine E, Barthelemy JC, De Filippis JP, Thibaudin D, Berthoux P, et al. Assessing renal graft function in clinical trials: can tests predicting glomerular filtration rate substitute for a reference method? Kidney Int. 2004;65(1):289–297. doi: 10.1111/j.1523-1755.2004.00350.x. [DOI] [PubMed] [Google Scholar]
- (7).Gaspari F, Ferrari S, Stucchi N, Centemeri E, Carrara F, Pellegrino M, et al. Performance of different prediction equations for estimating renal function in kidney transplantation. Am.J.Transplant. 2004;4(11):1826–1835. doi: 10.1111/j.1600-6143.2004.00579.x. [DOI] [PubMed] [Google Scholar]
- (8).Bosma RJ, Doorenbos CR, Stegeman CA, van der Heide JJ, Navis G. Predictive performance of renal function equations in renal transplant recipients: an analysis of patient factors in bias. Am.J.Transplant. 2005;5(9):2193–2203. doi: 10.1111/j.1600-6143.2005.00982.x. [DOI] [PubMed] [Google Scholar]
- (9).Pascual M, Vallhonrat H, Cosimi AB, Tolkoff-Rubin N, Colvin RB, Delmonico FL, et al. The clinical usefulness of the renal allograft biopsy in the cyclosporine era: a prospective study. Transplantation. 1999;67(5):737–741. doi: 10.1097/00007890-199903150-00016. [DOI] [PubMed] [Google Scholar]
- (10).Furness PN. Predicting allograft survival: abundant data, but insufficient knowledge? Transplantation. 2007;83(6):681. doi: 10.1097/01.tp.0000262005.84789.de. [DOI] [PubMed] [Google Scholar]
- (11).Rush D. Can protocol biopsy better inform our choices in renal transplantation? Transplant.Proc. 2009;41(6 Suppl):S6–8. doi: 10.1016/j.transproceed.2009.06.092. [DOI] [PubMed] [Google Scholar]
- (12).Gillespie A, Lee IJ. Biomarkers in renal transplantation. Biomark Med. 2008;2(6):603–612. doi: 10.2217/17520363.2.6.603. [DOI] [PubMed] [Google Scholar]
- (13).Fabian M, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu.Rev.Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- (14).Kasinath B, Feliers D, Sataranatarajan K, Choudhury G, Lee M, Mariappan M. Regulation of mRNA translation in renal physiology and disease. American journal of physiology.Renal physiology. 2009;297(5):F1153–F1165. doi: 10.1152/ajprenal.90748.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- (16).Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- (17).Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat.Rev.Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- (18).Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit.Rev.Oncol.Hematol. 2010 doi: 10.1016/j.critrevonc.2010.11.004. [DOI] [PubMed] [Google Scholar]
- (19).Li JY, Yong TY, Michael MZ, Gleadle JM. Review: The role of microRNAs in kidney disease. Nephrology (Carlton) 2010;15(6):599–608. doi: 10.1111/j.1440-1797.2010.01363.x. [DOI] [PubMed] [Google Scholar]
- (20).Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc.Natl.Acad.Sci.U.S.A. 2009;106(13):5330–5335. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Sui W, Dai Y, Huang Y, Lan H, Yan Q, Huang H. Microarray analysis of MicroRNA expression in acute rejection after renal transplantation. Transpl.Immunol. 2008;19(1):81–85. doi: 10.1016/j.trim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- (22).Halloran PF, Langone AJ, Helderman JH, Kaplan B. Assessing long-term nephron loss: is it time to kick the CAN grading system? Am.J.Transplant. 2004;4(11):1729–1730. doi: 10.1111/j.1600-6143.2004.00662.x. [DOI] [PubMed] [Google Scholar]
- (23).Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann.Intern.Med. 1999;130(6):461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- (24).Scian MJ, Maluf DG, Archer KJ, Suh JL, Massey D, Fassnacht RC, et al. Gene expression changes are associated with loss of kidney graft function and interstitial fibrosis and tubular atrophy: diagnosis versus prediction. Transplantation. 2011;91(6):657–665. doi: 10.1097/TP.0b013e3182094a5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Cunningham JM, Oberg AL, Borralho PM, Kren BT, French AJ, Wang L, et al. Evaluation of a new high-dimensional miRNA profiling platform. BMC Med.Genomics. 2009;2:57. doi: 10.1186/1755-8794-2-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–11. doi: 10.1093/nar/gkp427. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–10. doi: 10.1093/nar/gkn851. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Cherukuri A, Welberry-Smith MP, Tattersall JE, Ahmad N, Newstead CG, Lewington AJ, et al. The clinical significance of early proteinuria after renal transplantation. Transplantation. 2010;89(2):200–207. doi: 10.1097/TP.0b013e3181c352c5. [DOI] [PubMed] [Google Scholar]
- (29).Scherer A, Gwinner W, Mengel M, Kirsch T, Raulf F, Szustakowski JD, et al. Transcriptome changes in renal allograft protocol biopsies at 3 months precede the onset of interstitial fibrosis/tubular atrophy (IF/TA) at 6 months. Nephrol.Dial.Transplant. 2009;24(8):2567–2575. doi: 10.1093/ndt/gfp183. [DOI] [PubMed] [Google Scholar]
- (30).Mas VR, Archer KJ, Scian M, Maluf DG. Molecular pathways involved in loss of graft function in kidney transplant recipients. Expert Rev.Mol.Diagn. 2010;10(3):269–284. doi: 10.1586/erm.10.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Nankivell BJ, Chapman JR. Chronic allograft nephropathy: current concepts and future directions. Transplantation. 2006;81(5):643–654. doi: 10.1097/01.tp.0000190423.82154.01. [DOI] [PubMed] [Google Scholar]
- (32).Jevnikar AM, Mannon RB. Late kidney allograft loss: what we know about it, and what we can do about it. Clin.J.Am.Soc.Nephrol. 2008;3(Suppl 2):S56–67. doi: 10.2215/CJN.03040707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Li C, Yang CW. The pathogenesis and treatment of chronic allograft nephropathy. Nat.Rev.Nephrol. 2009;5(9):513–519. doi: 10.1038/nrneph.2009.113. [DOI] [PubMed] [Google Scholar]
- (34).Fletcher JT, Nankivell BJ, Alexander SI. Chronic allograft nephropathy. Pediatr.Nephrol. 2009;24(8):1465–1471. doi: 10.1007/s00467-008-0869-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Mueller TF, Reeve J, Jhangri GS, Mengel M, Jacaj Z, Cairo L, et al. The transcriptome of the implant biopsy identifies donor kidneys at increased risk of delayed graft function. Am.J.Transplant. 2008;8(1):78–85. doi: 10.1111/j.1600-6143.2007.02032.x. [DOI] [PubMed] [Google Scholar]
- (36).Hauser P, Schwarz C, Mitterbauer C, Regele HM, Muhlbacher F, Mayer G, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Lab.Invest. 2004;84(3):353–361. doi: 10.1038/labinvest.3700037. [DOI] [PubMed] [Google Scholar]
- (37).Cosio FG, Grande JP, Larson TS, Gloor JM, Velosa JA, Textor SC, et al. Kidney allograft fibrosis and atrophy early after living donor transplantation. Am J Transplant. 2005;5(5):1130–1136. doi: 10.1111/j.1600-6143.2005.00811.x. [DOI] [PubMed] [Google Scholar]
- (38).Nankivell BJ, Borrows RJ, Fung CL, O’Connell PJ, Allen RD, Chapman JR. Natural history, risk factors, and impact of subclinical rejection in kidney transplantation. Transplantation. 2004;78(2):242–249. doi: 10.1097/01.tp.0000128167.60172.cc. [DOI] [PubMed] [Google Scholar]
- (39).Nankivell BJ, Borrows RJ, Fung CL, O’Connell PJ, Chapman JR, Allen RD. Delta analysis of posttransplantation tubulointerstitial damage. Transplantation. 2004;78(3):434–441. doi: 10.1097/01.tp.0000128613.74683.d9. [DOI] [PubMed] [Google Scholar]
- (40).Moreso F, Ibernon M, Gomà M, Carrera M, Fulladosa X, Hueso M, et al. Subclinical rejection associated with chronic allograft nephropathy in protocol biopsies as a risk factor for late graft loss. Am J Transplant. 2006;6(4):747–752. doi: 10.1111/j.1600-6143.2005.01230.x. [DOI] [PubMed] [Google Scholar]
- (41).Huang B, Zhao J, Lei Z, Shen S, Li D, Shen GX, et al. miR-142-3p restricts cAMP production in CD4+CD25− T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 2009;10(2):180–185. doi: 10.1038/embor.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Aquino-Dias EC, Joelsons G, da Silva DM, Berdichevski RH, Ribeiro AR, Veronese FJ, et al. Non-invasive diagnosis of acute rejection in kidney transplants with delayed graft function. Kidney Int. 2008;73(7):877–884. doi: 10.1038/sj.ki.5002795. [DOI] [PubMed] [Google Scholar]
- (43).Afaneh C, Muthukumar T, Lubetzky M, Ding R, Snopkowski C, Sharma VK, et al. Urinary Cell Levels of mRNA for OX40, OX40L, PD-1, PD-L1, or PD-L2 and Acute Rejection of Human Renal Allografts. Transplantation. 2010;90(12):1381–1387. doi: 10.1097/TP.0b013e3181ffbadd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Muthukumar T, Dadhania D, Ding R, Snopkowski C, Naqvi R, Lee JB, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N.Engl.J.Med. 2005;353(22):2342–2351. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- (45).Bunnag S, Allanach K, Jhangri GS, Sis B, Einecke G, Mengel M, et al. FOXP3 expression in human kidney transplant biopsies is associated with rejection and time post transplant but not with favorable outcomes. Am.J.Transplant. 2008;8(7):1423–1433. doi: 10.1111/j.1600-6143.2008.02268.x. [DOI] [PubMed] [Google Scholar]
- (46).Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur.J.Haematol. 2008;81(4):304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- (47).Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- (48).Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68(15):6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc.Natl.Acad.Sci.U.S.A. 2008;105(35):12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, et al. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma -A miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48(7):615–623. doi: 10.1002/gcc.20669. [DOI] [PubMed] [Google Scholar]
- (51).Petillo D, Kort EJ, Anema J, Furge KA, Yang XJ, Teh BT. MicroRNA profiling of human kidney cancer subtypes. Int.J.Oncol. 2009;35(1):109–114. doi: 10.3892/ijo_00000318. [DOI] [PubMed] [Google Scholar]
- (52).Dacic S, Kelly L, Shuai Y, Nikiforova MN. miRNA expression profiling of lung adenocarcinomas: correlation with mutational status. Mod.Pathol. 2010;23(12):1577–1582. doi: 10.1038/modpathol.2010.152. [DOI] [PubMed] [Google Scholar]
- (53).Mascaux C, Laes JF, Anthoine G, Haller A, Ninane V, Burny A, et al. Evolution of microRNA expression during human bronchial squamous carcinogenesis. Eur.Respir.J. 2009;33(2):352–359. doi: 10.1183/09031936.00084108. [DOI] [PubMed] [Google Scholar]
- (54).Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33(4):1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol.Cell.Biol. 2007;27(6):2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4(8):e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, et al. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9(3):293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, et al. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J.Am.Soc.Nephrol. 2008;19(11):2159–2169. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J.Am.Soc.Nephrol. 2008;19(11):2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, et al. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J.Am.Soc.Nephrol. 2008;19(11):2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Nagalakshmi VK, Ren Q, Pugh MM, Valerius MT, McMahon AP, Yu J. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int. 2011;79(3):317–330. doi: 10.1038/ki.2010.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.