Abstract
Despite being a curable disease, tuberculosis (TB) killed more people in 2009 than during any previous year in history. Progress in TB research has been slow, and remains burdened by important gaps in our knowledge of the basic biology of Mycobacterium tuberculosis, the causative agent of TB, and its interaction with the human host. Fortunately, major systems biology initiatives have recently been launched that will help fill some of these gaps. However, to fully comprehend TB, and control this disease globally, current systems biological approaches will not suffice. The influence of host and pathogen diversity, changes in human demography, and socioeconomic and environmental factors will also need to be considered. Such a multidisciplinary approach might be best described as ‘systems epidemiology’ in an effort to overcome the traditional boundaries between basic biology and classical epidemiology.
New tools are needed to control tuberculosis globally
Tuberculosis (TB) is caused by a group of closely related Gram-positive bacilli, collectively known as the Mycobacterium tuberculosis complex (MTBC). MTBC comprises the typical human pathogens M. tuberculosis and Mycobacterium africanum, as well as variants affecting various animal species. These animal pathogens include Mycobacterium bovis (a pathogen of cattle), Mycobacterium caprae (goats and sheep), Mycobacterium microti (voles), and Mycobacterium pinnipedii (seals and sea lions) [1]. Contrary to many other pathogenic bacteria, MTBC does not have classical virulence factors such as recently acquired pathogenecity islands, nor does it produce any toxin. Yet, MTBC is able to efficiently infect, survive and transmit among hosts. According to estimates by the World Health Organization, one-third of the world’s human population is latently infected with MTBC, and 1.7 million people die of TB each year [2]. The outcome of TB infection and diseases is highly variable, ranging from complete elimination of the bacteria by innate immunity to classical pulmonary disease, disseminated TB, and death. In 90% of the cases, the infection remains latent, while 10% will develop active disease at some point during their lifetime [3]. The intimate cross-talk between the bacteria and the host immune system is one of the main complexities determining these variable outcomes [4]. In order to better control TB globally, new tools are urgently needed, in particular better diagnostics, new antibiotics, and better vaccines [5].
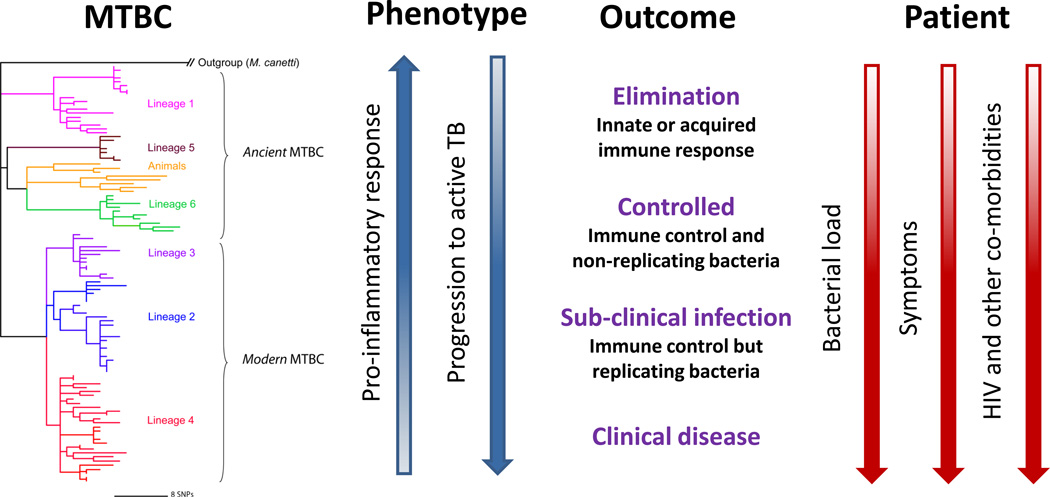
In most parts of the world, active TB is still being diagnosed by sputum microscopy [5]. However, this technique has a limited sensitivity, and up to 50% of cases are routinely missed. Although bacterial culture is the current gold standard for detecting TB, this technique takes up to 4 weeks, and requires skilled technicians and well-equipped laboratories, all of which are rarely available in developing countries. Fortunately, novel and highly sensitive molecular tools are being developed which show great promise for rapid detection of active TB [6]. One additional difficulty in diagnosing TB is to reliably differentiate between latent and active disease. Contrary to the traditional view considering TB as a simple binary state of active versus latent disease, the manifestation of TB is currently thought to represent a whole spectrum of infection [7] (Figure 1). Ideally, biomarkers should be available that allow classifying patients according to this spectrum [8]. Such biomarkers would be particularly valuable if they allowed identifying infected individuals most likely to progress to active TB. A recent study suggests this may become possible [9].
Figure 1. MTBC strain variation and the spectrum of responses to TB.
Instead of the traditional binary division into active and latent TB, a spectrum of patient responses to infection has been proposed to better describe the observed heterogeneity in TB patients and latently infected individuals [7]. HIV-coinfection and increased bacterial burden drive latency towards active disease (adapted from [7]). Likewise, pathogen genotype could also play a role in shaping the outcome of TB within this spectrum. For example, based on recent data, ‘modern’ MTBC lineages elicit a delayed innate immune response [59], and progress faster to active disease [62] compared to ‘ancient’ strains.
Apart from the difficulties in diagnosing TB, the standard treatment against TB is also complicated, as it involves a six month regimen with multiple antibiotics. Long-term treatments are inherently problematic, as patient non-adherence or drug shortages can lead to the development of drug resistance [10]. Drug-resistant strains of MTBC started to appear shortly after the introduction of streptomycin in 1943 [11]. Today, many regions of the world report increases in multidrug-resistant TB, and some MTBC strains are now resistant to all available drugs [12]. Adding to the problem of drug-resistant TB is that no new anti-TB drug has been licensed since the discovery of ethambutol in the 1960s. Following the onset of the HIV epidemic in the early 1980s, overconfidence regarding old antibiotics combined with long-term neglect in TB research and surveillance led to a re-emergence of the disease in the developed world [13]. While in the developing world TB presumably never had actually declined, HIV had a dramatic impact on TB incidence, particularly in sub-Saharan Africa. Fortunately, the development of new drugs and shorter treatment regimens against TB are back on the agenda [14].
In addition to new diagnostics and new antimicrobials, a better vaccine against TB is urgently needed. Considering the large pool of latently infected individuals [3], prevention of TB infection and disease through vaccination might be the only realistic way of controlling global TB in the long run. However, the Bacille Calmette-Guérin (BCG) vaccine is the only currently approved vaccine against TB and it has a questionable efficiency against pulmonary TB in adults, ranging from 0 to 80% [15]. Yet, BCG remains the most widely used vaccine in the world because it protects children against TB meningitis, the most severe form of the disease [16]. BCG was derived in the first quarter of the 20th century from a virulent strain of M. bovis. The reasons for the observed variation in protective efficacy of BCG are unclear, although differences among BCG strains, exposure to environmental mycobacteria, and human genetic diversity have been invoked [15]. Currently, several new vaccine candidates are at various stages of development [17]. Yet, despite significant progress over the past 20 years, a new and broadly effective vaccine against TB will not be available any time soon. A significant hurdle in TB vaccinology is our limited understanding of what constitutes protective immunity (Box 1). This, as well as many other important gaps of knowledge will need to be filled before an effective TB vaccine can become a reality [18].
BOX 1. Outstanding questions.
What is latent TB?
What constitutes protective immunity in TB?
How can individuals be categorized within the spectrum of latent and active TB?
How does infection with different strains affect the likelihood of being in a different state of the spectrum?
What is the role of CD4+ and CD8+ responses in promoting TB transmission?
How will the ongoing changes in human demography influence the evolution of MTBC in the future?
There is increasing consensus among the TB research community that systems biology will play an important role in generating new insights relevant to the development of new diagnostics, drugs, and vaccines against TB [5, 19]. The chronic nature of the disease, characterized by a complex dialog between the host immune system and the pathogen, combined with features such as latency, a complex mycobacterial cell wall, and the phenomenon of antimicrobial persistence, all call for more comprehensive approaches to study the biology of TB (Box 1). In this review, we start by briefly reviewing recent advances in applying systems biology to TB research. We then discuss why systems biology should be combined with complementary approaches to understand and control TB globally. Finally, we review recent data on the genetic diversity and evolution of MTBC, and end by proposing a new hypothesis on the evolution of virulence in MTBC, which, if confirmed, could impact the future spread and control of TB in the world..
Systems biology of TB: necessary but not sufficient
Two major systems biology initiatives to study TB have recently been funded by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) and the EU, respectively (Box 2). Systems biology relies on various high-throughput technologies combined with computational modeling to integrate complex biological data [20]. In particular, next-generation sequencing is revolutionizing biology at many levels. Thanks to these new technologies, transcriptomes and transcription factors binding sites will be studied to an extent never before possible [21]. Proteomics [22] and metabolomics [23] are also gaining momentum in TB research. Yet, significant analytical problems remain, including finding ways to incorporate all these different types of omics data into a form that not only makes biological sense, but also allows designing theoretical models that can be tested in the laboratory [24]. In TB, such models have been used to study the formation of lung granulomas [25]. These complex structures are formed by immune cells and are believed to encapsulate mycobacteria away from the rest of the body.
BOX 2. Online resources and links for systems biology and epidemiology.
TB Systems Biology Program (http://www.broadinstitute.org/annotation/tbsysbio): tuberculosis systems biology approach funded by the National Institute of allergy and infectious diseases (NIAID). The NIAID is also funding systems biology initiatives for other pathogens such as influenza, enteropathogens and emerging respiratory viruses [19].
SysteMTb (http://www.systemtb.org): a systems biology approach founded by the seventh framework program of the European Union. The project involves the activity of 13 partners and collaborates with the TB System Biology Program.
Immune Epitope Database and Analysis Resource (http://www.immuneepitope.org): contains data on B-cell and T-cell epitopes for different hosts and includes epitopes from pathogens such as M. tuberculosis but also information on autoimmune diseases [75].
The Allele Frequency Net Database (http://www.allelefrequencies.net): a resource that centralizes the information available regarding frequency and geographical distribution of different polymorphic areas of the human genome. The database includes available data on frequency of HLA alleles across many countries [76].
TB Database (http://www.tbdb.org): an online database, which centralizes updated experimental data relevant to tuberculosis research. It includes genomic databases to compare M. tuberculosis to other mycobacterial genomes, an analysis of mutations between different clinical strains representative of the MTBC lineages, and extended transcriptome data [77].
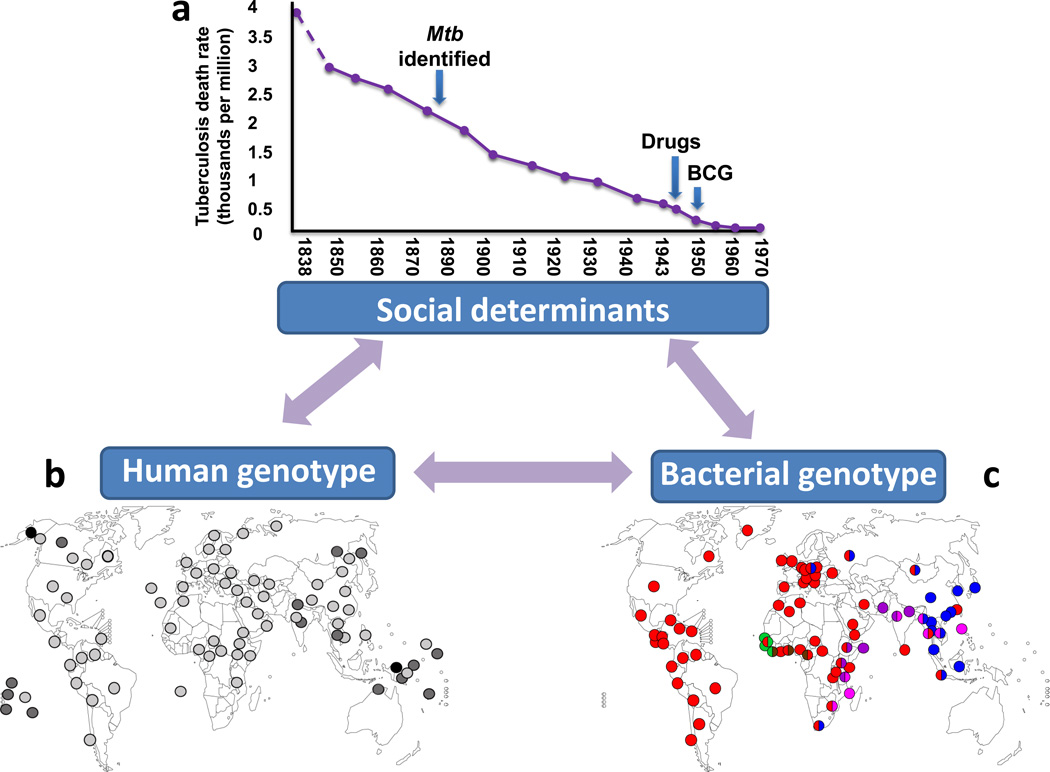
Even if systems biology gave as a complete picture of the cellular processes involved in host-pathogen interactions in TB, there are still many factors inherent to infectious diseases, and particularly to TB, that are not addressed by most current systems biological approaches [26]. These factors are nevertheless crucial for understanding, and ultimately controlling TB globally. TB in Europe started to decrease long before the introduction of biomedical interventions (Figure 2a). Improved living conditions, better nutrition and sanitation are believed to be responsible for this decline [27]. The fact that this trend continued well into the 20th century, raises questions as to the actual contribution of BCG vaccination and chemotherapy to the overall reduction of TB in Europe. Hence, in addition to the important questions addressed through systems biology, other factors related to the environmental, social and demographic contexts need to be studied using complementary approaches [28]. Moreover, substantial genetic variation exists both among humans and within MTBC. This diversity needs to be considered to ensure that novel TB diagnostics, drugs and vaccines will be universally effective [29, 30].
Figure 2. TB driven by biology and sociology.
Many environmental, socioeconomic and evolutionary factors are not generally considered when applying current systems biological approaches to infectious disease research. (a) Improved living conditions lead to a decline in TB deaths. In England and Wales TB mortality started to decrease long before the causative agent of TB (Mtb) was identified by Robert Koch and before BCG vaccination and chemotherapy became available (adapted from [27]). (b) Human genetic diversity impacts susceptibility to TB. The HLA II allele DQB1*0503 was the first HLA associated to increase TB risk [80] and along with other DQB1 alleles has been associated with increase susceptibility to TB in different places in Asia [31]. The DQB1*0503 allele is more common in Asian countries and pacific islands (dark grey and black dots) than other parts of the world (light grey) (data from http://www.allelefrequencies.net, Box 2). (c) Geographical distribution of the six human MTBC lineages. Each dot represents a country and its color indicates the most frequent lineage(s) within this country (adapted from [42]). The three ‘modern’ lineages (Lineage 4, red; Lineage 2, blue; Lineage 3, purple) are more globally widespread and hence more successful compared to the three ‘ancient’ lineages (M. africanum Lineage 5, green; M. africanum Lineage 6, brown; Lineage 1, pink), which are more geographically restricted. There is increasing evidence for a role of bacterial genotype in TB infection and disease [32].
Over the past 20 years, more than 100 studies have looked for possible associations between human gene polymorphisms and susceptibility to TB [31] (Figure 2b). The polymorphisms that were found reflect the influence of human genotype on disease, a factor which is not generally considered in current systems biology of infectious diseases. Similarly, at least 100 studies have explored the role of MTBC strain diversity in TB [32] (Figure 2c). Many experimental studies found clear evidence for strain effects on immune recognition and virulence, but only a few studies have reported consistent differences in clinical settings. To date, only five studies have explored possible interactions between human genotypes and MTBC genetic diversity [33–37]. More such studies are needed to understand the role of genetic diversity in host-pathogen interactions.
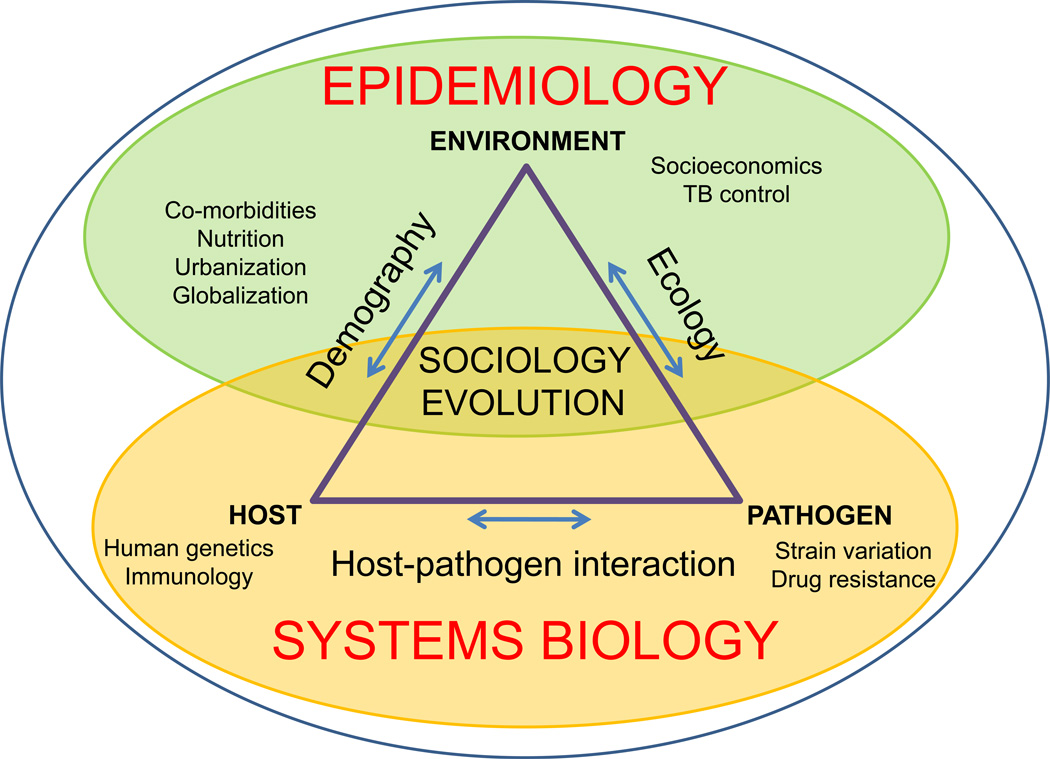
Given the diversity of factors driving TB in the world (Figure 2), a more comprehensive approach is needed, which complements and informs current TB research. We refer to this approach as ‘systems epidemiology’ (Figure 3), and envisage it as the combination of ‘classical’ systems biology, which addresses most of the biological aspects of TB, with epidemiology, sociology, evolutionary biology and ecology, which collectively cover the physical and social environments, as well as the evolution of the host and the pathogen [26]. An improved understanding of the interplay between these many factors is a prerequisite to develop and successfully implement more effective tools and strategies to control TB in the world. For the remainder of this review, we shall focus on work carried out during the past decade, which illustrates how a better grasp of the genetic diversity and evolution of MTBC contributes to our understanding of TB.
Figure 3. Systems epidemiology of TB.
Systems biology will be key to elucidate the biology of TB. In addition, complementary efforts need to be directed towards understanding the environmental, sociological and evolutionary factors driving global TB epidemics (adapted from [26]).
Not ‘yet another typing method’
Many methods have been used to genotype bacteria. The development of additional genotyping techniques has not always been justified [38]. As a consequence, classifying strain diversity among bacterial species has often been confusing. In MTBC, the first genotyping methods were used to study the epidemiology and transmission dynamics of TB. Therefore, these techniques targeted molecular markers with fast evolutionary rates, such as insertion sequences (IS6110 RFLP), variable number tandem repeats (VNTRs) or CRISPRs (spoligotyping) [39]. However, these methods have limited utility when applied to long-term evolutionary questions [40]. Hence, more slowly-evolving markers have been used to track the main phylogenetic lineages of the MTBC. Because of the low rate of DNA sequence diversity in MTBC compared to other bacteria [41], standard multilocus sequence typing (MLST), which is based on sequencing of a few housekeeping genes, does not generate sufficient phylogenetic information in MTBC [29]. In addition to being genetically monomorphic, MTBC exhibits a clonal population structure with no evidence of ongoing horizontal gene transfer. Hence, a novel mutation occurring in the ancestor of any given MTBC lineage will be inherited to all members of this lineage and thus serve as a phylogenetic marker for all strains belonging to this lineage. This phenomenon has been taken advantage of, first for genomic deletion analysis [42–45], then for genotyping based on single nucleotide polymorphisms (SNPs) [46–50], and most recently, for comparative whole genome sequencing [51]. Thus, rather than representing mere ‘yet another typing method’ [38], the application of the latter three genotyping approaches have shed new light onto the evolution and biology of MTBC.
One of the first genotyping methods applied to evolutionary questions in TB was based on genomic deletions. Two studies used genomic deletions as phylogenetic markers to revisit the classification of the different human and animal subspecies within MTBC [44, 45]. One of the important findings of these studies was that human MTBC did not, as previously assumed, originate from animal-adapted M. bovis during the initiation of agriculture and animal domestication. Instead, all human MTBC share a common ancestor with all animal MTBC, which most likely existed long before the Neolithic transition. Another study screened 100 clinical isolates of human MTBC by comparative genome hybridization and found that 68 different genome regions were deleted in one or more of these strains, corresponding to 4.2% of the coding capacity of the MTBC reference strain H37Rv [52]. Some of these genomic deletions grouped strains into discrete lineages [43]. A further screen based on genomic deletions in 875 strains from global sources revealed that human MTBC consists of six main lineages associated with different geographic regions and human populations (Figure 2c) [50]. A molecular epidemiological investigation in San Francisco reported differential transmission of these lineages in different patient populations, depending on whether the infected patient came from a geographic area associated with a particular MTBC lineage or not. Based on these data, the authors hypothesized that the different lineages of MTBC might have co-evolved with different human populations [42, 43].
Taken together, these evolutionary studies support an ancient origin of human TB, and suggest that the phylogeographic distribution of MTBC and possible host-pathogen co-evolution need to be considered when developing new tools to combat TB [29].
Human density and the evolution of virulence
Deletion analysis gave us insight into the phylogeographical population structure of MTBC, and pointed towards an association between the bacteria and different human populations. However, genomic deletions do not correlate with phylogenetic distances and therefore do not indicate how closely related one strain is to any other. In 2008, Hershberg et al. [50] published the results of DNA sequencing of 89 genes in each of 108 global strains of MTBC. This work resulted in a phylogeny that corroborated the deletion-based strain groupings, but now the genetic distances between strains could be evaluated. Based on the correlation between these genetic distances and the geographic distances between patient origins, the authors proposed a new evolutionary scenario for human MTBC. The so-called ‘out-of-and-back-to-Africa’ scenario postulated that MTBC originated in Africa. After dispersing around Africa, giving rise to two MTBC lineages also known as M. africanum [53], a first wave of phylogenetically ‘ancient’ MTBC accompanied the out-of-Africa migrations of modern humans and populated the south of India and Southeast Asia. Shortly thereafter or concomitantly, the phylogenetically 'modern' MTBC lineages spread out of Africa into Europe and Asia. These modern lineages then expanded as a result of the massive increases of human populations in Europe, India, and China during the past few centuries. Consistent with this scenario, a study by Wirth et al. based on VNTR analyses of a global collection of MTBC found signals of population expansion, which were more pronounced in MTBC strains originating from Europe or Asia compared to strains from Africa [54].
This new evolutionary scenario for MTBC is not just an interesting academic exercise, but leads to a new hypothesis with respect the evolution of virulence in MTBC and its possible impact on disease progression. This hypothesis builds on previous findings in evolutionary ecology suggesting that increased access to host or increased host density can select for increased virulence [55]. Based on the evolutionary scenario proposed by Hershberg et al. [50], 'modern' MTBC lineages have evolved in the context of high human population densities (i.e. easy access to susceptible hosts), whereas 'ancient' lineages evolved in regions where human host densities remained low until very recently. This new hypothesis postulates that 'modern' strains became more virulent as they adapted to large host populations, whiles 'ancient' strains remained less virulent in order to avoid exhausting the limited pool of susceptible hosts. When re-formulating this hypothesis in terms of the spectrum of latent versus active TB discussed above [7], we expect that a person infected with a 'modern' strain is more likely to experience the condition in which the infecting mycbacteria are able to survive the initial contact with the human immune system and replicate actively, leading to a faster disease progression (Figure 1). However, a person infected with an 'ancient' strain will be associated with the condition in which the infection is better controlled by the host immune system, increasing the likelihood of latent TB. As we shall discuss below, there are emerging data from both the laboratory and the field that are consistent with this idea.
Strain variation influences innate immune responses and disease progression
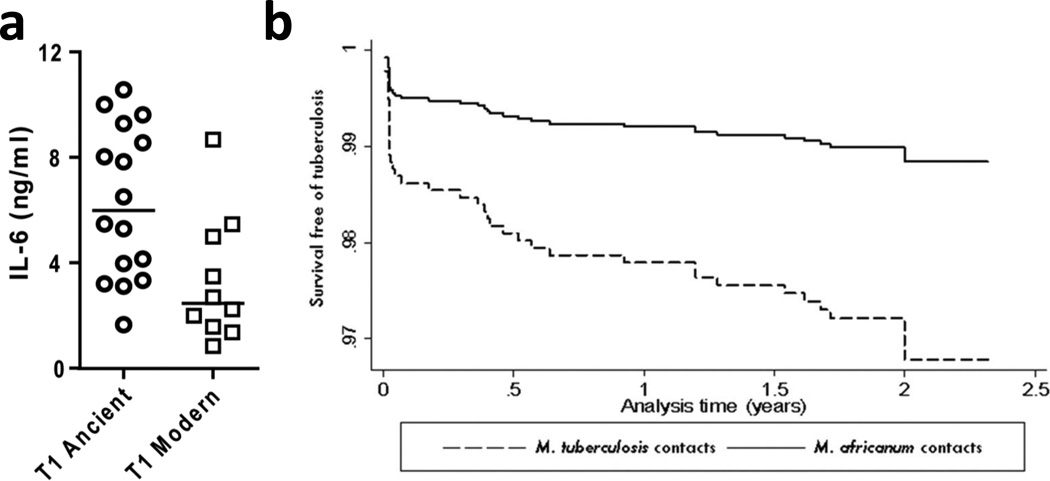
Research in immunology and cell biology has illuminated many aspects of host-pathogen interaction in TB [56, 57]. An increasing body of evidence from cellular and animal infection models demonstrate that MTBC strains differ in their immunogenicity and virulence [32, 58]. However, most studies to date have studied a limited number of MTBC strains. A more comprehensive effort was recently reported by Portevin et al. [59]. In that study, 26 strains representative of the global diversity of human MTBC were selected and used to infect monocyte-derived macrophages and dentritic cells from multiple human donors. The authors found that strains belonging to the ‘modern’ lineages induced lower pro-inflammatory responses in both cell types compared to the ‘ancient’ lineages (Figure 4a). Importantly, these differences were maintained across eight different donors. However, no such differences were observed when infecting unfractionated peripheral blood mononuclear cells. Considering that hypo-inflammatory innate immune responses have been associated with hypervirulence in animal models of TB [60, 61], the findings by Portevin et al. [59] support a model in which 'modern' lineages might be able to progress more rapidly to disease and transmit to new hosts by avoiding early immune recognition. If confirmed, this would be consistent with the idea that ‘modern’ lineages have evolved under higher host densities, which in turn might have selected for increased virulence and transmissibility.
Figure 4. Experimental and epidemiological evidence for the impact of bacterial genotype on TB disease.
(a) ‘Modern’ lineages elicit lower pro-inflammatory cytokines (e.g. IL-6) compared to ‘ancient’ lineages in human monocyte-derived macrophages [59]. The inhibition of innate immunity could allow ‘modern’ strains to replicate and establish an infection more efficiently before more efficient immune responses quick in. Inhibition of innate immunity has been associated with increased MTBC virulence in animal models of infection [60, 61]. (b) Strains of the ‘ancient’ Lineage 6, also known as M. africanum, were three times less likely to progress to active TB compared to other MTBC lineages [62].
A study by de Jong et al. [62] in the Gambia followed a cohort of TB patients and their household contacts over two years. The authors found no difference in the rate of transmission between the 'modern' lineages of MTBC and the 'ancient' lineage known as M. africanum. However, they found that patients infected with 'modern' strains were three times more likely to progress to active disease compared to M. africanum (Figure 4b). Taken together, these studies suggest that 'modern' MTBC strains differ from ‘ancient’ strains in a way consistent with the hypothesis on the evolution of virulence in MTBC presented above. However, more studies are needed to confirm these findings and determine how the current trends of globalisation, urbanisation and general population growth will impact the virulence of MTBC in the future (Box 3).
BOX 3. A hypothesis regarding the evolution of virulence in MTBC.
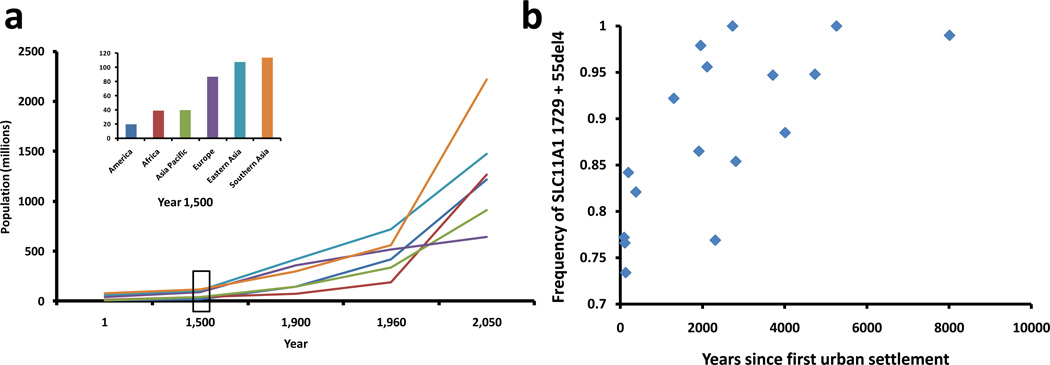
The main selective pressures acting on a pathogen include the host immune response, exposure to antibiotics, and the host population dynamics. As explained in the main text, the role of the immune system in shaping pathogen diversity is starting to be elucidated and pressure by antibiotics is well known to have resulted in the emergence of drug-resistant microbes. The impact of human population changes, however, has been rarely explored. Human population sizes have been changing from small hunter-gatherer populations to large urban settlements in Europe, Asia and America (Figure Ia). It is therefore expected that given the larger number of susceptible hosts available, more virulent strains will evolve in densely populated areas [50]. Recent reports suggest that this could be the case. It has been shown that strains from MTBC lineages associated with Europe and Asia (i.e. the 'modern' MTBC) are associated with a delayed pro-inflammatory immune response (Figure 4a) [59] and a shorter progression to active TB (Figure 4b) [62]. Increased pathogen pressure, associated with higher human densities, has been shown to select for higher frequencies of resistance alleles against infection. This is the case of a human genetic variant in the gene SLC11A1 known to provide resistance against intracellular pathogens. The frequency of this allele in human populations correlates with the time of first urbanization in different geographic areas (Figure Ib) [78]. The selection for human resistant variants associated with regions of increased human settlement suggests that the selective pressure of TB on humans might be higher in urban than rural areas, perhaps because of the emergence of more virulent strains. Likewise, the continued increase of human populations all over the world, and especially in Africa, suggests that more virulent strains of MTBC could emerge in the future [79].
Genome sequencing reveals immune subversion
One of the features of host-pathogen co-evolution is the ongoing evolutionary arms-race between the pathogen and the host immune system. T-cell based immunity is essential for the control of human TB [56]. This is illustrated by the fact that HIV-infected individuals with low CD4+ T-cell counts are at high risk of developing active TB. Many pathogens avoid immune recognition through the accumulation of genetic diversity in antigens; a phenomenon known as immune evasion [63]. Two early studies suggested that no immune evasion was occurring in MTBC [50, 64]. However, these studies were limited with respect to the number of antigens studied. By May 2010, the Immune Epitope Database and analysis resource (Box 2) comprised a total of 491 experimentally confirmed human T-cell epitopes of MTBC, corresponding to 78 antigens in the MTBC genome. This offered an opportunity to revisit the question of immune evasion in MTBC based on a larger dataset. Comas et al. [51] generated the nearly complete genome sequences of 21 clinical strains representative of the global diversity of human MTBC. They then inferred a new phylogeny based on the concatenation of all 9,037 phylogenetic informative SNPs identified among these strains. This new genome-based phylogeny of MTBC was congruent with the phylogenies published earlier, but had a higher resolution. For example, the basal position of the two M. africanum lineages corroborated the likely African origin of MTBC. In addition, this new phylogeny allowed reconstruction of the ancestral state of all the SNP positions identified in these 21 genomes, and the direction the SNP change was determined at each position. The authors then extracted all the known T-cell antigens and classified the remaining of the genome into essential and non-essential genes based on previous experimental work [65]. As expected, essential genes were more evolutionarily conserved than non-essential genes. Surprisingly however, the known T-cell antigens turned out to be equally conserved as essential genes. Because antigens consist of epitopes which are immunologically recognised, and non-epitope regions which are not, the authors studied these regions separately. The results showed that 95% of epitopes in MTBC had no amino acid change at all. Furthermore, epitopes were more evolutionarily conserved than non-epitope regions, and overall were the most conserved regions in the entire genome.
Based on these findings, the authors hypothesized that this hyperconservation of T-cell epitopes in MTBC might reflect the fact that the host immune responses to these epitopes are beneficial to the bacteria rather than to the host. In other words, MTBC does not seem to rely on immune evasion, but rather on some type of immune subversion. Some characteristics of the life history of TB support this view. For example, cavitary TB, the most contagious form of the disease [66], is primarily driven by immune-pathological processes which promote lung damage and thereby increase the transmissibility of TB [67, 68].
The hyperconservation of human T-cell epitopes in MTBC has important implications for the design of new TB diagnostics and vaccines. On the positive side, new diagnostics based on these epitopes will be universally applicable. By contrast, developing vaccines candidates targeting these epitopes could become problematic if the human immune responses elicited offer a net benefit to MTBC.
Genome sequencing informs epidemiology
In addition to generating new insights into the genetic diversity and evolution of MTBC, next-generation genome sequencing will increasingly be used to address more pragmatic public health-driven questions (see [69] and accompanying papers). The most obvious application in this respect is in the emerging field of genome epidemiology [70, 71]. In MTBC, next-generation sequencing has been used to highlight the limitations of current genotyping techniques for differentiating between closely related strains [72]. In a study from The Netherlands, a cluster of TB transmission comprising 104 patients was investigated by genome sequencing of three of the corresponding patient isolates. Eight SNPs were identified among these three isolates and used to genotype the remaining patient isolates of the cluster. This approach allowed identifying the index cases and clarifying the routes of transmission [73]. More recently, the genome sequences of 32 patient isolates of a transmission cluster in British Columbia was reported [74]. The SNP data was integrated with information about the social contacts of the patients. The results highlighted the role of ‘superspreaders’ in the transmission of TB. There is little doubt that next-generation sequencing will increasingly be used for routine molecular epidemiology of TB in the future [26].
Concluding remarks and future directions
In conclusion, systems biology approaches are crucial to address some of the most urgent questions in TB research (Box 1). Answering these questions will be essential to drive the development of new drugs and vaccines against TB. However, many more questions need to be addressed, and we think that an even more comprehensive approach will be necessary to understand TB in all its complexity. We have been referring to this multidisciplinary concept as systems epidemiology (Figure 3). Some of the new high-throughput technologies routinely used in systems biology are increasingly being introduced into other disciplines. In particular, high-throughput DNA sequencing, which continues to provide novel insights into biology, has the potential to revolutionize molecular and genetic epidemiology of infectious diseases, as the genome sequences of both the infecting agent and the patient will soon be determined routinely. The rationale for studying the genomic diversity of both MTBC and its human host is given, as mounting evidence supports the relevance of this diversity for understanding TB [31, 32]. In particular, the possible consequences of host-pathogen co-evolution in TB have only recently been addressed [51, 59, 62]. Some of the resulting observations seem contradictory at first glance. However, they can be reconciled in a model embedded in ecological theory on the evolution of virulence (Box 3). Following this model, the hyperconservation of T-cell epitopes in MTBC [51] suggest that all MTBC strains depend on some aspects of the host adaptive immune response for successful transmission. By contrast, the variation in innate immune responses reported by Portevin et al. [59] reflect subtle differences between strains directly or indirectly linked to different rates of progression to active disease [62], which perhaps have been selected as a consequence of changes in human host densities [50]. More work is needed to confirm this model and explore its implications for the global control of TB.
Figure I. Human demographic changes and the evolution of TB.
(a) For most of human history, the population of the world was less than one billion people. By the end of the 20th century, the population was six billion, and it is estimated that by 2050 it will reach 10 billion. This dramatic increase in human population occurred unevenly across different geographic regions, which could have influenced the evolution of the different MTBC lineages. ‘Modern’ lineages evolved in high populated areas whereas ‘ancient’ lineages have been evolving in regions where human densities remained low until very recently. However, these regions are catching up and expected to experience the strongest population growth in the future (data from http://www.sasi.group.shef.ac.uk/worldmapper, Box 2). (b) In addition to these historical and projected trends of population growth, changes in human behaviours that lead to higher population densities could also affect the evolution of the pathogen and the host. Humans with a deletion in the gene SLC11A1 are more resistant to intracellular infections. The frequency of this gene variant is positively correlated with regions of early urbanization [78]. Urbanization leads to an increase in human densities, which could favour the evolution of more virulent strains [79].
Acknowledgments
We thank Andrés Moya and Douglas Young as well as other members of our group for valuable comments on the manuscript. The work in our laboratory is supported by the Swiss National Science Foundation (grant no. PP00A-119205), the Medical Research Council, UK (MRC_U117588500), the Leverhulme-Royal Society Africa Award (AA080019), and the National Institutes of Health (AI090928 and HHSN266200700022C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith NH, et al. Ecotypes of the Mycobacterium tuberculosis complex. J Theor Biol. 2006;239:220–225. doi: 10.1016/j.jtbi.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Geneva: WHO; 2010. Global tuberculosis control - surveillance, planning, financing. [Google Scholar]
- 3.Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328:856–861. doi: 10.1126/science.1185449. [DOI] [PubMed] [Google Scholar]
- 4.Young D, et al. Systems biology of persistent infection: tuberculosis as a case study. Nat Rev Microbiol. 2008;6:520–528. doi: 10.1038/nrmicro1919. [DOI] [PubMed] [Google Scholar]
- 5.Young DB, et al. Confronting the scientific obstacles to global control of tuberculosis. J Clin Invest. 2008;118:1255–1265. doi: 10.1172/JCI34614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehme CC, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barry CE, 3rd, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pai M. Spectrum of latent tuberculosis - existing tests cannot resolve the underlying phenotypes. Nat Rev Microbiol. 2010;8:242. doi: 10.1038/nrmicro2236-c1. [DOI] [PubMed] [Google Scholar]
- 9.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zur Wiesch PA, et al. Population biological principles of drug-resistance evolution in infectious diseases. Lancet Infect Dis. 2011;11:236–247. doi: 10.1016/S1473-3099(10)70264-4. [DOI] [PubMed] [Google Scholar]
- 11.Pyle MM. Relative numbers of resistant tubercle bacilli in sputa of patients before and during treatment with streptomycin. Mayo Clin Proc. 1947;22:465–473. [PubMed] [Google Scholar]
- 12.Gandhi NR, et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet. 2010;375:1830–1843. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 13.Brudney K, Dobkin J. Resurgent tuberculosis in New York City. Human immunodeficiency virus, homelessness, and the decline of tuberculosis control programs. Am Rev Respir Dis. 1991;144:745–749. doi: 10.1164/ajrccm/144.4.745. [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann SH, Parida SK. Changing funding patterns in tuberculosis. Nat Med. 2007;13:299–303. doi: 10.1038/nm0307-299. [DOI] [PubMed] [Google Scholar]
- 15.Andersen P, Doherty TM. Opinion: The success and failure of BCG - implications for a novel tuberculosis vaccine. Nat Rev Microbiol. 2005;3:656–662. doi: 10.1038/nrmicro1211. [DOI] [PubMed] [Google Scholar]
- 16.Colditz GA, et al. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. JAMA. 1994;271:698–702. [PubMed] [Google Scholar]
- 17.Kaufmann SH, et al. New vaccines for tuberculosis. Lancet. 2010;375:2110–2119. doi: 10.1016/S0140-6736(10)60393-5. [DOI] [PubMed] [Google Scholar]
- 18.Russell DG, et al. Tuberculosis: what we don't know can, and does, hurt us. Science. 2010;328:852–856. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aderem A, et al. A systems biology approach to infectious disease research: innovating the pathogen-host research paradigm. MBio. 2011;2:e00325–e00310. doi: 10.1128/mBio.00325-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 21.Mardis ER. Next-generation DNA sequencing methods. Annual review of genomics and human genetics. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 22.Kunnath-Velayudhan S, et al. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc Natl Acad Sci U S A. 2010;107:14703–14708. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Carvalho LP, et al. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, et al. Integrating multiple 'omics' analysis for microbial biology: application and methodologies. Microbiology. 2010;156:287–301. doi: 10.1099/mic.0.034793-0. [DOI] [PubMed] [Google Scholar]
- 25.Ray JC, et al. Synergy between individual TNF-dependent functions determines granuloma performance for controlling Mycobacterium tuberculosis infection. J Immunol. 2009;182:3706–3717. doi: 10.4049/jimmunol.0802297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comas I, Gagneux S. The past and future of tuberculosis research. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000600. e1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKeown T. The modern rise of population. Academic Press; 1976. [Google Scholar]
- 28.Lonnroth K, et al. Drivers of tuberculosis epidemics: The role of risk factors and social determinants. Soc Sci Med. 2009;68:2240–2246. doi: 10.1016/j.socscimed.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 29.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 30.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moller M, et al. Past, present and future directions in human genetic susceptibility to tuberculosis. FEMS Immunol Med Microbiol. 2010;58:3–26. doi: 10.1111/j.1574-695X.2009.00600.x. [DOI] [PubMed] [Google Scholar]
- 32.Coscolla M, Gagneux S. Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov Today Dis Mech. 2010;7:e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caws M, et al. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000034. e1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thye T, et al. Variant G57E of mannose binding lectin associated with protection against tuberculosis caused by Mycobacterium africanum but not by M. tuberculosis. PLoS ONE. 2011;6:e20908. doi: 10.1371/journal.pone.0020908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Intemann CD, et al. Autophagy gene variant IRGM −261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000577. e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herb F, et al. ALOX5 variants associated with susceptibility to human pulmonary tuberculosis. Hum Mol Genet. 2008;17:1052–1060. doi: 10.1093/hmg/ddm378. [DOI] [PubMed] [Google Scholar]
- 37.van Crevel R, et al. Infection with Mycobacterium tuberculosis Beijing genotype strains is associated with polymorphisms in SLC11A1/NRAMP1 in Indonesian patients with tuberculosis. J Infect Dis. 2009;200:1671–1674. doi: 10.1086/648477. [DOI] [PubMed] [Google Scholar]
- 38.Achtman M. A surfeit of YATMs? J Clin Microbiol. 1996;34:1870. doi: 10.1128/jcm.34.7.1870-1870.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato-Maeda M, et al. Genotyping of Mycobacterium tuberculosis: application in epidemiologic studies. Future microbiology. 2011;6:203–216. doi: 10.2217/fmb.10.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Comas I, et al. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS ONE. 2009;4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 42.Gagneux S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirsh AE, et al. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci U S A. 2004;101:4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brosch R, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mostowy S, et al. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J Infect Dis. 2002;186:74–80. doi: 10.1086/341068. [DOI] [PubMed] [Google Scholar]
- 46.Baker L, et al. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg Infect Dis. 2004;10:1568–1577. doi: 10.3201/eid1009.040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutacker MM, et al. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J Infect Dis. 2006;193:121–128. doi: 10.1086/498574. [DOI] [PubMed] [Google Scholar]
- 48.Filliol I, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006;188:759–772. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dos Vultos T, et al. Evolution and diversity of clonal bacteria: the paradigm of Mycobacterium tuberculosis. PLoS ONE. 2008;3:e1538. doi: 10.1371/journal.pone.0001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hershberg R, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biology. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsolaki AG, et al. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci U S A. 2004;101:4865–4870. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Jong BC, et al. Mycobacterium africanum--review of an important cause of human tuberculosis in West Africa. PLoS Negl Trop Dis. 2010;4:e744. doi: 10.1371/journal.pntd.0000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirth T, et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000160. e1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.May RM, Anderson RM. Epidemiology and genetics in the coevolution of parasites and hosts. Proc R Soc Lond B Biol Sci. 1983;219:281–313. doi: 10.1098/rspb.1983.0075. [DOI] [PubMed] [Google Scholar]
- 56.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 57.Russell DG. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev. 2011;240:252–268. doi: 10.1111/j.1600-065X.2010.00984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicol MP, Wilkinson RJ. The clinical consequences of strain diversity in Mycobacterium tuberculosis. Trans R Soc Trop Med Hyg. 2008;102:955–965. doi: 10.1016/j.trstmh.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 59.Portevin D, et al. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001307. e1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reed MB, et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 61.Manca C, et al. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha /beta. Proc Natl Acad Sci U S A. 2001;98:5752–5757. doi: 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Jong BC, et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J Infect Dis. 2008;198:1037–1043. doi: 10.1086/591504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palmer GH, et al. 'Nothing is permanent but change'- antigenic variation in persistent bacterial pathogens. Cell Microbiol. 2009;11:1697–1705. doi: 10.1111/j.1462-5822.2009.01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Musser JM, et al. Negligible genetic diversity of mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics. 2000;155:7–16. doi: 10.1093/genetics/155.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodrigo T, et al. Characteristics of tuberculosis patients who generate secondary cases. Int J Tuberc Lung Dis. 1997;1:352–357. [PubMed] [Google Scholar]
- 67.Barber DL, et al. CD4 T cells promote rather than control tuberculosis in the absence of PD-1-mediated inhibition. J Immunol. 2011;186:1598–1607. doi: 10.4049/jimmunol.1003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flynn JL, Chan J. What's good for the host is good for the bug. Trends Microbiol. 2005;13:98–102. doi: 10.1016/j.tim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 69.Eisen JA, MacCallum CJ. Genomics of emerging infectious disease: A PLoS collection. PLoS Biology. 2009;7 doi: 10.1371/journal.pbio.1000224. e1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewis T, et al. High-throughput whole-genome sequencing to dissect the epidemiology of Acinetobacter baumannii isolates from a hospital outbreak. J Hosp Infect. 2010;75:37–41. doi: 10.1016/j.jhin.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 72.Niemann S, et al. Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS ONE. 2009;4:e7407. doi: 10.1371/journal.pone.0007407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schurch AC, et al. High resolution typing by integration of genome sequencing data in a large tuberculosis cluster. J Clin Microbiol. 2010;48:3403–3406. doi: 10.1128/JCM.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gardy JL, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Q, et al. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gonzalez-Galarza FF, et al. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39:D913–D919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reddy TB, et al. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37:D499–D508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barnes I, et al. Ancient urbanization predicts genetic resistance to tuberculosis. Evolution. 2011;65:842–848. doi: 10.1111/j.1558-5646.2010.01132.x. [DOI] [PubMed] [Google Scholar]
- 79.Cowley D, et al. Recent and rapid emergence of W-Beijing strains of Mycobacterium tuberculosis in Cape Town, South Africa. Clin Infect Dis. 2008;47:1252–1259. doi: 10.1086/592575. [DOI] [PubMed] [Google Scholar]
- 80.Goldfeld AE, et al. Association of an HLA-DQ allele with clinical tuberculosis. JAMA. 1998;279:226–228. doi: 10.1001/jama.279.3.226. [DOI] [PubMed] [Google Scholar]