Abstract
The collection and analysis of genomic data has the potential to reveal novel druggable targets by providing insight into the genetic basis of disease. However, the number of drugs, targeting new molecular entities, approved by the US Food and Drug Administration (FDA) has not increased in the years since the collection of genomic data has become commonplace. The paucity of translatable results can be partly attributed to conventional analysis methods that test one gene at a time in an effort to identify disease-associated factors as candidate drug targets. By disengaging genetic factors from their position within the genetic regulatory system, much of the information stored within the genomic data set is lost. Here we discuss how genomic data is used to identify disease-associated genes or genomic regions, how disease-associated regions are validated as functional targets, and the role network analysis can play in bridging the gap between data generation and effective drug target identification.
The role of human genomics in drug target discovery
The current paradigm of target-based drug design is limited, not by the synthesis of new chemical compounds, but by the identification of novel biological targets. All biological macromolecules (i.e., DNA, RNA, proteins, carbohydrates and lipids) are potential drug targets, although the vast majority of drugs target proteins [1, 2]. With an estimated 10,000 potentially druggable targets in humans, there are fewer than 400 distinct molecular targets of drugs approved by the US Food and Drug Administration (FDA)[1, 2, 3]. Over the past 50 years, the number of drugs sharing the same mechanism of action or target has averaged 4.1, ranging from 2–14 drugs per target [4]. There is potential to reveal new and better druggable targets by taking advantage of the recent trend in genomic research to comprehensively map the genetic basis of human disease [5]. Acquisition of such a map can provide a new perspective on disease etiology by revealing druggable targets that address the cause of disease as opposed to those that are palliative.
The landscape of human genomics research has undergone a profound transformation over the course of the last decade, but the fundamental goal of understanding the mapping relationship between a genotype and a phenotype has remained constant. The Human Genome Project (HGP) was completed in 2003, releasing, for the first time, a comprehensive map of the three billion nucleotides that make up the human genome [6]. This impressive feat was touted as the discovery of the `book of life,' carrying with it the promise to saturate the research community with disease-causing genes and novel drug targets. The expectation was that a simple mapping relationship would emerge in which each disease had one causal gene, and hence, one effective druggable target. Instead, it became apparent that the genome is more complex than predicted and that the DNA sequence alone has limited value for understanding the dynamic nature of gene expression. To date, the translatable results stemming from the HGP have been few, but perhaps more importantly, the HGP has served as a scaffold for the unbiased collection of data on the genomic scale.
Genomic data is collected for studies of evolution, population variation, gene function, and disease association. In the last ten years, billions of nucleotide bases have been sequenced, millions of functional elements have been identified, and thousands of chromosomal regions have been associated with disease contributing valuable insight into the genotype to phenotype mapping relationship. With the staggering volume of data being routinely collected, the utility of these genome-wide measures largely depends on the computational interventions used to transform the data into an interpretable form. For the purpose of identifying novel drug targets, it is essential to choose computational methods that can take into account the role that disease-associated genetic factors play within the broader context of the entire genetic regulatory system [7]. Here we discuss how genomic data is used to identify and interpret disease-associated genes or genomic regions and the role network analysis can play in bridging the gap between data generation and effective drug target discovery.
Genome-wide association studies (GWAS): mapping the genetic basis of disease
A byproduct of the Human Genome Project was the systematic identification of single nucleotide polymorphisms (SNPs). SNPs are individual base pair changes commonly found in the DNA sequence that can capture genetic variation between individuals [8, 9]. A database cataloging not only common SNPs but also the correlation structure between these SNPs has been assembled through the HapMap Project [10]. This correlation between SNPs, called linkage disequilibrium (LD), occurs because recombination rarely happens between sites of DNA that are in close proximity to one another, meaning that neighboring SNPs tend to be inherited together more often than would be expected if they were segregating independently. These neighborhoods are known as haplotype blocks, and due to LD, the entire neighborhood can be reconstructed by identifying only a few of its SNPs [11]. Based on this knowledge, it is feasible to measure genome-wide variation by sampling only a subset of all known SNPs. Together, the sequencing of the human genome, the generation of the HapMap, and high-throughput genotyping arrays have laid the foundation for population-based genome-wide association studies (GWAS) [6, 10, 12].
GWASs for the identification of disease-associated variants detect correlations between SNP alleles and the presence or absence of a disease. These studies are conducted by selecting one subset of the population to represent cases (i.e., individuals with disease), and another to represent matched controls (i.e., individuals without disease). High-throughput genotyping is done by hybridizing purified DNA samples from each individual in the study to high-density microarrays that contains allelle-specific oligonucleotide probes [13]. The probes are chosen as representatives of haplotype blocks uniformly distributed across the genome. DNA-probe pairs are detected by afluorescent signal that can be used to infer SNP genotypes.
Traditional statistical analysis (e.g., χ2 test of independence, logistic regression) and more recent data mining approaches (e.g., random forests, multifactor dimensionality reduction (MDR)) are used to establish significant differences between the SNP genotype profiles of cases and controls [14, 15]. If a SNP allele is detected more often in the presence of a disease, then that SNP allele is said to be associated with that disease and becomes a candidate for a replication study to validate the association [16]. An associated SNP is not necessarily responsible for conferring disease. Due to LD, the disease-associated SNP may instead be a proxy for a nearby causal variant [17].
Perhaps the best example of novel target discovery by a GWAS comes from research on age-related macular degeneration (AMD), the leading cause of severe vision loss in elderly populations. A SNP allele in the complement factor H (CFH) gene was found to have a statistically significant association with AMD [18]. The association between CFH and AMD was unexpected, yet it has been repeatedly observed [19, 20, 21]. Currently, new therapeutic strategies targeting the complement factor pathway are under development, including a recombinant CFH protein carrying the protective allele [22]. This example illustrates the impact GWASs can have on drug development by novel target discovery. However, AMD has been the exception rather than the rule.
GWASs have identified over 1200 genomic regions associated with common phenotypic traits and diseases [23]. However, unlike the case of AMD in which the CFH variant is associated with a multi-fold increase in risk (4.6–7.4 fold), the vast majority of GWASs have not revealed obvious disease mechanisms or drug targets [18]. In general, individual SNPs confer only incremental changes in relative risk (1.1–1.5 fold) and account for only a small proportion of heritability (<5%) in common diseases [24, 25]. Ongoing studies continue to search for this missing heritability. Some of the most common explanations include roles for unmeasured low-frequency and rare alleles, many variants each conferring small effects, and gene-gene and gene-environment interactions [26, 27, 28]. Furthermore, the functional role of the vast majority of variants is unknown, highlighted by the fact that over 80% of disease-associated SNPs are found in non-coding regions of the genome [25]. Readers are referred to existing literature for more information on GWASs and methods to resolve the missing heritability [13, 29, 30].
It is important to acknowledge that GWASs are designed to identify disease-associated variants based on statistical correlations without regard for prior biological knowledge. Amassing sets of disease-associated genetic variants does not directly translate into an understanding of the genetic basis of disease. Rather, disease-associated variants provide a foothold for generating testable hypotheses within the context of known biology.
From disease-associated genomic regions to functional targets
Biological functions arise from context-dependent interactions among cellular macromolecules [31]. To understand the genetic mechanisms that underlie common diseases it is necessary to validate the functional consequences of genetic variation within a relevant biological context. It is this functional insight into the dynamic nature of gene regulation and expression that can offer unparalleled resolution of biological processes, and by extension, disease processes that can be directly targeted.
Functional genetics
The paradigm of target-based drug discovery was founded on the principles of functional genetics. Functional assays driven by molecular biology enable the isolation and characterization of disease-associated genes and gene-products which can be molecularly targeted by specific drugs [32]. One of the earliest successes in this area was the discovery of imatinib for the treatment of chronic myelogenous leukemia (CML). Chromosomal staining and microscopy studies revealed a consistent reciprocal translocation between portions of chromosomes 9 and 22 in cells collected from CML patients [33, 34, 35]. This translocation results in a gene fusion to create the BCR-ABL oncogene [36].
The functional consequence of the BCR-ABL gene fusion is the production of the BCR-ABL protein product, which has elevated tyrosine kinase activity [37, 38]. Evidence for a causal relationship between BCR-ABL and leukemia was demonstrated in mouse models in which the introduction of the BCR-ABL transgene resulted in the onset of a leukemia-like disease [39, 40]. A chemical screen for the inhibition of BCR-ABL activity produced a lead compound that was used as the structural basis for the synthesis of imatinib [41].
Imatinib selectively inhibits the proliferation of BCR-ABL -expressing cells without affecting the growth of normal cells [42]. Imatinib has been FDA-approved for use in BCR-ABL -positive CML patients since 2001. This example embodies the integral role of functional genetics in elucidating the causal genetic component of a disease and using that knowledge to identify or design rational drug therapies.
Functional genomics
Many functional assays now draw on established DNA sequence information and advances in technology to collect functional data on the genomic scale. These assays have given rise to new fields of study including transcriptomics (i.e., gene expression profiling), epigenomics (i.e., DNA methylation and chromatin remodeling), cistromics (i.e., transcription factor binding profiling), and proteomics (i.e., protein expression profiling). Functional assays, in addition to validating the association between genes or DNA sequence variations and disease, are also used in unbiased screens to compare genetic variability between controls and disease cells or tissues to provide clues regarding disease mechanisms.
Functional genomics have played a crucial role in elucidating the molecular basis of cancers [43,44, 45]. For example, the distinct subtypes of breast cancer (i.e., basal-like, luminal A, luminal B, ERBB2+, and Claudin-low), in addition to normal breast-like tumors, are defined by unique patterns of gene expression which were discovered by transcriptional profiling with gene expression microarrays [46, 47]. These gene signatures can be used to predict patient relapse, overall survival, and treatment response to stratify patients for tailored therapies [48]. This use of functional genomics has significantly advanced our understanding of cancer by demonstrating that cancers, once thought to be single diseases, can have very different genetic mechanisms which explain variable treatment responses.
Functional genomics have also been used to identify druggable targets in cancer cell lines through the use of high-throughput RNA interference (RNAi) screening technologies [49]. This approach enables the silencing of genes in a sequence-dependent manner through the use of established short-interfering RNA (siRNA) libraries. Eligible targets are identified when gene silencing results in a loss of cell viability. Effective targets will be cell line-specific, as genes are differentially regulated under various biological conditions. This is just one example of the utility of functional genomics assays in druggable target identification. For a complete review of the role of functional genomics throughout the drug discovery process see Kramer et al. [50].
Bridging the gap with network analysis
There has been a decline in the approval rates of new drugs following the widespread adoption of genomic technologies in the drug development process [51]. The absence of translatable results can be partly attributed to the way in which genomic data are conventionally analyzed. Genomic data sets capture the global state of a biological system at a given moment in time. Through serial sampling and integration, genomic data has the potential to reveal systems level behavior. Paradoxically, this context-specific data is most often analyzed from a reductionist perspective with the intention of isolating independent disease-associated genetic factors as potential drug targets. However, even when functionally validated, the aberrant activity of a disease-associated gene or gene product alone does not necessarily make it the best target. Rather, the effectiveness of a potential target is determined by establishing its position within the hierarchy of the regulatory control mechanisms within the cell [7]. By disengaging disease-associated genetic factors from their position within the genetic regulatory system, much of the information stored within the genomic data set is lost. This paradox can be resolved by analyzing genomic data with new computational strategies that move away from the one gene or one variant at a time approach toward a more holistic approach that recognizes the complexity of the underlying genetic basis of a disease [30].
Network biology
Biological macromolecules that serve as drug targets are also the basic building blocks, or component parts, of biological systems. The interactions between these component parts give rise to biological functions [31]. Accordingly, biological systems can be modeled by networks to capture the global web of interactions that connect the individual component parts [52]. In this context, the network is used as an abstract representation of genomic data in which nodes represent one or more genomic entities (e.g., mRNA, gene, protein) and edges indicate an interaction or relationship such as co-expression, transcriptional regulation, or physical interaction between the pairs of nodes that they connect. This abstraction allows for versatility enabling network analysis to be applied to many types of empirical data. It is the high-throughput, genome-wide measurements of the cellular state at a given time that enable the assembly and analysis of genomic networks.
Genome-wide regulatory networks (GWRN): from disease-associated variants to disease-associated genes
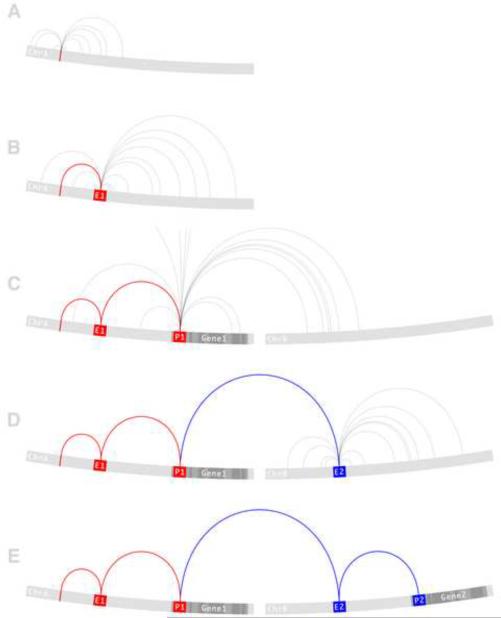
Network analysis provides a way to model the complex interactions within and between the multiple levels of genomic information that direct biological function. For example, genome-wide regulatory networks (GWRN) have recently been proposed as a framework for integrating prior biological knowledge with empirically gathered GWAS data into a single multidimensional network [53]. The nodes of a GWRN regulatory network can be genes, enhancers, promoters, insulators, and DNA sequence variants. The edges are based on linkage disequilibrium as reported by the HapMap, long-range physical interactions, or transcription factor binding sites. As illustrated in Figure 1, a GWRN is built from a defined starting point, an associated variant identified by a GWAS. The network is developed by adding edges based on empirical evidence gathered at the genome-wide level either independently or from publicly available databases. By following the edges between the various classes of nodes, regulatory mechanisms can be traced between genomic elements. This process guides the generation of novel mechanistic hypotheses and potentially leads to the assignment of functional causality to disease-associated variants.
Figure 1.
A proposed computational framework to predict functional consequences of genetic variation. The red edges track the path from a disease-associated variant to a disease-associated gene. The blue edges track the path from the disease-associated gene to a functional target. (a) A variant node on chromosome A (red) has been associated with a disease. The variant may be causal or may be serving as a proxy of nearby causal variant so the first set of edges connect to those variant nodes that are in linkage disequilibrium (LD) with the associated variant. (b) One of the LD edges connects to the E1 enhancer node. Because an enhancer is a functional entity that directly regulates gene transcription, the second set of edges connect the E1 enhancer node to its long-range interacting partners. (c) One of the long-range interactions connects the E1 enhancer node to the P1 promoter node of the Gene1 gene node. This path reflects known empirical evidence in support of a regulatory interaction between the E1 enhancer and Gene1 mediated by the P1 promoter. Gene1 is a known transcription factor but has not previously been linked to the disease. (d) The third set of edges connect the transcription factor Gene1 to all of its known binding sites. One of these transcription factor binding site edges connects Gene1 to the E2 enhancer node found on chromosome B. Continuing the pattern established here, a fourth set of edges are drawn to connect the E2 enhancer node to its long-range interacting partners. (e) One of these long-range interactions connects the E2 enhancer node to the P2 promoter node of the Gene2 gene node. Gene2 is known to affect the disease. This final link creates a complete path through the GWRN from the associated variant to the causal gene. Every edge along this path is supported by empirical evidence however, this process is meant to provide the most likely causal explanation given the available data and as such is best used to guide the generation of novel hypotheses that can be experimentally validated.
Our knowledge of genome-wide regulation is still in its infancy. What we do know however, is that the regulatory architecture of the cell is highly distributed and interconnected. The human genome does not behave like a rigid segment but folds and interacts with itself within the nucleus. These interactions involve both physical loops of chromatin and the diffusion of transcription factors (TFs) to enhancers. Based solely on loops, each Transcriptional Start Site (TSS) for a gene interacts on average with five distal regulatory elements. Conversely, each distal regulatory element interacts on average with eighteen TSSs (Job Dekker, personal communication). Given the pervasiveness of distal enhancers and chromatin loops, regulatory elements and their target genes are unlikely to be contiguous on the genomic segment. Thus, disease-associated variants need not be close to the genes they affect; they can even be on different chromosomes.
The assembly of GWRNs has only recently become feasible. First, the chromatin signatures of pivotal regulatory elements have now been discovered [54, 55]. With the advent of chromatin immunoprecipitation coupled with deep sequencing (ChIP-Seq), regulatory elements can now be mapped at the genome-wide level. Second, through ChIP-Seq, the binding sites for a given TF can be assessed genome-wide for a given cell-type. Third, the chromatin conformation capture family of techniques has allowed the genome-wide mapping of chromatin loops (see [56]). Through these technologies, the regulatory nodes and edges formed through TF diffusion and chromatin architecture are now being mapped extensively. The ENCODE project (ENCyclopedia Of DNA Elements) acts as a hub for such data sets and is growing rapidly [57].
The detailed mechanistic information provided by GWRNs, once experimentally validated, can greatly contribute to our understanding of the genetic basis of disease. It is this understanding of the regulatory control mechanisms that underlie a disease, within the broader context of the entire system, that can reveal the Achilles' heel of that disease as a potential target itself or as a marker to look for more appropriate upstream or downstream targets. Furthermore, given our current knowledge of genomic architecture, each disease-associated variant will likely yield dozens of disease-associated genes from which to select functional targets.
Network pharmacology: from disease-associated genes to functional targets
Network analysis also provides a way to model the complex interactions that occur between single levels of genomic information, (e.g., through protein-protein interaction networks or gene-gene co-expression networks). The nodes of these networks represent a single genomic entity. The edges are usually based on known physical interactions or statistical correlations. These types of networks are analyzed for their structural and dynamical properties which can be manipulated in silico to study the internal organization of the system [52]. For example, the connectivity of a network is a structural characteristic that describes the arrangement of edges connecting the various nodes within the graph. Comparative analysis between healthy and disease networks can identify changes in network connectivity provoked by the disease state that may reveal gains in function or vulnerabilities such as the loss of biological redundancy, loss of feedback regulation, or changes in the regulation of druggable targets [58]. Accordingly, target identification through network analysis involves prioritizing those nodes in the disease network that, when selectively targeted, will modify the network connectivity to reorganize the aberrant interactions reverting the network back to the healthy state or, when appropriate, leading to cell death (e.g., cancer). Effective target nodes can also be systematically identified by studying two network properties that are closely related to connectivity, information flow through networks and robustness (i.e., resilience to perturbations) of networks.
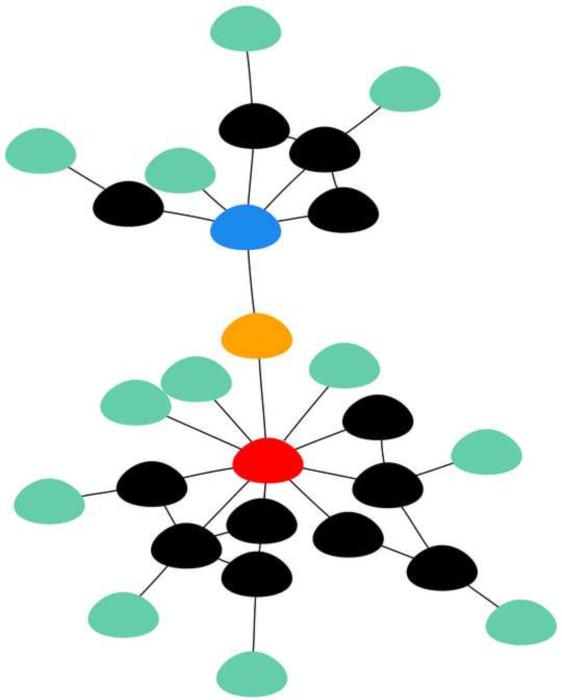
Information flow through interaction networks is based on the connectivity and position of individual nodes. Signals propagate through connected nodes to create paths through the graph. As information is passed from an affected node to its neighboring nodes, information cascades ripple across the network. Network models of social, technological, and biological data tend to have many nodes with few connections, termed peripheral nodes, and few nodes with many connections, termed hubs (see Figure 2) [52]. Hubs play an important anchoring role in maintaining the integrity of a network.
Figure 2.
A toy example of a biological network model. This network has undirected edges which are generally used to display co-expression or binding relationships. The connectivities described in the text are illustrated here. The red node is a hub, intermediate nodes are shown in black, and peripheral nodes are shown in teal. Hubs play an important role in maintaining network integrity and in information propagation. This makes targeting hubs complicated. Peripheral nodes tend to have limited influence on signal transduction because of their limited connectivity. Even if a peripheral node sends a message to a hub, that message is tempered by all of the other signals the hub is receiving which diminishes the chances that that message will be propagated. Intermediate nodes strike a fine balance between hubs and peripheral nodes making promising drug targets. In addition to hubs, intermediate, and peripheral nodes, a bridging nodes is shown in orange and a bottleneck node is shown in blue. Bridging nodes connect subnetworks that would otherwise be isolated. Bridging nodes have low connectivity but high centrality and they tend to be independently regulated. As such they may characterize disease conditions making them good drug targets. Bottleneck nodes are positioned in paths that are well-traveled. They generally have high connectivity and high centrality. They could be classified as hubs; however, they differ from hubs in that they are independently regulated. These properties make bottleneck nodes promising drug targets as well.
In the context of a protein-protein interaction network, a study of the connectivity of known protein targets of FDA-approved drugs revealed that the targeted proteins tend to have more connections on average than peripheral nodes but fewer connections on average than hubs [59]. This is a logical finding in terms of information flow because hubs are so well connected that their modulation may lead to cascading effects that cause unexpected side-effects or compromise the integrity of the network. Peripheral nodes, on the other hand, are generally on the fringe of the network and the effects of their modulation may have limited reach. This demonstrates that successful drug targets have intermediate connectivities and suggests a strategy based on node connectivity with which to prioritize potential drug targets.
Additional network metrics provide further nuance to this strategy. Studies in yeast have shown that nodes positioned at critical junctures of information flow within a network also make promising targets. Examples of these positional nodes are bridging nodes, (i.e. nodes connecting otherwise isolated sub-networks (Figure 2)). When knocked-out in yeast, bridging nodes have lower lethality than hubs, meaning that their modulation can alter the connectivity of a network while maintaining its integrity [60]. In addition, bridging nodes are independently regulated based on the biological context, meaning that the connectivity of bridging nodes may be specific to a disease network [60]. Together, these findings suggest that the modulation of bridging nodes can potentially abrogate aberrant interactions without destabilizing the entire system.
Other of positional nodes are bottlenecks or nodes with high centrality (i.e., the thoroughfare nodes of the network, see figure 2). Studies of yeast protein-protein interaction networks show that, like bridging nodes, bottleneck nodes are independently regulated, showing below average co-expression with their neighbors [61]. However, bottleneck nodes tend to be essential proteins. Therefore, unlike bridging nodes, their removal causes lethality in vivo and compromises network stability in silico [61]. This point highlights that the desired therapeutic goal can dictate the selection of either bridging nodes or bottlenecks. Although no single network property will universally reveal the optimal target, together the network properties of node connectivity and position can facilitate the efficient identification and prioritization of druggable targets in disease-specific networks.
In some occasions, however, a single functional target may not be the best solution. Biological systems respond to exogenous perturbations, such as drug treatments, through coordinated responses and interactions of cellular components. This type of response has been visualized in individual cancer cells where the temporal expression levels and localization of approximately 1000 tagged-proteins were tracked following treatment with a chemotherapeutic agent [62]. In this study, nearly all of the proteins showed a dynamic response. Many of these responses were cell-specific and correlated with survival outcomes, demonstrating both biological robustness and fragility in the face of a pharmacological perturbation. Biological systems have evolved buffering mechanisms through feedback regulation and functional redundancy to help them endure both endogenous and exogenous perturbations. However, the system is vulnerable when challenged with perturbations for which these mechanisms cannot cope [63].
Network analysis provides a computational framework for performing perturbation experiments in silico and to assess the global effects of targeted interventions [52]. These experiments measure the stability or the resilience of a network to targeted perturbations which simulate pharmacological inhibition. The removal of nodes or edges correspond to complete or partial inhibition, respectively [64]. Perturbation studies on the transcriptional regulatory networks of yeast and bacteria show that partial inhibition of multiple targets can have a larger effect on the structural properties of the network than the complete inhibition of a single, well-connected node [64]. This is consistent with the notion of biological robustness in that the partial inhibition of multiple targets can more effectively eliminate the functional redundancies and reorganize feedback loops to abrogate disease processes. This result has clear implications for the identification and prioritization of potential targets, and supports the recent trend toward polypharmacology, that is, the philosophy that multiple targeted therapies will be the most effective treatment strategies [32].
Concluding remarks
The paradigm of target-based drug design followed from advances in molecular biology and functional genetics. A surge in technology ushered-in the era of genomics, changing the way data is collected by enabling genome-wide surveys at the population and functional levels. This has lead to unbiased screens for disease causing genes or genetic variants in genomics and parallel unbiased screens for therapeutic targets in the drug discovery process. Limited success in identifying effective drug targets can potentially be overcome by using analysis strategies that embrace the complexity of biology. One such strategy is network analysis which is able to capture global properties of the system while preserving the molecular detail necessary for target identification. As the research community continues to be inundated by a data deluge, the next phase of genomics will be computationally motivated by the critical need to develop analytical methods that can reduce the total search space and prioritize both disease-associated variants and druggable targets in the most biologically meaningful way. The future of genomics will entail the integration of computational methods and experimental biology to create comprehensive maps of the complex genetic architectures underlying disease processes, thereby fulfilling the promise of systems genetics [65]. Combining systems genetics with molecular pharmacology will enable accurate modeling of targeted perturbations on a systems level, revolutionizing the drug target discovery process.
Acknowledgements
This work was supported by NIH grants LM009012, LM010098 and AI59694.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Drews J. Drug discovery: a historical perspective. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- [2].Hopkins A, Groom C. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- [3].Overington J, et al. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- [4].DiMasi J, Faden L. Competitiveness in follow-on drug R&D: a race or imitation? Nat Rev Drug Discov. 2011;10:23–27. doi: 10.1038/nrd3296. [DOI] [PubMed] [Google Scholar]
- [5].Altshuler D, et al. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- [7].Araujo R, et al. Proteins, drug targets and the mechanisms they control: the simple truth about complex networks. Nat Rev Drug Discov. 2007;6:871–880. doi: 10.1038/nrd2381. [DOI] [PubMed] [Google Scholar]
- [8].Venter J, et al. The sequence of the human genome. Science. 2001;291:1304. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- [9].Sachidanandam R, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- [10].International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gabriel S, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- [12].Wang D, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- [13].Kingsmore S, et al. Genome-wide association studies: progress and potential for drug discovery and development. Nat Rev Drug Discov. 2008;7:221–230. doi: 10.1038/nrd2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Clarke G, et al. Basic statistical analysis in genetic case-control studies. Nat Protoc. 2011;6:121–133. doi: 10.1038/nprot.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cordell H. Detecting gene--gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chanock S, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- [17].Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- [18].Klein R, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Edwards A, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- [20].Haines J, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- [21].Hageman G, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. PNAS. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gehrs K, et al. Complement, age-related macular degeneration and a vision of the future. Arch Ophthalmol. 2010;128:349–358. doi: 10.1001/archophthalmol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hindorff L, et al. A catalog of published genome-wide association studies. National Human Genome Research Institute. 2010 Available at http://www. genome. gov/gwastudies. [Google Scholar]
- [24].WTCCC A genome-wide scan of 14,000 non-synonymous coding SNPs in 5,500 individuals: The Wellcome Trust Case Control Consortium. Nat Genet. 2007;39:1329–37. [Google Scholar]
- [25].Hindorff L, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. PNAS. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Manolio T, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lohmueller K, et al. Meta-analysis of genetic association studies supports a contribution ofcommon variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- [28].Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Eichler E, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Moore J, et al. Bioinformatics challenges for genome-wide association studies. Bioinformatics. 2010;26:445. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hartwell LH, et al. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- [32].Hopkins A. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- [33].Nowell P, Hungerford D. Chromosome studies on normal and leukemic human leukocytes. JNCI. 1960;25:85. [PubMed] [Google Scholar]
- [34].Rowley J. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- [35].Watt J, Page B. Reciprocal translocation and the Philadelphia chromosome. Hum Genet. 1978;42:163–170. doi: 10.1007/BF00283636. [DOI] [PubMed] [Google Scholar]
- [36].Heisterkamp N, et al. Structural organization of the bcr gene and its role in the Ph translocation. Nature. 1985;315:758–761. doi: 10.1038/315758a0. [DOI] [PubMed] [Google Scholar]
- [37].Ben-Neriah Y, et al. The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science. 1986;233:212–214. doi: 10.1126/science.3460176. [DOI] [PubMed] [Google Scholar]
- [38].Konopka J, et al. An alteration of the human c-abl protein in K562 leukemia cells unmasks associated tyrosine kinase activity. Cell. 1984;37:1035–1042. doi: 10.1016/0092-8674(84)90438-0. [DOI] [PubMed] [Google Scholar]
- [39].Heisterkamp N, et al. Acute leukaemia in bcr/abl transgenic mice. Nature [Google Scholar]
- [40].Daley G, et al. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- [41].Buchdunger E, et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56:100–104. [PubMed] [Google Scholar]
- [42].Druker B, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- [43].Perou C, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. PNAS. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Golub T, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- [45].Alizadeh A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- [46].Sørlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. PNAS. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Herschkowitz J, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Perou C, Børresen-Dale A. Systems Biology and Genomics of Breast Cancer. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a003293. doi: 10.1101/cshperspect.a003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Iorns E, et al. Integrated Functional, Gene Expression and Genomic Analysis for the Identification of Cancer Targets. PLoS One. 2009;4:e5120. doi: 10.1371/journal.pone.0005120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kramer R, Cohen D. Functional genomics to new drug targets. Nat Rev Drug Discov. 2004;3:965–972. doi: 10.1038/nrd1552. [DOI] [PubMed] [Google Scholar]
- [51].Butcher E, et al. Systems biology in drug discovery. Nature Biotechnol. 2004;22:1253–1259. doi: 10.1038/nbt1017. [DOI] [PubMed] [Google Scholar]
- [52].Barabási A, Oltvai Z. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- [53].Cowper-Sal R, et al. Layers of epistasis: genome-wide regulatory networks and network approaches to genome-wide association studies. WIREs Syst Biol Med. doi: 10.1002/wsbm.132. doi:10.1002/wsbm.132 ( http://onlinelibrary.wiley.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lupien M, et al. FoxA1 Translates Epigenetic Signatures into Enhancer-Driven Lineage-Specific Transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Heintzman N, et al. Histone modifications at human enhancers reflect global cell-type specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- [57].Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Dixon S, Stockwell B. Identifying druggable disease-modifying gene products. Curr Opin Chem Biol. 2009;13:549–555. doi: 10.1016/j.cbpa.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yildirim M, et al. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- [60].Hwang W, et al. Identification of information flow-modulating drug targets: a novel bridging paradigm for drug discovery. Clin Pharmacol Ther. 2008;84:563–572. doi: 10.1038/clpt.2008.129. [DOI] [PubMed] [Google Scholar]
- [61].Yu H, et al. The importance of bottlenecks in protein networks: Correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3:0713–0720. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cohen A, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- [63].Kitano H. A robustness-based approach to systems-oriented drug design. Nat Rev Drug Discov. 2007;5:202–210. doi: 10.1038/nrd2195. [DOI] [PubMed] [Google Scholar]
- [64].Ágoston V, et al. Multiple weak hits confuse complex systems: a transcriptional regulatory network as an example. Phys Rev E. 2005;71:051909. doi: 10.1103/PhysRevE.71.051909. [DOI] [PubMed] [Google Scholar]
- [65].Nadeau J, Dudley A. Systems Genetics. Science. 2011;331:1015–1016. doi: 10.1126/science.1203869. [DOI] [PMC free article] [PubMed] [Google Scholar]